Abstract
The Wave proteins are major activators of the Arp2/3 complex. The ubiquitous Wave-2 is required for actin polymerization at the leading edge of migrating cells. Here we purify Wave-2 from HeLa cells. Five proteins, Sra, Nap, Wave-2, Abi, and Hspc, are copurified, indicating that they form a tight complex. These proteins are only present in the complexed form, with the exception of Hspc, which displays a free pool. We reconstitute the Wave-2 complex by cotranslating in vitro the five subunits and use this system together with specific immunoprecipitations to study the molecular architecture of the complex. The complex is organized around a core of Nap and Abi. Sra is a peripheral subunit recruited on the Nap side, whereas the Wave and Hspc subunits are recruited on the Abi side of the core.
The actin cytoskeleton is required to ensure the proper shape of the eukaryotic cell. The dynamics of the actin monomer-polymer equilibrium are under tight control from the cell and are regulated by signal transduction cascades. Cell migration is typically initiated by growth factor stimulation. An essential structure for cell migration is the lamellipodium, which is the projection of the plasma membrane at the leading edge of a migrating cell (1). Actin polymerization provides the driving force for this plasma membrane projection. The molecular processes responsible for localized actin polymerization are the subject of intense investigation because of their critical functions in the physiology of animal cells, for example, during cell shape changes and motility in development, and in pathology, for example, during tumorigenesis when tumor cells acquire an invasive behavior.
The Arp2/3 complex, which contains two actin-related proteins, is a major regulator of actin polymerization. In particular, the Arp2/3 complex stimulates actin polymerization by actin filament branching on existing filaments (1). In animal cells, the two major classes of Arp2/3 activators are the WASP proteins and the Wave proteins. WASP proteins are dispensable for the formation of lamellipodia (2, 3), whereas Wave proteins have emerged as a critical regulator of this structure. In fact, Wave proteins are localized at the tip of the protruding lamellipodium and delocalized whenever this structure retracts (4); genetic inactivation of the ubiquitously expressed Wave-2 gene causes defects in the formation of lamellipodia (5, 6). These results highlight the importance of the Wave–Arp2/3 pathway in the formation of lamellipodia in migrating cells.
The small GTPase Rac controls the formation of lamellipodia (7). The formation of lamellipodia by active Rac is blocked by dominant negative constructs of Wave, suggesting that Wave is a downstream Rac effector (8). However, no direct binding was detected, suggesting other intermediaries in this signaling pathway. The first identified link was IRSp53, which was shown to bind to active Rac and to the ubiquitous Wave-2 protein but very poorly to the other Wave proteins (9). Recently, our group has purified a multiprotein complex containing the brain-specific Wave-1 (10). This complex maintained Wave-1 in an inactive conformation. An active form of Wave-1 was released on binding to active Rac (10). We show here that the ubiquitously expressed Wave-2 protein is in a similar large complex as the brain-specific Wave-1. We investigated the substructure of the Wave-2 complex by using reconstitution of partial complexes and specific immunoprecipitations. This architecture, which may be general for Wave proteins, shows the complex built around a core of the Nap and Abi subunits.
Materials and Methods
Purification of the Wave-2 Complex by Chromatography and Protein Identification by Mass Spectrometry. HeLa-S3 cell pellets were obtained from the National Cell Culture Center (Minneapolis, MN). All chromatography was performed at 4°C by using resins purchased from Amersham Pharmacia Biotech. Cells were resuspended in 3-fold their volume of XB buffer (20 mM Hepes/100 mM KCl/1 mM MgCl2/0.1 mM EDTA/1mMDTT, pH 7.7) and lysed by nitrogen cavitation [500 psi for 20 min (1 psi = 6.89 kPa); Parr Instruments, Moline, IL]. The extract was clarified by centrifugation (15,000 rpm for 15 min; Beckman Coulter SA 600 rotor) and ultracentrifugation (40,000 rpm for 2 h; Beckman Coulter 45Ti rotor). The Wave complex was precipitated by an ammonium sulfate cut from 20% to 30% saturation, resuspended in 20 mM Pipes/50 mM KCl/1 mM MgCl2/0.1 mM EDTA/1 mM DTT, pH 6.8, and loaded onto a 6-ml Resource S column. The Wave complex was eluted with a linear gradient of KCl and came off at ≈130 mM KCl. The fractions then were dialyzed against 20 mM Tris/50 mM KCl/1 mM MgCl2/0.1 mM EDTA/1 mM DTT, pH 8.0, and loaded onto a 1-ml Mono Q column. The Wave complex was eluted with a linear gradient of KCl and came off at ≈330 mM KCl. The complex was then purified on a 12-ml 5–20% linear sucrose gradient (see below).
