Abstract
Intestinal bacterial metabolites are an important communication tool between the host immune system and the commensal microbiota to establish mutualism. In a recent paper published in Science, Wendy Garrett and her colleagues report an exciting role of the three most abundant microbial-derived short-chain fatty acids (SCFA), acetic acid, propionic acid and butyric acid, in colonic regulatory T cell (cTreg) homeostasis.
A number of studies have shown that increased cTreg numbers and their immunoregulatory function are promoted by the presence of commensal intestinal microbes (either individual species such as Bacteroides fragilis1, defined benign consortia of bacteria such as the altered Schaedler flora2 or groups of Clostridia3). In a recent paper in Science, Garrett and colleagues report how these effects are generated through molecular exchanges between the host and the enormous load of microbes carried in the lower intestine4.
Smith et al.4 investigated the role of SCFA, which are bacterial fermentation products produced by a wide variety of bacteria through anaerobic acidogenic pathways. SCFA released by colonic bacteria have long been known to be important as a carbon source for colonic epithelial cells5. From this new work we can now see that signaling effects of SCFA also regulate cTreg homeostasis.
Microbiota-derived SCFA were found to increase total (thymic-derived) cTreg numbers. The homing characteristics to the colon and the regulatory functions of these cells (such as IL-10 production) were also enhanced through SCFA treatment.
These effects are mediated by the G-protein-coupled free fatty acid receptor 43 (GPR43). Using mice that are genetically deficient in this receptor, Smith et al. showed that this signaling pathway is responsible for the increased cTreg numbers in vivo and that signaling by SCFA reduces the susceptibility to chronic intestinal inflammation. As they found GPR43 expression on cTreg (compared with lower GPR43 expression on Treg from other sites) this may be a direct effect, e.g. alterations in histone deacetylation. However, other cell types in the GI tract also express GPR43, including enteroendocrine cells and other leukocytes, therefore indirect effects are not yet excluded. In fact, Atarashi and colleagues have recently published their studies of how Clostridial species induce cTreg6. They found that bacterial-derived SCFA stimulate epithelial cells to produce TGFβ, contributing to Treg differentiation and expansion.
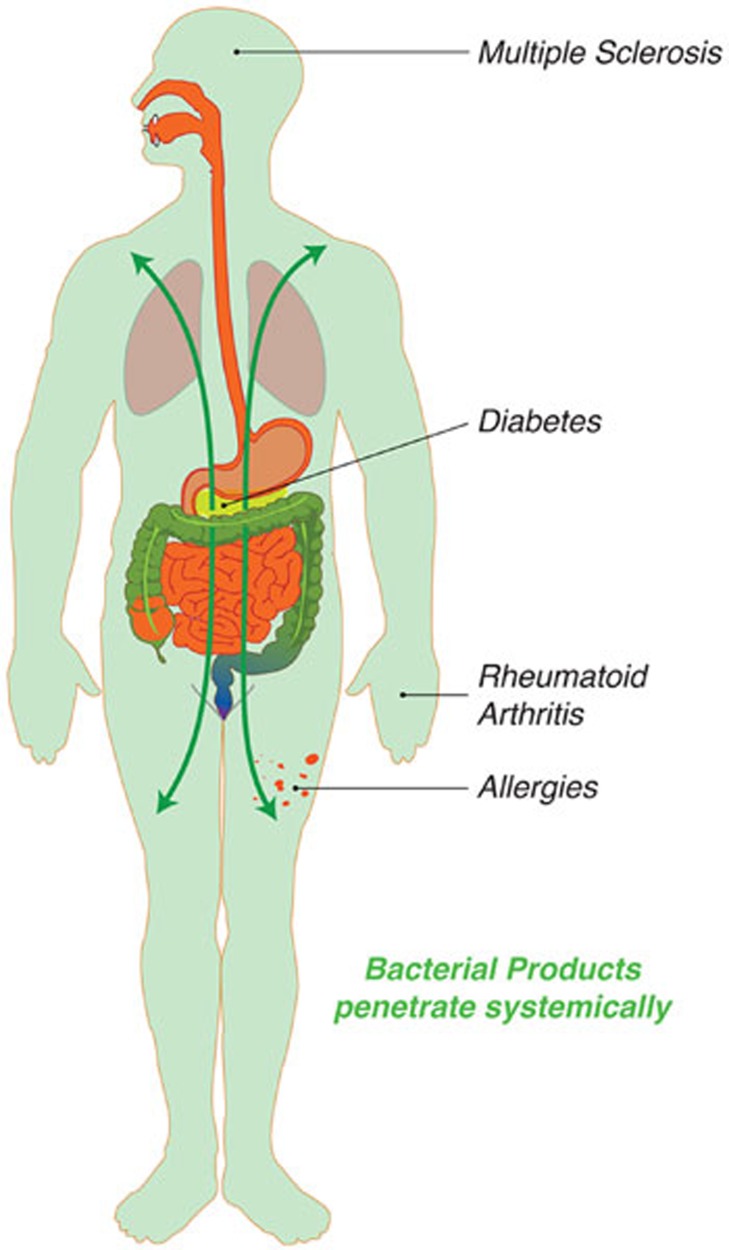
Whereas other species-specific bacterial molecules, such as B. fragilis-derived PSA, have previously been demonstrated to have immunomodulatory functions2, the report by Smith et al. is an elegant demonstration of the ubiquitous and pervasive bacterial metabolites that impact on the mucosal immune system. There is really a rather promiscuous exchange of metabolites between the microbiota and the host, with metabolic pathways that require components of both eukaryotic and prokaryotic cells. Bile acids are a great example of such a mixed pathway, where a dysbiosis caused by obesity promotes liver cancer through alterations in the microbial bile acid metabolism7. Although Smith et al. do not see any SCFA-mediated effects on central Treg compartments (outside the colon), other bacterial metabolites that reach systemic sites likely modulate adaptive or innate immune cell function at systemic sites. This may eventually rationalize the observed increased incidence of intestinal inflammation and systemic immune-mediated disorders such as autoimmune or allergic diseases (Figure 1), which are often linked to changes within the microbiota due to diet or antibiotic use8.
