Abstract
Chloroeremomycin, a vancomycin family glycopeptide antibiotic has three sugars, one d-glucose and two l-4-epi-vancosamines, attached to the crosslinked heptapeptide backbone by three glycosyltransferases, GtfA, -B, and -C. Prior efforts have revealed that GtfB and -C in tandem build an epivancosaminyl-1,2-glucosyldisaccharide chain on residue 4 of the aglycone; however, the characterization of GtfA and glycosylation sequence of chloroeremomycin have been lacking. Here, we report the expression and purification of GtfA, as well as synthesis of its sugar donor, 2′-deoxy-thymidine 5′-diphosphate (dTDP)-β-l-4-epi-vancosamine. GtfA transfers 4-epi-vancosamine from the chemically synthesized dTDP-4-epi-vancosamine to the β-OH-Tyr6 residue of the aglycone, preferentially after GtfB action, to generate chloroorienticin B. With the preferred kinetic order of GtfB, then GtfA, then GtfC established, we have succeeded in reconstitution of chloroeremomycin from the heptapeptide aglycone by the sequential actions of the three enzymes.
Two glycopeptide antibiotics, vancomycin and teicoplanin, are clinically important therapeutics for the treatment of life-threatening bacterial infections caused by Gram-positive pathogens, such as methicillin-resistant Staphylococcus aureus and Enterococci. With the global emergence of vancomycin-resistant Enterococci (VRE) and, more recently, vancomycin-resistant S. aureus (VRSA) (1, 2), research has been undertaken to search for a new generation of glycopeptide antibiotics to combat these vancomycin-resistant pathogens (3–6).
The heptapeptide skeletons of glycopeptide antibiotics are assembled by nonribosomal peptide synthetases (7). The heptapeptide backbone is then extensively modified by postassembly tailoring enzymes, including oxidative cross-linking of the electron-rich aromatic side chains, generating three crosslinks in the vancomycin subfamily and four crosslinks in the teicoplanin subgroup (8). The cross-linking modifications rigidify the peptide scaffold, creating the binding pocket for the drug target, the terminal d-Ala–d-Ala moiety of peptidoglycan (9).
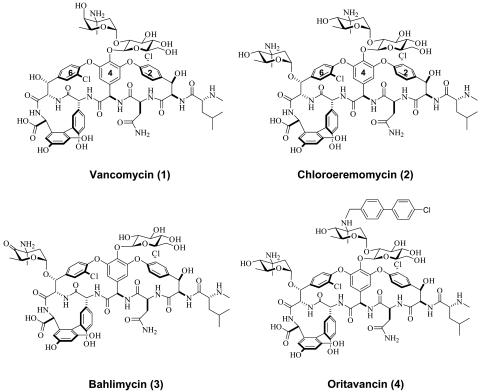
Other important postassembly modifications include glycosylation for both vancomycin and teicoplanin groups and sugar acylation for the teicoplanin subfamily. Within the vancomycin family, a disaccharide of l-aminodeoxyhexosyl-1,2-d-glucose is assembled and tethered to the phenolic hydroxyl of 4-hydroxyphenylglycine residue 4 (4-OH-PheGly4) in the crosslinked heptapeptide (Fig. 1). In vancomycin 1, the l-aminodeoxysugar is l-vancosamine, a 2,3,6-trideoxy-3-methyl-3-amino hexose. In chloroeremomycin 2, the terminal aminodeoxysugar of the disaccharide moiety is l-4-epi-vancosamine, the 4-OH equatorial epimer of vancosamine. In addition, 2 contains a second 4-epi-vancosaminyl unit tethered to the β-OH of the β-hydroxy-tyrosine residue 6 (β-OH-Tyr6) of the heptapeptide aglycone. In balhimycin 3, there is a single glucosylmoiety on residue 4 and a 4-oxo-vancosaminyl unit on residue 6. The 4-epi-vancosaminyl sugar of the disaccharide moiety in chloroeremomycin has been reductively N-alkylated by a chlorobiphenyl group to create a semisynthetic lipoglycopeptide, known as oritavancin 4. This compound displays encouraging activity against VRE, and is in late-stage clinical trials for the treatment of VRE infections (10). Consequently, there is both basic and applied interest in understanding specificity, mechanism, and structure of these antibiotic glycosyltransferases to understand the basis of glycosyl diversity and differential activity against resistant pathogens.
Fig. 1.
Natural and semisynthetic vancomycin family glycopeptide antibiotics.