Protein identification by tandem mass spectrometry was as described (11). Briefly, protein bands were excised from SDS/PAGE gels and digested with sequencing-grade trypsin (Promega). Digested samples were pressure-loaded onto a fused silica microcapillary C18 column (Magic beads; Michrom BioResources, Auburn, CA) packed in-house. An Agilent 1100 high-pressure liquid chromatography (HPLC) system (Agilent Technologies, Palo Alto, CA) was used to deliver a gradient across a flow splitter to the column. Eluting peptides from the column were ionized by electrospray ionization and analyzed by an LCQ-Deca XP ion-trap mass spectrometer (Thermo Finnigan, Woburn, MA). Peptide ions were dynamically selected by the operating software for fragmentation. The peptide fragmentation spectra were searched against the nonredundant protein database by using the sequest computer algorithm. The lists of peptides unambiguously identifying the subunits are available on request.
cDNAs and Antibodies. Human Sra (GenBank accession no. XM_039225), Nap (GenBank accession no. AB011159), Wave (GenBank accession no. AB026542), Abi (GenBank accession no. BC024254), and Hspc (GenBank accession no. BC019303) were obtained by PCR of cDNA or the corresponding ESTs and cloned into pCS2 to express untagged proteins or into pCS2-HA to express fusion proteins with a triple hemagglutinin (HA) tag in the N terminus. ORFs were sequenced to ensure that no mutation was introduced. Specific polyclonal sera were obtained after immunization of rabbits with the following KLH-conjugated peptides (Zymed): Sra, (C)LVHPTDKYSNKDCPDSA; Nap, (C)KKSKKQTGKKGEPEREK; Wave-2, (C)NQRGSGLAGPKRSS; Abi, (C)SSGGYRRTPSV; and Hspc, (C)IEYIEARVTKGETLT. All of the antibodies were affinity-purified against the immobilized peptides.
In Vitro Translation and Immunoprecipitations. In vitro translation of pairs of plasmids or of an equimolar mixture of the five plasmids was performed by using the SP6 polymerase and 40 μCi of [35S]methionine in a total reaction volume of 50 μl with the TnT kit according to the manufacturer's instructions (Promega). Four hundred fifty microliters of XB, 10 μl of protein G ultralink beads (Pierce), and 1 μg of 12CA5 mAb then were added. The mixture was incubated for 2 h at 4°C with rocking. The beads then were washed four times with 1 ml of XB and resuspended in SDS loading buffer. The inputs (1/25th) and the HA immunoprecipitates (1/3rd) were analyzed by SDS/PAGE on 4–12% Bis-Tris gels with the Mes buffer (Invitrogen). After migration, the gel was fixed in for 20 min, rinsed in water, incubated in 1 M of sodium salicylate, dried, and exposed to films or phosphorimager screens (Bio-Rad).
Sucrose Gradient. Linear 5–20% sucrose gradients were poured in XB by using the Auto Densi Flow from Labconco (Kansas City, MO). Markers were chymotrypsinogen A 2.6 S, albumin 4.6 S, aldolase 7.3 S, catalase 11.3 S, thyroglobulin 19.2 S (Amersham Pharmacia Biotech). Two hundred microliters of markers, high-speed extract, or the in vitro translation reaction was loaded on top of the 12-ml gradient. The gradients were run for 17 h at 40,000 rpm in a SW40 rotor (Beckman Coulter). Fractions of ≈0.5 ml were harvested from the top by using the Auto Densi Flow. Fractions of the high-speed extract then were concentrated by using trichloroacetic acid precipitation with insulin as a carrier.
