Abstract
MicroRNAs (miRNAs) play pivotal roles in numerous biological processes, and their dysregulation is a common feature of human cancer. Thanks to recent advances in the analysis of the cancer epigenome, we now know that epigenetic alterations, including aberrant DNA methylation and histone modifications, are major causes of miRNA dysregulation in cancer. Moreover, the list of miRNA genes silenced in association with CpG island hypermethylation is rapidly growing, and various oncogenic miRNAs are now known to be upregulated via DNA hypomethylation. Histone modifications also play important roles in the dysregulation of miRNAs, and histone deacetylation and gain of repressive histone marks are strongly associated with miRNA gene silencing. Conversely, miRNA dysregulation is causally related to epigenetic alterations in cancer. Thus aberrant methylation of miRNA genes is a potentially useful biomarker for detecting cancer and predicting its outcome. Given that many of the silenced miRNAs appear to act as tumor suppressors through the targeting of oncogenes, re-expression of the miRNAs could be an effective approach to cancer therapy, and unraveling the relationship between epigenetic alteration and miRNA dysregulation may lead to the discovery of new therapeutic targets.
Keywords: microRNA, tumor suppressor, oncomir, CpG island methylation, histone modification, biomarker, EZH2
INTRODUCTION
MicroRNAs (miRNAs) are endogenous, small, non-coding single-stranded RNAs about 22 nucleotides in length, which function at the post-transcriptional level as negative regulators of gene expression (He and Hannon, 2004). Each miRNA negatively regulates its target genes in one of two ways, depending on the degree of complementarity between itself and its target messenger RNAs (mRNAs). miRNAs that bind to mRNA sequences with perfect or nearly perfect complementarity induce the RNA-mediated interference (RNAi) pathway, in which mRNA transcripts are cleaved by a miRNA-associated RNA-induced silencing complex (miRISC). This mechanism is mainly observed in plants, though miRNA-directed mRNA cleavage does occur in animals. Most animal miRNAs are thought to act by binding to imperfectly complementary sites within the 3′ untranslated regions (UTRs) of target mRNAs, thus inhibiting the initiation of translation via the miRISC.
Annotation of their genomic locations suggests that many miRNA genes are located within intergenic regions, though they are also found within exonic and intronic regions in either the sense or antisense orientation. Like genes encoding proteins, miRNA genes are mainly transcribed by RNA polymerase II. They are initially transcribed as large precursors, called primary miRNAs (pri-miRNAs), which may encode multiple miRNAs in a polycistronic arrangement. Pri-miRNAs are then processed by the RNase III enzyme Drosha and its cofactor DGCR8/Pasha to produce ~70-nt hairpin-structured second precursors, called pre-miRNAs. The pre-miRNAs are transported to the cytoplasm by the nuclear export protein Exportin-5 (XPO5), after which they are processed by another RNase III enzyme, DICER, to generate mature miRNA products.
Sequences of miRNAs are highly conserved among species, and play critical roles in a variety of biological processes, including cell proliferation, development, differentiation, and apoptosis. In addition, subsets of miRNAs are thought to act as tumor suppressor genes or oncogenes, and their dysregulation is a common feature of human cancers (Esquela-Kerscher and Slack, 2006; Croce, 2009). More specifically, expression of miRNAs is generally downregulated in tumor tissues, as compared to corresponding healthy tissues, which suggests some miRNAs behave as tumor suppressors in some tumors. Although the mechanism underlying the alteration of miRNA expression in cancer is still not fully understood, recent studies have shown that cancer affects multiple mechanisms involved in regulating miRNA levels. For example, a significant number of miRNAs are located within cancer-associated genomic regions or in fragile sites (Calin et al., 2004). The first report of altered miRNA expression in cancer was related to the frequent chromosomal deletion and downregulated expression of miR-15 and miR-16, two miRNAs thought to target the antiapoptotic factor B cell lymphoma 2 (BCL2) in chronic lymphocytic leukemia (CLL; Calin et al., 2002). More recent studies indicate that genetic mutations affecting proteins involved in the processing and maturation of miRNA, such as TARBP2 and XPO5, can also lead to overall reductions in miRNA expression (Melo et al., 2009, 2010). In addition, epigenetic alterations, including aberrant DNA methylation and histone modifications, appear to be a major mechanism by which the normal patterns of miRNA expression are disrupted in cancer. In this review, we will highlight the contribution made by epigenetic alteration of miRNAs to cancer, and discuss their clinical application as biomarkers and therapeutic targets.
IDENTIFICATION OF EPIGENETICALLY DYSREGULATED miRNAs IN CANCER
The first evidence that epigenetic mechanisms are involved in silencing miRNAs in cancer came from a pharmacological unmasking experiment. Using a miRNA microarray, Saito et al. (2006) analyzed the expression profiles of miRNAs in T24 human bladder cancer cells and LD419 human normal fibroblasts treated with or without the DNA methyltransferase (DNMT) inhibitor 5-aza-2′-deoxycytidine (5-aza-dC) and the histone deacetylase (HDAC) inhibitor 4-phenylbutyric acid (4-PBA). Among the genes upregulated in the cancer cells was miR-127, which was embedded within a CpG island and was upregulated in association with DNA demethylation, acetylation of histone H3 and trimethylation of histone H3 lysine 4 (H3K4me3), which are marks of active transcription. Experimental evidence confirmed that the proto-oncogene BCL6 is a target of miR-127, suggesting miR-127 acts as a tumor suppressor (Saito et al., 2006).
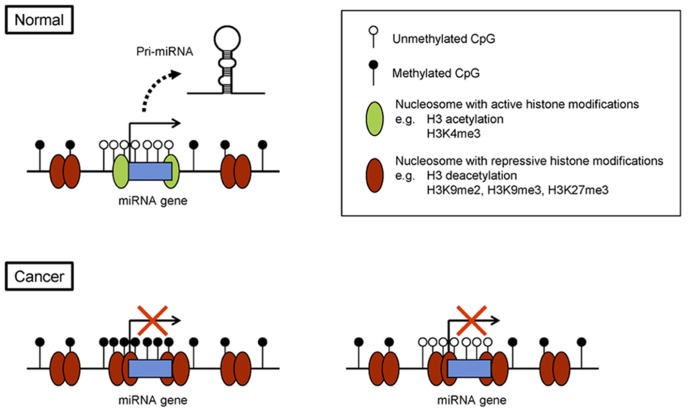
As with protein-coding genes, epigenetic regulation of miRNA genes is tightly associated with histone modification (Figure 1). As mentioned, H3K4me3 and acetylation of histone H3 lysine 9/14 are hallmarks of active miRNA gene promoters in embryonic stem cells and in cancer cells (Marson et al., 2008; Ozsolak et al., 2008; Suzuki et al., 2011). By contrast, di- or trimethylation of histone H3 lysine 9 (H3K9me2 or H3K9me3) and trimethylation of lysine 27 (H3K27me3) are marks of repression. For instance, a combination of chromatin immunoprecipitation (ChIP)-on-chip and miRNA microarray analyses in prostate cancer cells revealed that miRNA expression correlates positively with H3K4me3 and correlates inversely with H3K27me3 in miRNA promoter regions (Ke et al., 2009). In addition, genome-wide screening for miRNA genes with reduced levels of H3K4me3 and increased levels of H3K9me2 led to the identification of 13 miRNA genes, including the miR-124 family, miR-9 family, and miR-34b/c, that are epigenetically silenced in acute lymphoblastic leukemia (ALL; Roman-Gomez et al., 2009). To assess genome-wide histone modifications, we recently performed deep sequencing (ChIP-seq) in colorectal cancer (CRC) cells and identified the putative promoter regions of 174 pri-miRNA genes (Suzuki et al., 2011). By searching for miRNAs that showed upregulated expression and increases in H3K4me3 marks upon DNA demethylation, we identified 37 miRNA genes as potential targets of epigenetic silencing in CRC cells.