Consistent with the presence of three sugars in antibiotic 2, the biosynthetic cluster for chloroeremomycin producing strain Amycolatopsis orientalis (A82846) contains three glycosyltransferases (Gtfs), GtfA, -B, and -C, along with the nonribosomal peptide synthetases genes, genes encoding nonproteinogenic amino acids, 4-OH-PheGly and 3,5-dihydroxyphenylglycine (3,5-(OH)2-PheGly), in the peptide backbone and genes for conversion of dTDP-d-glucose to dTDP-l-4-epi-vancosamine (11, 12). GtfB attaches the d-glucosyl moiety to 4-OH-PheGly4 of the vancomycin aglycone (AGV), generating the monoglucosylheptapeptide desvancosaminyl-vancomycin (DVV), and has a homologue GtfE in vancomycin biosynthesis (13). GtfC completes the disaccharide chain by using dTDP-l-4-epi-vancosamine and DVV to create epivancomycin. In the vancomycin biosynthesis pathway, the GtfC homologue, GtfD, transfers dTDP-l-vancosamine to DVV to produce vancomycin 1 (13). It has been proposed that GtfA, like GtfC, would use dTDP-l-4-epi-vancosamine as a sugar donor but show distinct regiospecificity by glycosylating the benzylic hydroxyl of the β-OH-Tyr6 residue to complete the biosynthetic pathway (12).
Here we report the expression and purification of GtfA, the synthesis of dTDP-β-l-4-epi-vancosamine, and the kinetics of GtfA action on AGV, DVV, and epivancomycin to establish the preferred pathway for successive glycosyl transfers in the maturation of chloroeremomycin.
Materials and Methods
Materials. Escherichia coli Top10 and BL21(DE3) competent cells were purchased from Invitrogen. The pET-22b E. coli expression vector was purchased from Novagen. Primers were ordered from Intergrated DNA Technology. Pfu DNA polymerase was purchased from Stratagene. Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs. Other chemicals were purchased from Sigma.
GtfA Expression and Purification. The cloning, expression, and purification of GtfA is modified from described procedures (14). Briefly, the GtfA gene from A. orientialis A82846 genomic DNA was cloned into a pET-22b vector, and was transformed into BL21(DE3) cells. The expression of C-terminal His6-tagged GtfA was induced with 300 μM isopropyl-1-thio-β-d-galactoside (IPTG) at 16°C for 20 h. The protein was purified by nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography (Qiagen, Valencia, CA).
Preparation of AGV and DVV. DVV and AGV were obtained by controlled acid hydrolysis of vancomycin as described (13).
Synthesis of dTDP-l-4-epi-Vancosamine. dTDP-l-4-epi-vancosamine was synthesized from known 4-O-Ac-3-N-Cbz-epi-vancosamine (13) as described in supporting information, which is published on the PNAS web site.
Synthesis of UDP-d-2-Amino-Glucose. The synthesis of UDP-d-2-amino-glucose follows a literature protocol. d-glucosamine was converted to its monophosphate derivative (15, 16) followed by coupling with UMP-morpholidate to yield the desired product (17, 18).
Initial Analysis of GtfA Reaction. To determine the optimal acceptor substrate for GtfA, 500 μM AGV, DVV, or epivancomycin (13) and 1 mM dTDP-4-epi-vancosamine were incubated with 10 μM GtfA in the reaction buffer [75 mM Tris, pH 7.5, 2.5 mM Tris-(2-carboxyethyl)-phosphine (TCEP), 12 mM MgCl2, 1 mg/ml BSA, and 10% (vol/vol) dimethyl sulfoxide] at 37°C for 1–3 h. The reactions were quenched with 100 μl of 50:50 MeOH/H2O (50 mM ammonium formate, pH 3.5), and were analyzed by HPLC with a Vydac small-pore C18 column using a gradient of 5–40% CH3CN in 0.1% trifluoroacetic acid (TFA)/H2O over 20 min. The products were monitored at 280 nm. The peaks for chloroeremomycin (tR = 12.2 min), epivancomycin (tR = 12.8 min), chloroorienticin B (tR = 13.0 min), DVV (tR = 13.8 min), chloroorienticin C (tR = 16.7 min), and AGV (tR = 17.7 min) were integrated, and the concentrations of products were calculated from its percentage of the total peak area. The molecular weights of products were determined by matrix-assisted laser desorption ionization–time-of-f light (MALDI-TOF) mass spectroscopy.