Results
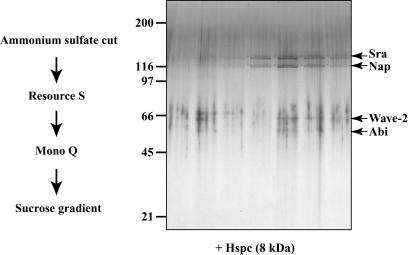
To examine whether Wave-2 was in a complex, as described for the brain-specific Wave-1 protein, we purified Wave-2 from HeLa cells after its immunoreactivity. We prepared a high-speed supernatant from a culture of 20 liters of cells. This extract then was sequentially fractionated by ammonium sulfate precipitation, two ion exchange columns, and a final sucrose gradient (Fig. 1). After these four steps, Wave-2 had been purified together with four other proteins. These five proteins subsequently were shown to form a tight complex (see below), indicating that the procedure we describe here leads to the purification to homogeneity of the Wave-2 complex.
Fig. 1.
Purification of the Wave-2-containing complex from HeLa cells. After four steps of chromatography (depicted on the left), the final fractions obtained after the sucrose gradient were loaded on 5–15% gradient polyacrylamide gel. After SDS/PAGE, the gel was silver-stained, and the bands were excised and analyzed by mass spectrometry. In addition to the four clear bands, Hspc was not detected by the staining of the gel but was clearly detected when the complete fraction was analyzed by mass spectrometry.
Mass spectrometry was used to identify the proteins present in the complex. The four detected bands, in decreasing size order, consisted of specifically Rac1-associated protein, or p140Sra-1 (12); Nck-associated protein 1, or p125Nap1 (13); Wave-2 (14); and Abl-interating protein 1, or Abi-1 (15). In addition, when the final fraction was analyzed directly by mass spectrometry, a fifth protein of ≈8 kDa, called HSPC300 (10), was unambiguously identified, even though a corresponding band after SDS/PAGE was too weak to be detected. These proteins, or homologous proteins, already were found to be complexed with Wave-1 purified from brain (10, 16). For all subunits except HSPC300, paralogous genes exist in mammalian genomes, three for Wave-2 and Abi-1 and two for p140Sra-1 and p125Nap1. Moreover, multiple splice variants of Abi-1 have been described (17). In addition to the combinatorial complexity typically displayed by different tissues, it is likely that even a single cell type possesses slightly different Wave complexes, because we could detect a few peptides from the homologous PIR121 protein in the p140Sra-1 band. For the rest of this article, the cDNAs corresponding to the major subunits found in the Wave-2 complex of HeLa cells, together with specific antibodies for these proteins, were used. For the sake of simplicity, these five proteins are referred to as Sra, Nap, Wave, Abi, and Hspc.
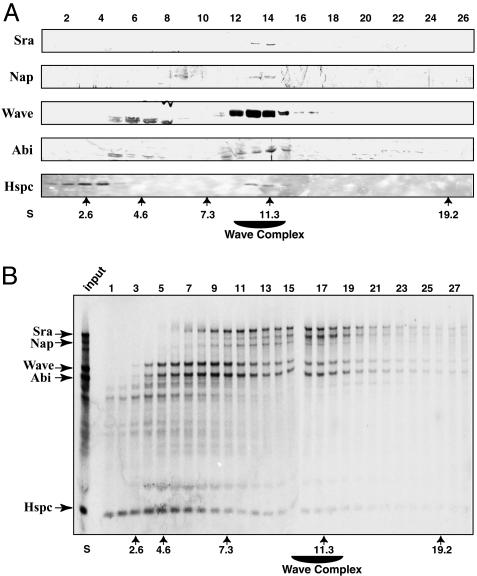
To improve our understanding of Wave regulation, we attempted to devise experiments revealing the architecture of the Wave complex. First, we asked whether partial complexes or free pools of subunits exist. We fractionated the HeLa extract on a sucrose gradient and analyzed the distribution of the subunits by Western blot. The complex peaks in fractions 13 and 14 at ≈11 Swedbergs where all five proteins are detected (Fig. 2A). Importantly, Sra and Nap were present only in the complex because no other migrating forms could be detected. Wave and Abi are mostly present in the complex, but some faster migrating bands also were observed between fractions 5 and 8. These additional bands might correspond to crossreacting proteins or degradation products of Abi and Wave. In contrast to the four other subunits, the major pool of Hspc was in the first fractions of the gradient, which correspond to a free form, with only the remaining portion being complexed in the 11 S complex. We then attempted to reconstitute the Wave complex to show that all of the subunits of the complex were identified. We expressed together the five cDNAs corresponding to the subunits in an in vitro translation system. The subunits were cotranslated in presence of [35S]methionine, and the reticulocyte lysate was loaded on a sucrose gradient. Importantly, a complex of all five proteins peaked in fractions 16 and 17, corresponding to 11 S, like the native complex (Fig. 2B). This peak was particularly well seen with the Nap protein, which was produced in limiting amount, whereas the excess of the other subunits trailed behind the 11 S complex as free forms or partial complexes. This experiment suggested that all of the components of the Wave complex were identified and provided us with a system to study the interactions between subunits.