FIGURE 1.
Epigenetic silencing of miRNA genes in cancer. Example of a miRNA gene with its CpG island. In normal cells, the CpG island is unmethylated, and the chromatin is associated with active histone modifications. In cancer cells, CpG island hypermethylation and repressive histone marks lead to epigenetic silencing of the miRNA gene (left). Alternatively, some miRNA genes are silenced through repressive histone modifications, rather than DNA methylation (right).
Epigenetically silenced miRNA genes were also identified through genome-wide DNA methylation analysis. For instance, methylation microarray analysis using the Infinium BeadChip revealed miR-10b to be a target of DNA methylation in gastric cancer (GC; Kim et al., 2011). In addition, Yan et al. (2011) performed a genome-wide methylome analysis entailing deep sequencing of MBD (methylated DNA binding domain)-isolated DNA in HCT116 cells, and identified a number of methylated genes, including miR-941, miR-1237, and miR-1247. And Baer et al. (2012) carried out an integrative analysis of genome-wide DNA methylation and histone modification (H3K4me3) in CLL and identified 128 miRNAs that carried aberrant DNA methylation at their promoters. Interestingly, of those 128 miRNA promoters, 38 exhibited hypermethylation, while 90 showed hypomethylation, which are indicative of epigenetically silenced and activated miRNAs, respectively. In fact, the hypermethylated regions included a number of well-defined epigenetically silenced miRNA genes, including miR-9-2, miR-124-2, and miR-129-2, while the hypomethylation was accompanied by upregulation of several miRNAs, including miR-21, miR-34a, and miR-155.
ABERRANT DNA METHYLATION OF miRNA GENES IN CANCER
Among the rapidly growing list of miRNAs that are now known to be aberrantly methylated in cancer, many are downregulated in association with CpG island hypermethylation, while some are upregulated via hypomethylation of their CpG island. Here we describe well-characterized miRNA genes showing aberrant DNA methylation in cancer.
Epigenetic silencing of miR-124 family genes was first discovered in CRC (Lujambio et al., 2007), and they are now known to be methylated in several other types of malignancy, including ALL (Agirre et al., 2009), non-Hodgkin’s lymphoma (Wong et al., 2011a), and liver (Furuta et al., 2010), pancreatic (Wang et al., 2013), renal (Gebauer et al., 2013), and cervical cancer (Wilting et al., 2010). Within the human genome, three independent loci (miR-124-1, miR-124-2, and miR-124-3) encode identical mature forms of miR-124, and all are associated with CpG islands, which may be targets of hypermethylation in cancer (Lujambio et al., 2007). miR-124 exerts its tumor suppressor function by targeting cyclin-dependent kinase 6 (CDK6), and epigenetic silencing of miR-124 reportedly results in CDK6 activation and Rb phosphorylation (Lujambio et al., 2007; Agirre et al., 2009). In ALL, miR-124 methylation is associated with higher recurrence and mortality rates, and may be an independent prognostic factor for disease-free and overall survival (Agirre et al., 2009). miR-124 family genes are also frequently methylated in the gastric mucosa of Helicobacter pylori-positive healthy individuals, suggesting their methylation could be induced by chronic inflammation (Ando et al., 2009).
Members of the miR-34 gene family (miR-34a, miR-34b, and miR-34c) are direct targets of p53, and their ectopic expression in cancer cells induces cell cycle arrest and apoptosis (Bommer et al., 2007; He et al., 2007). Within the human genome, miR-34a is located on chromosome 1p36, while miR-34b and miR-34c are co-transcribed from a single transcription unit on chromosome 11q23. The promoters of both genes are targets of CpG island hypermethylation in multiple malignancies including oral (Kozaki et al., 2008), esophageal (Chen et al., 2012), gastric (Suzuki et al., 2010), colorectal (Toyota et al., 2008), lung (Wang et al., 2011b), breast and renal cancer (Lodygin et al., 2008; Vogt et al., 2011), and hematological malignancies (Roman-Gomez et al., 2009; Wong et al., 2011b). Methylation of miR-34b/c has also been linked to cancer metastasis (Lujambio et al., 2008) and invasion (Watanabe et al., 2012). In addition, methylation-associated silencing of miR-34c was recently shown to promote self-renewal and epithelial–mesenchymal transition (EMT) in breast tumor-initiating cells (Yu et al., 2012). These findings, as well as their contribution to the p53 network, strongly imply that miR-34 family members act as tumor suppressors in cancer. Introduction of miR-34b/c into cancer cells leads to the downregulation of candidate target genes, including MET, CDK4, cyclin E2 (CCNE2), and MYC (Lujambio et al., 2008; Toyota et al., 2008). Likewise, restoration of endogenous miRNA expression through demethylation also downregulates target genes, suggesting miRNAs could be important targets for epigenetic cancer therapy (Toyota et al., 2008).
The CpG islands of miR-9 family genes (miR-9-1, miR-9-2, and miR-9-3) are also frequently methylated in various types of malignancies, including ALL (Roman-Gomez et al., 2009) and colorectal (Bandres et al., 2009), breast (Lehmann et al., 2008), pancreatic (Omura et al., 2008), and GCs (Tsai et al., 2011). Moreover, a screen for miRNA gene methylation in metastatic cancer cell lines identified miR-9 family genes as being methylated (Lujambio et al., 2008). Consistent with that finding, methylation of miR-9-1 is reportedly associated with lymph node metastasis in CRC (Bandres et al., 2009), while methylation of miR-9-1 and miR-9-3 correlates with metastatic recurrence of renal cell carcinoma (Hildebrandt et al., 2010). miR-9 has been shown to target fibroblast growth factor receptor 1 (FGFR1) and CDK6 in ALL (Rodriguez-Otero et al., 2011) and caudal-type homeobox 2 (CDX2) in GC (Rotkrua et al., 2011), suggesting a tumor-suppressive function.