Kinetic Analysis of GtfA Reaction. dTDP-4-epi-vancosamine and DVV were mixed in 50 μl of the reaction buffer. The reactions were initiated by adding 5 μM GtfA, and were kept at 37°C for 3–6 min before they were quenched with 100 μl of 50:50 MeOH/50 mM ammonium formate (pH 3.5). To determine the kinetics of GtfA with its authentic substrates, 5 μM enzyme was used in each reaction. For the determination of the Km value of DVV, its concentration was varied between 62.5 and 1,000 μM, whereas the concentration of dTDP-4-epi-vancosamine was fixed at 1 mM. To measure the Km value of dTDP-4-epi-vancosamine, its concentration was varied from 62.5 to 1,000 μM, and the DVV concentration was maintained at 500 μM. The reaction mixtures were analyzed by HPLC with a Vydac small-pore C18 column (10–30% CH3CN in 0.1% TFA/H2O over 20 min, 1 ml/min). The peaks for the product 6 (tR = 11.3 min) and remaining DVV 5 (tR = 12.0 min), as monitored at 280 nm, were integrated, and the concentration of product formed was calculated from its percentage of the total peak area. The molecular weights of products were determined by MALDI-TOF analysis.
Reconstitution of Chloroeremomycin by Tandem Glycosylation with GtfA and GtfC. The chloroeremomycin reconstitution consisted of two steps. In the first step, 1 mM DVV and 1 mM dTDP-4-epi-vancosamine were incubated with 10 μM of GtfA in reaction buffer (160 μl) at 37°C for 6 h. In the second step, 10 μM GtfC or H2O (as control) was added to 80 μl of GtfA the reaction mixture, respectively. The reactions were maintained at 37°C for 5 h. The reaction mixtures were analyzed by HPLC using a gradient of 10–30% CH3CN in 0.1% TFA/H2O over 20 min. The molecular weights of desired fractions were analyzed by MALDI-TOF.
Results
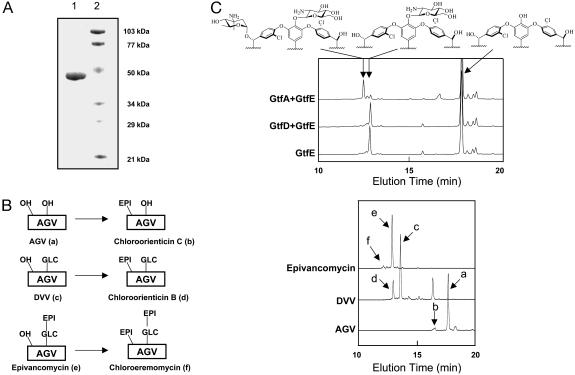
Overexpression and Purification of GtfA. Analysis of the biosynthetic cluster for chloroeremomycin-producing A. orientalis reveals three putative glycosyltransferases, GtfA, -B, and -C. Unlike GtfB and GtfC (13), the biochemical characterization of GtfA has not been established. The GtfA gene was subcloned from A. orientalis A82846 genomic DNA into an E. coli expression vector pET-22b, and GtfA was heterologously expressed as a C-terminal His6-tagged protein in E. coli. GtfA proteins (44 kDa) were subsequently purified by nickel-chelating chromatography to >90% homogeneity (Fig. 2A, lane 1), with an overall yield of 20 mg per liter of culture.
Fig. 2.
GtfA transfers l-4-epi-vancosamine to the hydroxyl group of the β-OH-Tyr6 of DVV. (A) SDS/12% PAGE of purified glycosyltransferase GtfA (lane 1) and molecular mass standard (lane 2). (B) GtfA prefers DVV as the acceptor substrate. The potential acceptors and the predicted products are depicted in simplified manner (Left). OH represents the hydroxyl groups at 4-OH-PheGly4 or β-OH-Tyr-6. Glc and epi stand for glucose and epivancosamine, respectively. The reaction mixtures were analyzed by RP-HPLC (Right). (C) GtfA, but not GtfD, transfers 4-epi-vancosamine to the 2-amino-DVV analogue prepared from AGV and UDP-2-amino-glucose. The reaction mixtures were analyzed by analytical RP-HPLC, and the predicted structure of each fraction is shown on the top.
Synthesis of TDP-β-l-4-epi-Vancosamine. We previously developed a chemical synthesis of UDP-β-l-4-epi-vancosamine to allow the characterization of GtfC and GtfD (13). However, GtfA was unable to use UDP-4-epi-vancosamine as the sugar substrate (19), making it necessary to obtain the natural dTDP-deoxy sugar donor. The dTDP-l-4-epi-vancosamine substrate has been prepared in small quantities via the in vitro reconstitution of the biochemical pathway (20), but the amounts are not sufficient for the characterization of GtfA. Although nucleotidyltransferases have been used previously to make a variety of TDP-derivatives for studying antibiotic glycosyltransferases, the enzymes that have been explored thus far are useful only for the preparation of dTDP-d-sugars (21). Therefore, we developed a chemical synthesis of dTDP-l-4-epi-vancosamine to obtain sufficient material for our biochemical studies. The 4-epi-4-O-acetyl-3-azidovancosaminyl monophosphate was prepared from the corresponding anomeric chloride in a 4:1 ratio favoring the desired β-isomer (see supporting information) and then coupled with dTMP morpholidate to give, upon deprotection and reduction of the azido group, dTDP-4-epi-vancosamine. A detailed report covering the development and the general applicability of the methodology for β-2-deoxy glycosyl phosphate formation is available (M.O., C.L., and D.K., unpublished data).