Fig. 2.
Distribution of the subunits and reconstitution of the Wave-2 complex. (A) A HeLa extract was loaded on a sucrose gradient, and the fractions were immunoblotted with antibodies targeting the different subunits. The complex peaks at ≈11 Swedbergs (fractions 13 and 14) where all five proteins could be detected. (B) Reconstitution of the complex by in vitro translation. The genes encoding the five subunits were cotranslated in vitro in the presence of [35S]methionine, and the mixture was separated on a sucrose gradient. Nap is the limiting component in the mixture and is highly enriched at the size of the native complex, 11 S. The other subunits also were incorporated in the reconstituted complex at 11 S.
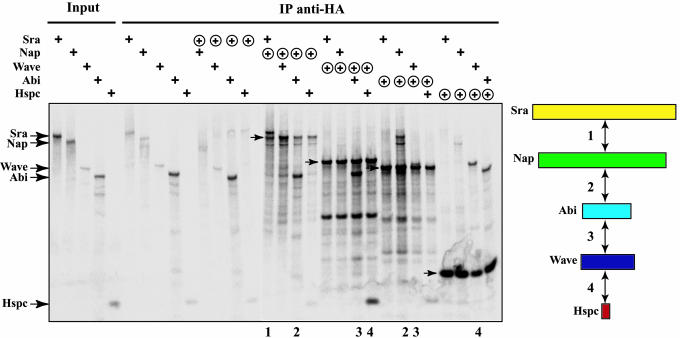
To examine all of the possible interactions between pairs of subunits, we transferred the five cDNAs into a plasmid encoding three HA epitopes in fusion with the N terminus of proteins. We tested the interaction for a pair by coimmunoprecipitating an untagged subunit with a HA-tagged subunit by using HA antibodies (Fig. 3). This method of cotranslating in vitro the subunits with [35S]methionine proved to be highly sensitive, because many interactions were detected between subunits even though this system produces only a very small amount of proteins. First, we controlled for the background amount of protein we retrieved in the HA immunoprecipitation when no HA protein was expressed. This background varies according to the expressed subunit and probably reflects the propensity of each isolated subunit to precipitate and stick to the beads. An interaction was scored positive when the amount of untagged protein bound to the HA-tagged protein was significantly higher than this background amount. The HA-tagged Nap, Wave, Abi, and Hspc were efficiently immunoprecipitated. Surprisingly, HA-tagged Sra could not be significantly immunoprecipitated, even though it was produced (data not shown) and was not analyzed further. The following interactions were found significant: HA-tagged Nap bound to both Sra and Abi; HA-tagged Wave bound to both Hspc and Abi; HA-tagged Abi bound to both Nap and Wave; and HA-tagged Hspc bound to Wave. This system thus detected four interactions between the five subunits, suggesting a model for the architecture of the Wave complex as a stack of Sra, Nap, Abi, Wave, and Hspc, in that order.
Fig. 3.
Interactions between subunits. All possible pairs of subunits were cotranslated in vitro in the presence of [35S]methionine, one subunit being untagged (+) and the other being tagged at the N terminus by HA epitopes (⊕). The input and HA immunoprecipitates were resolved by SDS/PAGE and autoradiographed. The immunoprecipitated subunits are indicated on the autoradiograph by arrows. The lanes demonstrating a significant interaction (over the negative control) are numbered, and the interactions are depicted on a diagram on the right.