The miR-200 gene family (miR-200a, miR-200b, miR-200c, miR-141, and miR-429) and miR-205 encode key regulators of EMT that act by directly targeting zinc finger E-box binding homeobox 1 (ZEB1) and ZEB2, which are transcriptional repressors that downregulate E-cadherin (CDH1; Gregory et al., 2008; Korpal et al., 2008; Park et al., 2008). Within the human genome, miR-200 family genes are grouped into two polycistronic units, miR-200b/200a/429 and miR-200c/141, located on chromosomes 1 and 12, respectively (Davalos et al., 2012). In normal mammary epithelial cells and fibroblasts, expression of miR-200 family and miR-205 genes is regulated by DNA methylation, histone modifications, or a combination of the two (Vrba et al., 2010), but aberrant DNA methylation leads to the silencing of these miRNAs in cancer (Ceppi et al., 2010; Neves et al., 2010; Wiklund et al., 2011). For instance, methylation of miR-200c/141 is tightly correlated with the invasive capacity of breast cancer cells (Neves et al., 2010). Similarly, in non-small cell lung cancer, promoter methylation is associated with loss of miR-200c expression, which is in turn associated with poor differentiation, lymph node metastasis, and weaker E-cadherin expression (Ceppi et al., 2010). Davalos et al. (2012) demonstrated that the upstream CpG islands of both units (miR-200b/200a/429 and miR-200c/141) are unmethylated in cancer cells with epithelial features, but are both methylated and silenced in transformed cells with mesenchymal characteristics.
In addition to its therapeutic implications, miRNA gene methylation could be a useful molecular marker for detecting cancer and/or predicting its outcome. For instance, the CpG island of miR-34b/c is methylated in more than 90% of primary CRCs, and methylation was detected in 75% of fecal specimens from CRC patients and in 16% of specimens from high-grade dysplasia patients, suggesting miR-34b/c methylation could be a useful feces-based screening marker (Kalimutho et al., 2011). It was also recently shown that miR-34a methylation and high levels of c-MET and β-catenin expression may be powerful predictive markers of liver metastasis in CRC (Siemens et al., 2013). In addition, miR-34b/c methylation was found to be elevated in the background gastric mucosa of multiple GC patients (Suzuki et al., 2010), and a subsequent study revealed that miR-34b/c methylation could be a marker for predicting the risk of metachronous GC (Suzuki et al., 2013). miRNA gene methylation is also detectable in urine specimens, and could be a useful marker of urinary tract cancer. A recent screening for epigenetically silenced miRNAs in bladder cancer cells identified methylation of four miRNA genes (miR-137, miR-124-2, miR-124-3, and miR-9-3), and their methylation in urinary DNA was found to be a useful biomarker of bladder cancer (Shimizu et al., 2013).
Many miRNA genes are reportedly downregulated in association with DNA hypermethylation in cancer, but some are epigenetically activated via DNA hypomethylation. As mentioned, a recent comprehensive analysis of miRNA in CLL revealed that approximately 60% of aberrantly methylated miRNA genes exhibited hypomethylation (Baer et al., 2012). For instance, the CpG island of let-7a-3 is heavily methylated in normal cells but is hypomethylated in lung adenocarcinoma, leading to its elevated expression (Brueckner et al., 2007). In lung cancer cells, let-7a-3 exerts oncogenic effects through actions on several genes involved in cell proliferation, adhesion, and differentiation. In addition, miR-200a and miR-200b are overexpressed in pancreatic cancer due to their hypomethylation, and their elevation in the serum of pancreatic cancer patients means they could potentially serve as diagnostic biomarkers (Li et al., 2010). miR-196 family genes (miR-196a and miR-196b) are located within the HOX gene cluster and are often overexpressed in tumors, which is indicative of their oncogenic functions (Luthra et al., 2008; Maru et al., 2009; Popovic et al., 2009; Guan et al., 2010). miR-196b is embedded within a CpG island, and its overexpression in GC is associated with its hypomethylation (Tsai et al., 2010).
Recently, Fornari et al. (2012) reported that miR-519d is upregulated due to DNA hypomethylation in hepatocellular carcinoma (HCC). miR-519d belongs to the chromosome 19 miRNA cluster (C19MC), which is the largest miRNA cluster in the human genome. miRNAs in the C19MC are normally expressed specifically in placenta (Bentwich et al., 2005), but DNA demethylation leads to their re-expression in cancer cells, which is indicative of their epigenetic repression in healthy tissue (Tsai et al., 2009; Suzuki et al., 2011). Upregulation of miR-519d, which is observed in approximately 50% of HCCs, is positively associated with CpG island hypomethylation and wild-type p53 (Fornari et al., 2012). miR-519d is thought to act as an oncogenic miRNA (oncomir) through its targeting of p21, PTEN, AKT3, and TIMP2.
miRNA DYSREGULATION CAUSES ABERRANT DNA METHYLATION
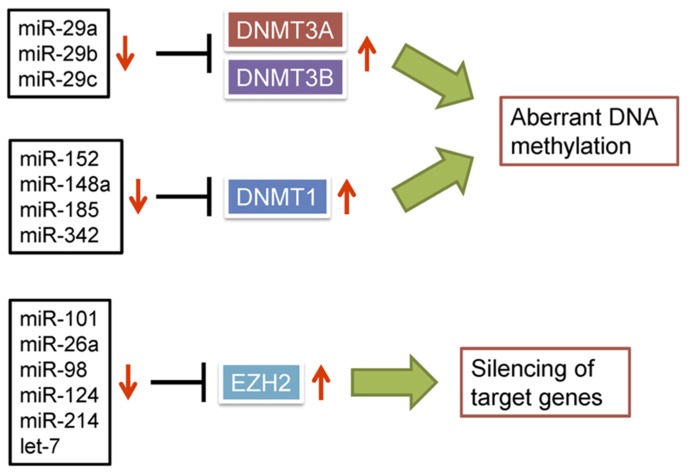
Several lines of evidence support the idea that dysregulation of miRNAs can lead to aberrant DNA methylation in cancer. For instance, the miR-29 family (miR-29a, miR-29b, and miR-29c), which is downregulated in lung cancer, directly targets DNMT3A and DNMT3B (Fabbri et al., 2007; Figure 2). Ectopic expression of the miR-29 family in lung cancer cells restores expression of methylation-silenced tumor suppressor genes, including fragile histidine triad (FHIT) and WW domain containing oxidoreductase (WWOX). In addition, miR-143 is frequently downregulated in CRC cells, where it normally targets DNMT3A (Ng et al., 2009), and downregulated expression of miR-152 in HBV-related HCC correlates with increased expression of DNMT1 (Huang et al., 2010). Forced expression of miR-152 in liver cell lines reduces DNMT1 expression and global DNA methylation, whereas inhibition of miR-152 causes global DNA hypermethylation and increased methylation of the glutathione S-transferase pi 1 (GSTP1) and CDH1 promoter regions. Similarly, DNMT1 is targeted by miR-148a and miR-152 in cholangiocarcinoma cells, and their ectopic expression suppresses DNMT1 and induces expression of the tumor suppressor genes Ras association domain family 1A (RASSF1A) and p16 (Braconi et al., 2010). miR-342 was found to be downregulated in CRC cells, and restoration of its expression downregulated DNMT1 and reactivated expression of cancer-related genes through demethylation of their promoter regions (Wang et al., 2011a). miR-185 is downregulated in glioma cells in association with loss of heterozygosity (LOH), and its restoration reduces global DNA methylation and leads to re-expression of hypermethylated genes through targeting DNMT1 (Zhang et al., 2011b).