GtfA Catalyzes the Addition of 4-epi-Vancosamine to the Hydroxyl Group of β-OH-Tyr-6 of DVV. Although sequence analysis from the A. orientalis gene cluster has suggested that GtfA is one of three tandemly arranged Gtfs, the regiospecificity and timing of GtfA action has not been defined. In principle, GtfA could use any of the three scaffolds, the heptapeptide aglycone AGV, monoglycosylated DVV or disaccharide-containing epivancomycin, as a potential acceptor substrate. To find the preferred acceptor, GtfA activities were measured by using all three scaffolds. Overnight incubation of AGV, DVV, or epivancomycin with dTDP-4-epi-vancosamine in the presence of GtfA led to, in each case, the formation of a new product (Fig. 2B), whose molecular weight analyzed by MALDI-TOF agrees with that of chloroorienticin C (calculated M+H+ = 1,288.1; measured M+H+ = 1,287.5), chloroorienticin B (calculated M+H+ = 1,450.3; measured M+H+ = 1,450.2), and chloroeremomycin (calculated M+H+ = 1,592.4; measured M+H+ = 1,592.9), respectively. However, kinetic analysis of these reactions indicated that GtfA catalyzed the addition of epivancosamine to DVV with a turnover rate >1 per min, whereas it transferred 4-epi-vancosamine to AGV or epivancomycin at much reduced rates of <0.05 min–1 (data not shown), indicating that DVV is the preferred acceptor for GtfA. Beside the formation of chloroorienticin B, incubation of DVV and dTDP-4-epi-vancosamine in the presence of GtfA resulted in a side product (tR = 16.7 min Fig. 2B; measured M+H+ = 1,289.0), most likely to be chloroorienticin C. The mechanism for the formation of this side product has yet to be determined.
GtfA can potentially transfer the 4-epi-vancosaminyl moiety to the hydroxyl group of β-OH-Tyr2 or β-OH-Tyr6 residue or to the 2-hydroxy group of glucosyl unit of DVV. Because our previous report shows that GtfC and GtfD transfer 4-epi-vancosamine to the 2-hydroxy of the glucosyl unit of DVV (13), the following experiments were carried out to validate the regiospecificity of GtfA. 2-amino-glucosyl-AGV, prepared by GtfE from AGV and UDP-2-amino-glucose (18), was incubated with dTDP-4-epi-vancosamine in the presence of GtfA or GtfD. As expected, GtfD failed to transfer 4-epi-vancosamine to 2-amino-glucosyl-AGV presumably because of the replacement of the hydroxyl group by the amino group; in contrast, GtfA transferred 4-epi-vancosamine to the 2-amino analogue of DVV to produce the 2-amino analogue of chloroorienticin B (calculated M+H+ = 1,148.3; measured M+H+ = 1,148.1, Fig. 2C). Moreover, postsource-decay mass analysis of reconstituted chloroeremomycin 2 (The synthesis of compound 2 is detailed below) gave a fragment (M+H+ = 1,287.5) with mass close to that of chloroorienticin C (M+H+ = 1,288.1) (see Fig. 4C). These experiments suggest that GtfA transfers 4-epi-vancosamine to β-OH-Tyr2 or β-OH-Tyr6 residue, but not to the 2-OH of the lycosyl unit. The β-OH of residue 6 is more accessible in the x-ray structure of glycopeptide (22), and chloroeremomycin is known to possess an epivancosamine unit at residue 6. The crystal structure of GtfA indicates the hydroxyl group of β-OH-Tyr6 is much closer (3.8 Å) to the active site than that of β-OH-Tyr-2 (19.3 Å) (14). Therefore, GtfA is highly likely to transfer the 4-epi-vancosamine moiety to the hydroxyl group of β-OH-Tyr6 regiospecifically.