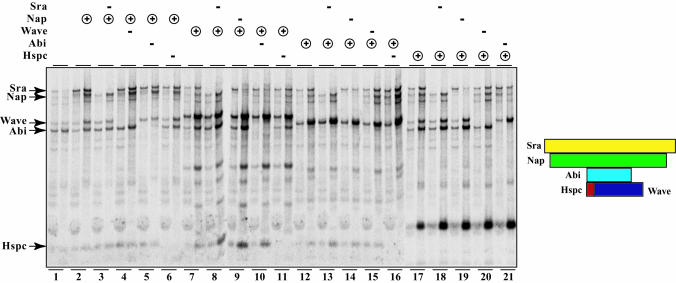
Such a linear model predicts that all of the more peripheral components would be lost when one central subunit is omitted from the reaction. Thus, to gain a better insight into the architecture of the Wave complex, we decided to assess the integrity of the complex when specific subunits were omitted. All possible combinations of omitted and immunoprecipitated subunits were tested (Fig. 4). In this experiment, the reaction where one subunit is omitted was compared to the complete reaction by using the same immunoprecipitated subunit. This experiment permits the classification of subunits into peripheral or core subunits depending on whether its omission leads to a destabilization of the complex formed by the remaining subunits.
Fig. 4.
Reconstitution of all possible complexes lacking one subunit. The five subunits were cotranslated in vitro in the presence of [35S]methionine. The HA-tagged subunit is indicated by ⊕, and the omitted subunit is indicated by –. The presence of all of the untagged subunits is not indicated for the sake of clarity. For each reaction, two lanes are shown, the input on the left and the HA immunoprecipitate on the right.
Sra is a peripheral subunit of the complex. When Sra was omitted, a complex of Nap, Wave, Abi, and Hspc still formed, and to the same extent as when Sra was included. This result was seen when Nap was immunoprecipitated (see reaction 3 vs. 2), when Wave was immunoprecipitated (8 vs. 7), when Abi was immunoprecipitated (13 vs. 12), and when Hspc was immunoprecipitated (18 vs. 17).
In contrast, Nap is a core component of the complex that is essential for the recruitment of Sra. When Nap was omitted, a complex of Wave, Abi, and Hspc still formed but no longer recruited Sra. This result was most clearly seen when Hspc was immunoprecipitated (19 vs. 17) but also when Wave was immunoprecipitated (9 vs. 7) and when Abi was immunoprecipitated (14 vs. 12).
Abi is also a core component. On the one hand, Abi recruits Nap and Sra. This result was seen when Hspc was immunoprecipitated (21 vs. 17) and when Wave was immunoprecipitated (10 vs. 7). On the other hand, Abi strongly enhanced the recruitment of Wave and Hspc when Nap was immunoprecipitated (5 vs. 2).
Wave is a peripheral subunit. When Wave was omitted, a complex of Sra, Nap, Abi, and Hspc still formed. This was most clearly seen when Hspc was immunoprecipitated (20 vs. 17) but also when Nap was immunoprecipitated (4 vs. 2) and when Abi was immunoprecipitated (15 vs. 12). The fact that Hspc was not lost on Wave omission indicates that Hspc does not only interact with Wave, as detected in Fig. 3, but also with another subunit. This other partner of Hspc is likely to be Abi, by elimination of the other candidates, because the immunoprecipitated Hspc did not coimmunoprecipitate Nap and Sra when Abi was omitted (21 vs. 17).
Hspc is also a peripheral subunit. When Hspc was omitted, a complex of Sra, Nap, Wave, and Abi still formed. This was most clearly seen when Nap was immunoprecipitated (6 vs. 2) but also when Wave was immunoprecipitated (11 vs. 7) and when Abi was immunoprecipitated (16 vs. 12).
In conclusion, the Wave complex is organized around a central core of Nap and Abi, which recruits the peripheral subunits, Sra on the Nap side and the Wave-Hspc unit on the other side, as depicted in Fig. 4.