FIGURE 2.
Dysregulation of miRNAs is causally related to epigenetic alterations in cancer. miRNAs able to negatively regulate DNMTs or EZH2 are frequently downregulated in cancer, which leads to epigenetic silencing of target genes.
miR-34b was recently shown to target both DNMTs and HDACs in prostate cancer cells (Majid et al., 2013). As in other malignancies, miR-34b is silenced in association with CpG island methylation in prostate cancer, and low miR-34b expression is strongly associated with poor survival. Interestingly, ectopic expression of miR-34b in prostate cancer cells suppressed DNMTs and HDACs and induced partial demethylation and active chromatin modification of the endogenous miR-34b gene, which suggests a positive feedback loop. Collectively, these results indicate that dysregulation of specific miRNAs may be causally related to aberrant methylation of promoter CpG islands.
HISTONE MODIFICATIONS AND miRNA DYSREGULATION
It is now evident that histone modifications also play a major role in the dysregulation of miRNAs in cancer. For example, treating a breast cancer cell line with the HDAC inhibitor LAQ824 induced upregulation or downregulation of a number of miRNAs within as little 5 h (Scott et al., 2006). Not only does this suggest the involvement of epigenetic mechanisms in the regulation of miRNAs in cancer cells, it also highlights the importance of secondary effects driven by miRNAs induced or downregulated through drug treatment. In addition, recent studies have shown that HDAC silences tumor-suppressive miRNAs in cancer. It is well documented that loss of miR-15a and miR-16 in CLL is associated with 13q loss; however, these miRNAs are also often downregulated in CLL samples without observable deletions in 13q, and Sampath et al. (2012) found that overexpression of HDACs (HDAC1, HDAC2, and HDAC3) is associated with downregulation of miR-15a, miR-16, and miR-29b. Furthermore, inhibition of the HDACs induced robust accumulation of active histone marks at the promoters of the miRNAs and increased their expression, which in turn led to downregulation of their target genes, BCL2 and MCL1. In another study, MYC interacted with HDAC3, which then colocalized to the promoters of miR-15a/miR-16-1 and their host gene DLEU2, resulting in MYC-induced suppression of these miRNAs in mantle cell lymphoma (Zhang et al., 2012a). And in HCC, upregulation of HDACs (HDAC1–3) was associated with repression of miR-449, which led to activation of the putative miR-449 target gene c-MET (Buurman et al., 2012).
In other settings, histone acetylation is involved in the activation of oncomirs in cancer. For example, miR-224 is commonly upregulated in HCC, and there is reportedly a positive correlation between miR-224 expression and histone acetylase protein EP300 in HCC tumors (Wang et al., 2012). It is well documented that the breast cancer susceptibility gene BRCA1 is involved in DNA damage repair and cell cycle regulation, but a recent study revealed an interesting link between BRCA1 and the epigenetic regulation of oncomirs. Chang et al. (2011) showed that wild-type BRCA1 epigenetically represses miR-155 by recruiting HDAC2 to the miR-155 promoter, while a BRCA1 R1699Q mutant relieves the repression and causes miR-155 to be overexpressed.
As mentioned, miRNA gene transcription is closely associated with histone modifications; thus some miRNA genes are silenced without DNA hypermethylation in cancer cells (Figure 1). For example, downregulation of miR-212 in lung cancer cells is reportedly associated with H3K9me2 and H3K27me3 but not DNA hypermethylation (Incoronato et al., 2011; Figure 1). miR-212 exerts a pro-apoptotic effect in lung cancer cells by targeting the anti-apoptotic gene PED, and inhibition of HDAC and the histone methyltransferase EZH2 strongly reactivates miR-212 expression in lung cancer cells. It was also recently found that miR-708 is repressed by H3K27me3 in metastatic breast cancer (Ryu et al., 2013). miR-708 targets neuronatin (NNAT), a regulator of intracellular Ca2+, and silencing miR-708 leads to elevation of intracellular Ca2+ levels and increased cell migration and metastasis.
miRNA DYSREGULATION CAUSES ABERRANT HISTONE MODIFICATIONS
Dysregulation of miRNAs can also lead to aberrant histone modifications. EZH2 is a member of the polycomb group (PcG) of proteins, which are key regulators that silence numerous developmental genes (Schuettengruber et al., 2007). EZH2 functions as a catalytic subunit of polycomb repressive complex 2 (PRC2), which trimethylates H3K27. The available evidence suggests that EZH2 has oncogenic properties, and its overexpression in prostate and breast cancers promotes tumorigenesis, invasiveness and metastasis (Varambally et al., 2002; Kleer et al., 2003). Varambally et al. (2008) reported that EZH2 is a target of miR-101, and genomic loss of miR-101 is an important cause of EZH2 overexpression in cancer. Reduced expression of miR-101 and upregulation of EZH2 occur in parallel during the progression of prostate cancer, and genomic loss of miR-101 is more frequently seen in metastatic disease than localized cancers. Moreover, the loss of miR-101 and resultant overexpression of EZH2 appears to alter the global chromatin structure in cancer (Friedman et al., 2009). The inverse association between miR-101 and EZH2 has now been seen in bladder, gastric, lung, and renal cancer (Friedman et al., 2009; Wang et al., 2010; Zhang et al., 2011a; Sakurai et al., 2012). In addition, several other miRNAs, including miR-26a (Wong and Tellam, 2008), miR-98 (Alajez et al., 2010), miR-124 (Zheng et al., 2012), miR-144 (Guo et al., 2013), miR-214 (Derfoul et al., 2011), and let-7 (Kong et al., 2012) are also reported to negatively regulate EZH2 (Figure 2). Thus, dysregulation of miRNAs appears to be one of the major causes of EZH2 overexpression in cancer.
Overexpression of EZH2 also leads to the silencing of multiple miRNAs in cancer. It was recently demonstrated that EZH2 is frequently upregulated in primary HCCs, and miRNA expression profiling in HCC cells with EZH2-knockdown revealed that a set of miRNAs, including miR-139-5p, miR-125b, let-7c, miR-101, and miR-200b, are epigenetically suppressed by EZH2 in HCC (Au et al., 2012). Interestingly, miR-200b reportedly targets another PRC2 subunit, SUZ12, in breast cancer stem cells (Iliopoulos et al., 2010), suggesting a possible feedback loop between EZH2 overexpression and miRNA silencing in cancer. In another study, Cao et al. (2011) demonstrated that in prostate and breast cancer cell lines, EZH2 represses a set of miRNAs (miR-181c, miR-181b, miR-200b, miR-200c, and miR-203), which in turn negatively regulate the PRC1 subcomponents BMI1 and RING2. The inverse correlation between miRNA and PRC protein levels were further confirmed in prostate cancer tissues. These results are indicative of an integral regulatory axis involving PRC1, PRC2, and the epigenetic silencing of miRNAs in cancer.