Figure 1.
Bacterial metabolites that reach systemic sites likely modulate adaptive or innate immune cell function at systemic sites. This may eventually rationalize the observed correlation of microbiota composition and susceptibility to systemic immune-mediated disorders such as autoimmune or allergic diseases.
A clinical situation in which the colon faces a deficiency of SCFA happens after surgery that diverts the fecal stream into a stoma bag, leaving the distal colon without its normal contents. This operation may be carried out to protect a low surgical anastomosis after removal of a tumor. The result is that the defunctioned colon frequently becomes inflamed, a condition recognized as 'diversion colitis'. In some cases, treatment with SCFA has been able to treat the condition successfully9. The lack of SCFA as a carbon source for colonocytes was previously considered as a key factor in the aetiopathogenesis of the condition, although this will need to be reviewed in the light of the new data on the effects of SFCA on colonic Treg numbers and function.
Our colonic health depends on our intestinal microbiota and what we feed them. Changes in Western dietary patterns, e.g., due to reduced intake of plant fibers, might drastically impact the production of SCFA within the intestine. Furthermore, Smith et al. demonstrate a direct effect of antibiotic (vancomycin) treatment on SCFA levels, which in turn affects intestinal immune regulation by reducing the number of cTreg.
Taken together, this draws a picture of a superorganism composed of the host (us) and our microbiota, with the metabolic interface as an important communication tool. This allows the host and the microbiota to adapt to and communicate with each other. Originally, germ-free animals were derived to challenge the notion that the existence of higher organisms was irrevocably linked to their associated microbiotas10. Although the germ-free program succeeded11, it has provided us with powerful tools to show that the original notion was justified: pervasive metabolic interactions and signaling make us the sum of our prokaryotic and eukaryotic cellular components.
References
- Round JL, Mazmanian SK. Proc Natl Acad Sci USA. 2010. pp. 12204–12209. [DOI] [PMC free article] [PubMed]
- Geuking MB, Cahenzli J, Lawson MA, et al. Immunity. 2011. pp. 794–806. [DOI] [PubMed]
- Atarashi K, Tanoue T, Shima T, et al. Science. 2011. pp. 337–341. [DOI] [PMC free article] [PubMed]
- Smith PM, Howitt MR, Panikov N, et al. Science. 2013. pp. 569–573. [DOI] [PMC free article] [PubMed]
- Roediger WE. Gastroenterology. 1982. pp. 424–429. [PubMed]
- Atarashi K, Tanoue T, Oshima K, et al. Nature. 2013. pp. 232–236. [DOI] [PubMed]
- Yoshimoto S, Loo TM, Atarashi K, et al. Nature. 2013. pp. 97–101. [DOI] [PubMed]
- Markle JG, Frank DN, Mortin-Toth S, et al. Science. 2013. pp. 1084–1088. [DOI] [PubMed]
- Kiely EM, Ajayi NA, Wheeler RA, et al. J Pediatr Surg. 2001. pp. 1514–1517. [DOI] [PubMed]
- Pasteur L. Comp Rend. 1885. p. 69.
- Glimstedt G. Acta Pathol Microbiol Scand Suppl. 1936. pp. 1–295.