Discussion
Our purification of the ubiquitous Wave-2 confirms the generality of the Wave complex. The same or homologous subunits previously were found in complex with the brain-specific Wave-1 (10). IRSp53, which previously was proposed to control specifically the activity of Wave-2 (9), was not part of the complex. It thus is likely that IRSp53 is only transiently recruited in response to an activating signal. Our reconstitution provides evidence that all of the subunits of the complex actually have been identified. Importantly, recent genetic evidence indicates that this structure of complex is also important for the regulation of invertebrate Wave proteins, known as Scar proteins. Indeed, inactivation of the Sra homolog in Dictyostelium or of Sra, Nap, and Abi homologs in Drosophila leads in both cases to Scar/Wave phenotypes (18–21). In addition, in both systems, on genetic removal of the subunits, Scar/Wave is degraded. In line with our finding that Hspc is present in excess in the HeLa cell extract, RNA interference depletion of the Hspc homologous protein in Drosophila cells results in a milder phenotype than the corresponding depletion of the other subunits (19, 20). Together with our biochemical analysis of subunit distribution, these results suggest that the Wave complex is the only stable entity in vertebrate and invertebrate cells. This evidence suggests that the architecture of the human Wave-2 complex defined here is likely to hold true for the invertebrate Scar/Wave complexes.
We have found that the interacting subunits Nap and Abi provide the core of the complex, because they are essential for the recruitment of peripheral subunits to the complex. The Sra subunit is recruited by Nap to the core, and the interacting Wave and Hspc subunits are recruited by Abi to the core. This protein complex maintains Wave in an inactive conformation, because the isolated recombinant Wave protein, but not the Wave complex, activates the Arp2/3 complex (10, 22). We found that Abi and Hspc are the only two subunits interacting directly with Wave within the complex and are thus the likely mediators of this inhibition of Wave. However, Hspc is not sufficient to inhibit Wave because the active form of Wave is a complex of Wave and Hspc (10). Abi is also not sufficient for this inhibition (A.G., unpublished data). It is likely that the inhibition of Wave requires not only the direct interactors Abi and Hspc but also the folding constraints provided by the complex that is the stable biochemical entity. On activation of the complex, the active unit composed of the Wave and Hspc is released (10). The GTP-loaded Rac activator binds to the Sra subunit (12). This suggests that Rac binding to Sra on the Nap side of the complex triggers a conformational change that is transferred to the Abi side and eventually results in the dissociation of the Wave-Hspc unit.
The reconstitution of a multiprotein complex by using in vitro translation is a general approach to study the interactions among subunits. We have used this system to test, in a systematic manner, all possible interactions between the subunits of the ubiquitous Wave complex by coimmunoprecipitations. These experiments allowed us to draw a map of interactions. This map constitutes a working model to integrate the architecture and the regulation of the Wave complex. It also provides essential information to form partial complexes in bacteria and insect cells by coexpressing interacting subunits to reconstitute a large amount of Wave complex for structural studies. In contrast to the Arp2/3 complex (23), this reconstitution of the Wave complex is likely required for crystallography, given the combinatorial complexity of the native mammalian Wave complexes.
Acknowledgments
We thank Ethan Lee, Olaf Stemmann, Sharon Eden, Ann Marie Pendergast, Annette Schenck, and Jean-Louis Mandel for reagents and the National Cell Culture Center for providing HeLa cells. This work was supported by grants from the National Institutes of Health, a National Institute of General Medical Sciences grant entitled “Biochemical Studies of Mitosis” (2-R01-GM026875-27), and a postdoctoral fellowship from the Human Frontier Science Program (to A.G.).
Abbreviation: HA, hemagglutinin.