Another recent study demonstrated the involvement of EZH2 and HDAC3 in MYC-mediated miRNA repression. In aggressive B cell lymphoma, miR-29a is repressed by MYC within a co-repressor complex that also includes HDAC3 and EZH2 (Zhang et al., 2012b). Interestingly, MYC contributes to EZH2 upregulation through repression of miR-26a, which targets EZH2, while EZH2 upregulates MYC by inhibiting miR-494, which targets MYC. It thus appears a positive MYC-miRNA-EZH2 feedback loop may mediate persistent overexpression of MYC and EZH2. Combined inhibition of HDAC3 and EZH2 induced restoration of miR-29 and suppressed lymphoma cell growth, suggesting the MYC–EZH2–miRNA axis could be a promising target for epigenetic therapy in B cell lymphoma.
CONCLUDING REMARKS
In this review, we highlighted the relationship between epigenetic alteration of miRNAs and cancer. Aberrant DNA methylation commonly underlies miRNA dysregulation in cancer, and methylation of a subset of miRNA genes may be a useful biomarker for detecting cancer and/or predicting clinical outcome. Alteration of the histone modification pattern also leads to abnormal miRNA expression. In addition, recent findings suggest that miRNA dysregulation is causally related to aberrant DNA methylation and histone modifications that leads to genome-wide epigenetic abnormalities. It is anticipated that additional study of the relationship between epigenetic regulation and miRNAs will lead to the discovery of new biomarkers as well as therapeutic targets.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr William Goldman for editing the manuscript. Hiromu Suzuki is supported by a Grant-in-Aid for Scientific Research (C) from the Japan Society for Promotion of Science, a Grant-in-Aid for the Third-term Comprehensive 10-year Strategy for Cancer Control from the Ministry of Health, Labor, and Welfare, Japan, and the A3 Foresight Program from the Japan Society for Promotion of Science.
REFERENCES
- Agirre X., Vilas-Zornoza A., Jimenez-Velasco A., Martin-Subero J. I., Cordeu L., Garate L., et al. (2009). Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res. 69 4443–4453 10.1158/0008-5472.CAN-08-4025 [DOI] [PubMed] [Google Scholar]
- Alajez N. M., Shi W., Hui A. B., Bruce J., Lenarduzzi M., Ito E., et al. (2010). Enhancer of Zeste homolog 2 (EZH2) is overexpressed in recurrent nasopharyngeal carcinoma and is regulated by miR-26a, miR-101, and miR-98. Cell Death Dis. 1 e85 10.1038/cddis.2010.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando T., Yoshida T., Enomoto S., Asada K., Tatematsu M., Ichinose M., et al. (2009). DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int. J. Cancer 124 2367–2374 10.1002/ijc.24219 [DOI] [PubMed] [Google Scholar]
- Au S. L., Wong C. C., Lee J. M., Fan D. N., Tsang F. H., Ng I. O., et al. (2012). Enhancer of zeste homolog 2 epigenetically silences multiple tumor suppressor microRNAs to promote liver cancer metastasis. Hepatology 56 622–631 10.1002/hep.25679 [DOI] [PubMed] [Google Scholar]
- Baer C., Claus R., Frenzel L. P., Zucknick M., Park Y. J., Gu L., et al. (2012). Extensive promoter DNA hypermethylation and hypomethylation is associated with aberrant microRNA expression in chronic lymphocytic leukemia. Cancer Res. 72 3775–3785 10.1158/0008-5472.CAN-12-0803 [DOI] [PubMed] [Google Scholar]
- Bandres E., Agirre X., Bitarte N., Ramirez N., Zarate R., Roman-Gomez J., et al. (2009). Epigenetic regulation of microRNA expression in colorectal cancer. Int. J. Cancer 125 2737–2743 10.1002/ijc.24638 [DOI] [PubMed] [Google Scholar]
- Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., et al. (2005). Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 37 766–770 10.1038/ng1590 [DOI] [PubMed] [Google Scholar]
- Bommer G. T., Gerin I., Feng Y., Kaczorowski A. J., Kuick R., Love R. E., et al. (2007). p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 17 1298–1307 10.1016/j.cub.2007.06.068 [DOI] [PubMed] [Google Scholar]
- Braconi C., Huang N., Patel T. (2010). MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology 51 881–890 10.1002/hep.23381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueckner B., Stresemann C., Kuner R., Mund C., Musch T., Meister M., et al. (2007). The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 67 1419–1423 10.1158/0008-5472.CAN-06-4074 [DOI] [PubMed] [Google Scholar]
- Buurman R., Gurlevik E., Schaffer V., Eilers M., Sandbothe M., Kreipe H., et al. (2012). Histone deacetylases activate hepatocyte growth factor signaling by repressing microRNA-449 in hepatocellular carcinoma cells. Gastroenterology 143 811–820e15. 10.1053/j.gastro.2012.05.033 [DOI] [PubMed] [Google Scholar]
- Calin G. A., Dumitru C. D., Shimizu M., Bichi R., Zupo S., Noch E., et al. (2002). Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 99 15524–15529 10.1073/pnas.242606799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G. A., Sevignani C., Dumitru C. D., Hyslop T., Noch E., Yendamuri S., et al. (2004). Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. U.S.A. 101 2999–3004 10.1073/pnas.0307323101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q., Mani R. S., Ateeq B., Dhanasekaran S. M., Asangani I. A., Prensner J. R., et al. (2011). Coordinated regulation of polycomb group complexes through microRNAs in cancer. Cancer Cell 20 187–199 10.1016/j.ccr.2011.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceppi P., Mudduluru G., Kumarswamy R., Rapa I., Scagliotti G. V., Papotti M., et al. (2010). Loss of miR-200c expression induces an aggressive, invasive, and chemoresistant phenotype in non-small cell lung cancer. Mol. Cancer Res. 8 1207–1216 10.1158/1541-7786.MCR-10-0052 [DOI] [PubMed] [Google Scholar]
- Chang S., Wang R. H., Akagi K., Kim K. A., Martin B. K., Cavallone L., et al. (2011). Tumor suppressor BRCA1 epigenetically controls oncogenic microRNA-155. Nat. Med. 17 1275–1282 10.1038/nm.2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Hu H., Guan X., Xiong G., Wang Y., Wang K., et al. (2012). CpG island methylation status of miRNAs in esophageal squamous cell carcinoma. Int. J. Cancer 130 1607–1613 10.1002/ijc.26171 [DOI] [PubMed] [Google Scholar]
- Croce C. M. (2009). Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 10 704–714 10.1038/nrg2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos V., Moutinho C., Villanueva A., Boque R., Silva P., Carneiro F., et al. (2012). Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene 31 2062–2074 10.1038/onc.2011.383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derfoul A., Juan A. H., Difilippantonio M. J., Palanisamy N., Ried T., Sartorelli V. (2011). Decreased microRNA-214 levels in breast cancer cells coincides with increased cell proliferation, invasion and accumulation of the Polycomb Ezh2 methyltransferase. Carcinogenesis 32 1607–1614 10.1093/carcin/bgr184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A., Slack F. J. (2006). Oncomirs – microRNAs with a role in cancer. Nat. Rev. Cancer 6 259–269 10.1038/nrc1840 [DOI] [PubMed] [Google Scholar]
- Fabbri M., Garzon R., Cimmino A., Liu Z., Zanesi N., Callegari E., et al. (2007). MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. U.S.A. 104 15805–15810 10.1073/pnas.0707628104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornari F., Milazzo M., Chieco P., Negrini M., Marasco E., Capranico G., et al. (2012). In hepatocellular carcinoma miR-519d is up-regulated by p53 and DNA hypomethylation and targets CDKN1A/p21, PTEN, AKT3 and TIMP2. J. Pathol. 227 275–285 10.1002/path.3995 [DOI] [PubMed] [Google Scholar]
- Friedman J. M., Liang G., Liu C. C., Wolff E. M., Tsai Y. C., Ye W., et al. (2009). The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 69 2623–2629 10.1158/0008-5472.CAN-08-3114 [DOI] [PubMed] [Google Scholar]
- Furuta M., Kozaki K. I., Tanaka S., Arii S., Imoto I., Inazawa J. (2010). miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis 31 766–776 10.1093/carcin/bgp250 [DOI] [PubMed] [Google Scholar]
- Gebauer K., Peters I., Dubrowinskaja N., Hennenlotter J., Abbas M., Scherer R., et al. (2013). Hsa-mir-124-3 CpG island methylation is associated with advanced tumours and disease recurrence of patients with clear cell renal cell carcinoma. Br. J. Cancer 108 131–138 10.1038/bjc.2012.537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory P. A., Bert A. G., Paterson E. L., Barry S. C., Tsykin A., Farshid G., et al. (2008). The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10 593–601 10.1038/ncb1722 [DOI] [PubMed] [Google Scholar]
- Guan Y., Mizoguchi M., Yoshimoto K., Hata N., Shono T., Suzuki S. O., et al. (2010). MiRNA-196 is upregulated in glioblastoma but not in anaplastic astrocytoma and has prognostic significance. Clin. Cancer Res. 16 4289–4297 10.1158/1078-0432.CCR-10-0207 [DOI] [PubMed] [Google Scholar]
- Guo Y., Ying L., Tian Y., Yang P., Zhu Y., Wang Z., et al. (2013). miR-144 downregulation increases bladder cancer cell proliferation by targeting EZH2 and regulating Wnt signaling. FEBS J. 280 4531–4538 10.1111/febs.12417 [DOI] [PubMed] [Google Scholar]
- He L., Hannon G. J. (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5 522–531 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- He L., He X., Lim L. P., De Stanchina E., Xuan Z., Liang Y., et al. (2007). A microRNA component of the p53 tumour suppressor network. Nature 447 1130–1134 10.1038/nature05939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt M. A., Gu J., Lin J., Ye Y., Tan W., Tamboli P., et al. (2010). Hsa-miR-9 methylation status is associated with cancer development and metastatic recurrence in patients with clear cell renal cell carcinoma. Oncogene 29 5724–5728 10.1038/onc.2010.305 [DOI] [PubMed] [Google Scholar]
- Huang J., Wang Y., Guo Y., Sun S. (2010). Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology 52 60–70 10.1002/hep.23660 [DOI] [PubMed] [Google Scholar]
- Iliopoulos D., Lindahl-Allen M., Polytarchou C., Hirsch H. A., Tsichlis P. N., Struhl K. (2010). Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol. Cell 39 761–772 10.1016/j.molcel.2010.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incoronato M., Urso L., Portela A., Laukkanen M. O., Soini Y., Quintavalle C., et al. (2011). Epigenetic regulation of miR-212 expression in lung cancer. PLoS ONE 6: e27722 10.1371/journal.pone.0027722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalimutho M., Di Cecilia S., Del Vecchio Blanco G., Roviello F., Sileri P., Cretella M., et al. (2011). Epigenetically silenced miR-34b/c as a novel faecal-based screening marker for colorectal cancer. Br. J. Cancer 104 1770–1778 10.1038/bjc.2011.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke X. S., Qu Y., Rostad K., Li W. C., Lin B., Halvorsen O. J., et al. (2009). Genome-wide profiling of histone h3 lysine 4 and lysine 27 trimethylation reveals an epigenetic signature in prostate carcinogenesis. PLoS ONE 4:e4687 10.1371/journal.pone.0004687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Lee H. C., Park J. L., Kim M., Kim S. Y., Noh S. M., et al. (2011). Epigenetic regulation of microRNA-10b and targeting of oncogenic MAPRE1 in gastric cancer. Epigenetics 6 740–751 10.4161/epi.6.6.15874 [DOI] [PubMed] [Google Scholar]
- Kleer C. G., Cao Q., Varambally S., Shen R., Ota I., Tomlins S. A., et al. (2003). EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 100 11606–11611 10.1073/pnas.1933744100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D., Heath E., Chen W., Cher M. L., Powell I., Heilbrun L., et al. (2012). Loss of let-7 up-regulates EZH2 in prostate cancer consistent with the acquisition of cancer stem cell signatures that are attenuated by BR-DIM. PLoS ONE 7:e33729 10.1371/journal.pone.0033729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M., Lee E. S., Hu G., Kang Y. (2008). The miR-200 family inhibits epithelial–mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 283 14910–14914 10.1074/jbc.C800074200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozaki K., Imoto I., Mogi S., Omura K., Inazawa J. (2008). Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 68 2094–2105 10.1158/0008-5472.CAN-07-5194 [DOI] [PubMed] [Google Scholar]
- Lehmann U., Hasemeier B., Christgen M., Muller M., Romermann D., Langer F., et al. (2008). Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J. Pathol. 214 17–24 10.1002/path.2251 [DOI] [PubMed] [Google Scholar]
- Li A., Omura N., Hong S. M., Vincent A., Walter K., Griffith M., et al. (2010). Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res. 70 5226–5237 10.1158/0008-5472.CAN-09-4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodygin D., Tarasov V., Epanchintsev A., Berking C., Knyazeva T., Korner H., et al. (2008). Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle 7 2591–2600 10.4161/cc.7.16.6533 [DOI] [PubMed] [Google Scholar]
- Lujambio A., Calin G. A., Villanueva A., Ropero S., Sanchez-Cespedes M., Blanco D., et al. (2008). A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. U.S.A. 105 13556–13561 10.1073/pnas.0803055105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A., Ropero S., Ballestar E., Fraga M. F., Cerrato C., Setien F., et al. (2007). Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 67 1424–1429 10.1158/0008-5472.CAN-06-4218 [DOI] [PubMed] [Google Scholar]