References
- 1.Pollard, T. D. & Borisy, G. G. (2003) Cell 112, 453–465. [DOI] [PubMed] [Google Scholar]
- 2.Lommel, S., Benesch, S., Rottner, K., Franz, T., Wehland, J. & Kuhn, R. (2001) EMBO Report 2, 850–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snapper, S. B., Takeshima, F., Anton, I., Liu, C. H., Thomas, S. M., Nguyen, D., Dudley, D., Fraser, H., Purich, D., Lopez-Ilasaca, M., et al. (2001) Nat. Cell Biol. 3, 897–904. [DOI] [PubMed] [Google Scholar]
- 4.Hahne, P., Sechi, A., Benesch, S. & Small, J. V. (2001) FEBS Lett. 492, 215–220. [DOI] [PubMed] [Google Scholar]
- 5.Yan, C., Martinez-Quiles, N., Eden, S., Shibata, T., Takeshima, F., Shinkura, R., Fujiwara, Y., Bronson, R., Snapper, S. B., Kirschner, M. W., et al. (2003) EMBO J. 22, 3602–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamazaki, D., Suetsugu, S., Miki, H., Kataoka, Y., Nishikawa, S., Fujiwara, T., Yoshida, N. & Takenawa, T. (2003) Nature 424, 452–456. [DOI] [PubMed] [Google Scholar]
- 7.Etienne-Manneville, S. & Hall, A. (2002) Nature 420, 629–635. [DOI] [PubMed] [Google Scholar]
- 8.Miki, H., Suetsugu, S. & Takenawa, T. (1998) EMBO J. 17, 6932–6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miki, H., Yamaguchi, H., Suetsugu, S. & Takenawa, T. (2000) Nature 408, 732–735. [DOI] [PubMed] [Google Scholar]
- 10.Eden, S., Rohatgi, R., Podtelejnikov, A. V., Mann, M. & Kirschner, M. W. (2002) Nature 418, 790–793. [DOI] [PubMed] [Google Scholar]
- 11.Gygi, S. P., Rochon, Y., Franza, B. R. & Aebersold, R. (1999) Mol. Cell. Biol. 19, 1720–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi, K., Kuroda, S., Fukata, M., Nakamura, T., Nagase, T., Nomura, N., Matsuura, Y., Yoshida-Kubomura, N., Iwamatsu, A. & Kaibuchi, K. (1998) J. Biol. Chem. 273, 291–295. [DOI] [PubMed] [Google Scholar]
- 13.Kitamura, T., Kitamura, Y., Yonezawa, K., Totty, N. F., Gout, I., Hara, K., Waterfield, M. D., Sakaue, M., Ogawa, W. & Kasuga, M. (1996) Biochem. Biophys. Res. Commun. 219, 509–514. [DOI] [PubMed] [Google Scholar]
- 14.Suetsugu, S., Miki, H. & Takenawa, T. (1999) Biochem. Biophys. Res. Commun. 260, 296–302. [DOI] [PubMed] [Google Scholar]
- 15.Shi, Y., Alin, K. & Goff, S. P. (1995) Genes Dev. 9, 2583–2597. [DOI] [PubMed] [Google Scholar]
- 16.Soderling, S. H., Binns, K. L., Wayman, G. A., Davee, S. M., Ong, S. H., Pawson, T. & Scott, J. D. (2002) Nat. Cell Biol. 4, 970–975. [DOI] [PubMed] [Google Scholar]
- 17.Ziemnicka-Kotula, D., Xu, J., Gu, H., Potempska, A., Kim, K. S., Jenkins, E. C., Trenkner, E. & Kotula, L. (1998) J. Biol. Chem. 273, 13681–13692. [DOI] [PubMed] [Google Scholar]
- 18.Blagg, S. L., Stewart, M., Sambles, C. & Insall, R. H. (2003) Curr. Biol. 13, 1480–1487. [DOI] [PubMed] [Google Scholar]
- 19.Kunda, P., Craig, G., Dominguez, V. & Baum, B. (2003) Curr. Biol. 13, 1867–1875. [DOI] [PubMed] [Google Scholar]
- 20.Rogers, S. L., Wiedemann, U., Stuurman, N. & Vale, R. D. (2003) J. Cell Biol. 162, 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bogdan, S. & Klambt, C. (2003) Development (Cambridge, U.K.) 130, 4427–4437. [DOI] [PubMed] [Google Scholar]
- 22.Machesky, L. M., Mullins, R. D., Higgs, H. N., Kaiser, D. A., Blanchoin, L., May, R. C., Hall, M. E. & Pollard, T. D. (1999) Proc. Natl. Acad. Sci. USA 96, 3739–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson, R. C., Turbedsky, K., Kaiser, D. A., Marchand, J. B., Higgs, H. N., Choe, S. & Pollard, T. D. (2001) Science 294, 1679–1684. [DOI] [PubMed] [Google Scholar]