- Luthra R., Singh R. R., Luthra M. G., Li Y. X., Hannah C., Romans A. M., et al. (2008). MicroRNA-196a targets annexin A1: a microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene 27 6667–6678 10.1038/onc.2008.256 [DOI] [PubMed] [Google Scholar]
- Majid S., Dar A. A., Saini S., Shahryari V., Arora S., Zaman M. S., et al. (2013). miRNA-34b inhibits prostate cancer through demethylation, active chromatin modifications, and AKT pathways. Clin. Cancer Res. 19 73–84 10.1158/1078-0432.CCR-12-2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A., Levine S. S., Cole M. F., Frampton G. M., Brambrink T., Johnstone S., et al. (2008). Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134 521–533 10.1016/j.cell.2008.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maru D. M., Singh R. R., Hannah C., Albarracin C. T., Li Y. X., Abraham R., et al. (2009). MicroRNA-196a is a potential marker of progression during Barrett’s metaplasia–dysplasia-invasive adenocarcinoma sequence in esophagus. Am. J. Pathol. 174 1940–1948 10.2353/ajpath.2009.080718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo S. A., Moutinho C., Ropero S., Calin G. A., Rossi S., Spizzo R., et al. (2010). A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell 18 303–315 10.1016/j.ccr.2010.09.007 [DOI] [PubMed] [Google Scholar]
- Melo S. A., Ropero S., Moutinho C., Aaltonen L. A., Yamamoto H., Calin G. A., et al. (2009). A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat. Genet. 41 365–370 10.1038/ng.317 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Neves R., Scheel C., Weinhold S., Honisch E., Iwaniuk K. M., Trompeter H. I., et al. (2010). Role of DNA methylation in miR-200c/141 cluster silencing in invasive breast cancer cells. BMC Res. Notes 3:219 10.1186/1756-0500-3-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng E. K., Tsang W. P., Ng S. S., Jin H. C., Yu J., Li J. J., et al. (2009). MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br. J. Cancer 101 699–706 10.1038/sj.bjc.6605195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura N., Li C. P., Li A., Hong S. M., Walter K., Jimeno A., et al. (2008). Genome-wide profiling of methylated promoters in pancreatic adenocarcinoma. Cancer Biol. Ther. 7 1146–1156 10.4161/cbt.7.7.6208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F., Poling L. L., Wang Z., Liu H., Liu X. S., Roeder R. G., et al. (2008). Chromatin structure analyses identify miRNA promoters. Genes Dev. 22 3172–3183 10.1101/gad.1706508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. M., Gaur A. B., Lengyel E., Peter M. E. (2008). The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22 894–907 10.1101/gad.1640608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic R., Riesbeck L. E., Velu C. S., Chaubey A., Zhang J., Achille N. J., et al. (2009). Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood 113 3314–3322 10.1182/blood-2008-04-154310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Otero P., Roman-Gomez J., Vilas-Zornoza A., Jose-Eneriz E. S., Martin-Palanco V., Rifon J., et al. (2011). Deregulation of FGFR1 and CDK6 oncogenic pathways in acute lymphoblastic leukaemia harbouring epigenetic modifications of the MIR9 family. Br. J. Haematol. 155 73–83 10.1111/j.1365-2141.2011.08812.x [DOI] [PubMed] [Google Scholar]
- Roman-Gomez J., Agirre X., Jimenez-Velasco A., Arqueros V., Vilas-Zornoza A., Rodriguez-Otero P., et al. (2009). Epigenetic regulation of microRNAs in acute lymphoblastic leukemia. J. Clin. Oncol. 27 1316–1322 10.1200/JCO.2008.19.3441 [DOI] [PubMed] [Google Scholar]
- Rotkrua P., Akiyama Y., Hashimoto Y., Otsubo T., Yuasa Y. (2011). MiR-9 downregulates CDX2 expression in gastric cancer cells. Int. J. Cancer 129 2611–2620 10.1002/ijc.25923 [DOI] [PubMed] [Google Scholar]
- Ryu S., Mcdonnell K., Choi H., Gao D., Hahn M., Joshi N., et al. (2013). Suppression of miRNA-708 by polycomb group promotes metastases by calcium-induced cell migration. Cancer Cell 23 63–76 10.1016/j.ccr.2012.11.019 [DOI] [PubMed] [Google Scholar]
- Saito Y., Liang G., Egger G., Friedman J. M., Chuang J. C., Coetzee G. A., et al. (2006). Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 9 435–443 10.1016/j.ccr.2006.04.020 [DOI] [PubMed] [Google Scholar]
- Sakurai T., Bilim V. N., Ugolkov A. V., Yuuki K., Tsukigi M., Motoyama T., et al. (2012). The enhancer of zeste homolog 2 (EZH2), a potential therapeutic target, is regulated by miR-101 in renal cancer cells. Biochem. Biophys. Res. Commun. 422 607–614 10.1016/j.bbrc.2012.05.035 [DOI] [PubMed] [Google Scholar]
- Sampath D., Liu C., Vasan K., Sulda M., Puduvalli V. K., Wierda W. G., et al. (2012). Histone deacetylases mediate the silencing of miR-15a, miR-16, and miR-29b in chronic lymphocytic leukemia. Blood 119 1162–1172 10.1182/blood-2011-05-351510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., Cavalli G. (2007). Genome regulation by polycomb and trithorax proteins. Cell 128 735–745 10.1016/j.cell.2007.02.009 [DOI] [PubMed] [Google Scholar]
- Scott G. K., Mattie M. D., Berger C. E., Benz S. C., Benz C. C. (2006). Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 66 1277–1281 10.1158/0008-5472.CAN-05-3632 [DOI] [PubMed] [Google Scholar]
- Shimizu T., Suzuki H., Nojima M., Kitamura H., Yamamoto E., Maruyama R., et al. (2013). Methylation of a panel of microRNA genes is a novel biomarker for detection of bladder cancer. Eur. Urol. 63 1091–1100 10.1016/j.eururo.2012.11.030 [DOI] [PubMed] [Google Scholar]
- Siemens H., Neumann J., Jackstadt R., Mansmann U., Horst D., Kirchner T., et al. (2013). Detection of miR-34a promoter methylation in combination with elevated expression of c-Met and beta-catenin predicts distant metastasis of colon cancer. Clin. Cancer Res. 19 710–720 10.1158/1078-0432.CCR-12-1703 [DOI] [PubMed] [Google Scholar]
- Suzuki H., Takatsuka S., Akashi H., Yamamoto E., Nojima M., Maruyama R., et al. (2011). Genome-wide profiling of chromatin signatures reveals epigenetic regulation of microRNA genes in colorectal cancer. Cancer Res. 71 5646–5658 10.1158/0008-5472.CAN-11-1076 [DOI] [PubMed] [Google Scholar]
- Suzuki H., Yamamoto E., Nojima M., Kai M., Yamano H. O., Yoshikawa K., et al. (2010). Methylation-associated silencing of microRNA-34b/c in gastric cancer and its involvement in an epigenetic field defect. Carcinogenesis 31 2066–2073 10.1093/carcin/bgq203 [DOI] [PubMed] [Google Scholar]
- Suzuki R., Yamamoto E., Nojima M., Maruyama R., Yamano H. O., Yoshikawa K., et al. (2013). Aberrant methylation of microRNA-34b/c is a predictive marker of metachronous gastric cancer risk. J. Gastroenterol. 10.1007/s00535-013-0861-7 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M., Suzuki H., Sasaki Y., Maruyama R., Imai K., Shinomura Y., et al. (2008). Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 68 4123–4132 10.1158/0008-5472.CAN-08-0325 [DOI] [PubMed] [Google Scholar]
- Tsai K. W., Hu L. Y., Wu C. W., Li S. C., Lai C. H., Kao H. W., et al. (2010). Epigenetic regulation of miR-196b expression in gastric cancer. Genes Chromosomes Cancer 49 969–980 10.1002/gcc.20804 [DOI] [PubMed] [Google Scholar]
- Tsai K. W., Kao H. W., Chen H. C., Chen S. J., Lin W. C. (2009). Epigenetic control of the expression of a primate-specific microRNA cluster in human cancer cells. Epigenetics 4 587–592 10.4161/epi.4.8.10230 [DOI] [PubMed] [Google Scholar]
- Tsai K. W., Liao Y. L., Wu C. W., Hu L. Y., Li S. C., Chan W. C., et al. (2011). Aberrant hypermethylation of miR-9 genes in gastric cancer. Epigenetics 6 1189–1197 10.4161/epi.6.10.16535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S., Cao Q., Mani R. S., Shankar S., Wang X., Ateeq B., et al. (2008). Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 322 1695–1699 10.1126/science.1165395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S., Dhanasekaran S. M., Zhou M., Barrette T. R., Kumar-Sinha C., Sanda M. G., et al. (2002). The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419 624–629 10.1038/nature01075 [DOI] [PubMed] [Google Scholar]
- Vogt M., Munding J., Gruner M., Liffers S. T., Verdoodt B., Hauk J., et al. (2011). Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 458 313–322 10.1007/s00428-010-1030-5 [DOI] [PubMed] [Google Scholar]
- Vrba L., Jensen T. J., Garbe J. C., Heimark R. L., Cress A. E., Dickinson S., et al. (2010). Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS ONE 5:e8697 10.1371/journal.pone.0008697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wu J., Meng X., Ying X., Zuo Y., Liu R., et al. (2011a). MicroRNA-342 inhibits colorectal cancer cell proliferation and invasion by directly targeting DNA methyltransferase 1. Carcinogenesis 32 1033–1042 10.1093/carcin/bgr081 [DOI] [PubMed] [Google Scholar]
- Wang Z., Chen Z., Gao Y., Li N., Li B., Tan F., et al. (2011b). DNA hypermethylation of microRNA-34b/c has prognostic value for stage non-small cell lung cancer. Cancer Biol. Ther. 11 490–496 10.4161/cbt.11.5.14550 [DOI] [PubMed] [Google Scholar]
- Wang H. J., Ruan H. J., He X. J., Ma Y. Y., Jiang X. T., Xia Y. J., et al. (2010). MicroRNA-101 is down-regulated in gastric cancer and involved in cell migration and invasion. Eur. J. Cancer 46 2295–2303 10.1016/j.ejca.2010.05.012 [DOI] [PubMed] [Google Scholar]
- Wang P., Chen L., Zhang J., Chen H., Fan J., Wang K., et al. (2013). Methylation-mediated silencing of the miR-124 genes facilitates pancreatic cancer progression and metastasis by targeting Rac1. Oncogene. 10.1038/onc.2012.598 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Wang Y., Toh H. C., Chow P., Chung A. Y., Meyers D. J., Cole P. A., et al. (2012). MicroRNA-224 is up-regulated in hepatocellular carcinoma through epigenetic mechanisms. FASEB J. 26 3032–3041 10.1096/fj.11-201855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Emoto N., Hamano E., Sunohara M., Kawakami M., Kage H., et al. (2012). Genome structure-based screening identified epigenetically silenced microRNA associated with invasiveness in non-small-cell lung cancer. Int. J. Cancer 130 2580–2590 10.1002/ijc.26254 [DOI] [PubMed] [Google Scholar]
- Wiklund E. D., Bramsen J. B., Hulf T., Dyrskjot L., Ramanathan R., Hansen T. B., et al. (2011). Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int. J. Cancer 128 1327–1334 10.1002/ijc.25461 [DOI] [PubMed] [Google Scholar]
- Wilting S. M., Van Boerdonk R. A., Henken F. E., Meijer C. J., Diosdado B., Meijer G. A., et al. (2010). Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Mol. Cancer 9 167 10.1186/1476-4598-9-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C. F., Tellam R. L. (2008). MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J. Biol. Chem. 283 9836–9843 10.1074/jbc.M709614200 [DOI] [PubMed] [Google Scholar]
- Wong K. Y., So C. C., Loong F., Chung L. P., Lam W. W., Liang R., et al. (2011a). Epigenetic inactivation of the miR-124-1 in haematological malignancies. PLoS ONE 6:e19027 10.1371/journal.pone.0019027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K. Y., Yim R. L., So C. C., Jin D. Y., Liang R., Chim C. S. (2011b). Epigenetic inactivation of the MIR34B/C in multiple myeloma. Blood 118 5901–5904 10.1182/blood-2011-06-361022 [DOI] [PubMed] [Google Scholar]
- Yan H., Choi A. J., Lee B. H., Ting A. H. (2011). Identification and functional analysis of epigenetically silenced microRNAs in colorectal cancer cells. PLoS ONE 6:e20628 10.1371/journal.pone.0020628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Jiao Y., Zhu Y., Wang Y., Zhu J., Cui X., et al. (2012). MicroRNA 34c gene down-regulation via DNA methylation promotes self-renewal and epithelial–mesenchymal transition in breast tumor-initiating cells. J. Biol. Chem. 287 465–473 10.1074/jbc.M111.280768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. G., Guo J. F., Liu D. L., Liu Q., Wang J. J. (2011a). MicroRNA-101 exerts tumor-suppressive functions in non-small cell lung cancer through directly targeting enhancer of zeste homolog 2. J. Thorac. Oncol. 6 671–678 10.1097/JTO.0b013e318208eb35 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Tang H., Wang Z., Zhang B., Liu W., Lu H., et al. (2011b). MiR-185 targets the DNA methyltransferases 1 and regulates global DNA methylation in human glioma. Mol. Cancer 10 124 10.1186/1476-4598-10-124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Chen X., Lin J., Lwin T., Wright G., Moscinski L. C., et al. (2012a). Myc represses miR-15a/miR-16-1 expression through recruitment of HDAC3 in mantle cell and other non-Hodgkin B-cell lymphomas. Oncogene 31 3002–3008 10.1038/onc.2011.470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhao X., Fiskus W., Lin J., Lwin T., Rao R., et al. (2012b). Coordinated silencing of MYC-mediated miR-29 by HDAC3 and EZH2 as a therapeutic target of histone modification in aggressive B-cell lymphomas. Cancer Cell 22 506–523 10.1016/j.ccr.2012.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zheng F., Liao Y. J., Cai M. Y., Liu Y. H., Liu T. H., Chen S. P., et al. (2012). The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut 61 278–289 10.1136/gut.2011.239145 [DOI] [PubMed] [Google Scholar]