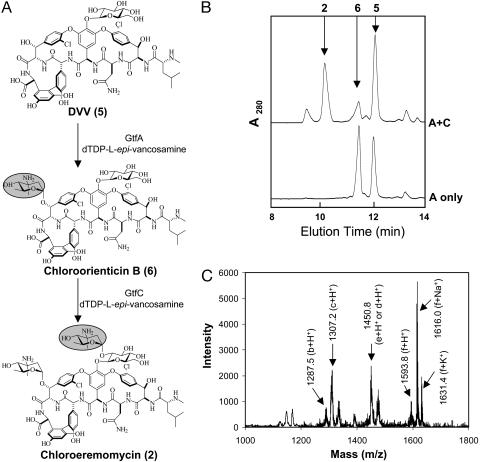
Fig. 4.
Enzymatic reconstitution of chloroeremomycin 2. (A) Scheme for the chloroeremomycin reconstitution. (B) RP-HPLC profile monitoring the enzymatic reconstitution of chloroeremomycin from DVV. (C) Postsource-decay mass analysis of chloroeremomycin prepared from sequential glycosylation of DVV by GtfA and GtfC. The fragments are labeled by letters defined as follows: b, chloroorienticin; c, DVV; d, chloroorienticin B; e, epivancomycin; f, chloroeremomycin.
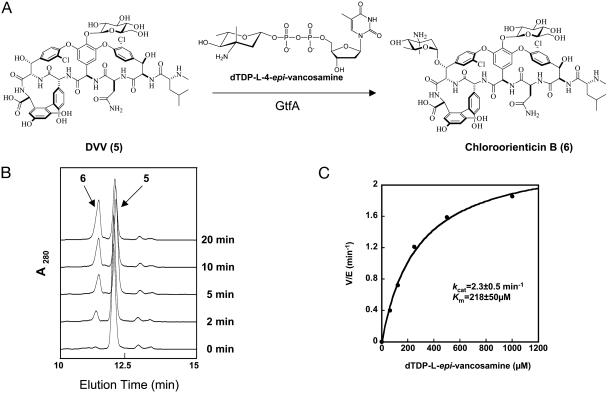
Steady-State Analysis of GtfA Activity. We then characterized the GtfA activity by using its preferred substrates, DVV and dTDP-4-epi-vancosamine (Fig. 3A). HPLC analysis of the reaction mixtures showed the appearance of a new peak (Fig. 3B). MALDI-TOF analysis indicated that the peak contained a compound (M+H+ = 1,450.3) with mass corresponding to the product chloroorienticin B 6 (M+H+ = 1,450.2). The enzyme exhibits optimal activity at pH = 7.5, in contrast to GtfB and GtfE, which had their pH optima at 9.0 (13). GtfA produces chloroorienticin B 6 at the maximal velocity of 2.3 per min (Fig. 3C). The Km for TDP-l-epivancosamine and DVV were 218 μM and 107 μM, respectively.
Fig. 3.
GtfA transfers l-4-epi-vancosamine to DVV 5 to yield chloroorienticin B 6.(A) Schematic representation of the conversion of 5 to 6.(B) Formation of 6 over 20 min analyzed by analytical RP-HPLC. (C) Michaelis–Menten plot for the measurement of the kinetic parameters of GtfA by varying dTDP-l-epi-vancosamine concentration. The DVV concentration was maintained at 500 μM.
Reconstitution of Chloroeremomycin 2 by Tandem Reaction of GtfA and GtfC. With the active GtfA, GtfB, and GtfC from the chloroeremomycin gene cluster, we attempted the reconstitution of chloroeremomycin from the AGV heptapeptide aglycone scaffold by tandem actions of the three Gtfs (Fig. 4A). Because our previous studies had already shown that GtfB converts AGV to DVV (13), we started the reconstitution experiment from DVV. As expected, incubation of GtfA with DVV and dTDP-4-epi-vancosamine over 6 h led to >50% conversion of DVV to chloroorienticin B 6. Subsequent incubation of this GtfA product with GtfC resulted in the appearance of a new product 2 as detected by HPLC (Fig. 4B). MALDI-TOF analysis indicated that the new peak contained molecules (M+H+ = 1,592.9) with the mass expected for chloroeremomycin (M+H+ = 1,592.4), proving that chloroeremomycin can be assembled from regiospecific triple glycosylation of the heptapeptide scaffold in vitro by the sequential reactions mediated by GtfB, GtfA, and GtfC.
Discussion
Several classes of natural products with antibiotic activities against different targets, including cell wall synthesis, protein synthesis, and DNA replication, are glycosylated in the late stages of their biosynthetic pathways. These include the vancomycin and teicoplanin classes of glycopeptides, the erythromycin group of the macrolides, and the novobiocin type of aminocoumarin antibiotics (5, 23, 24). The sugar moieties are added by dedicated glycosyltransferases encoded within the biosynthetic gene clusters. Many of the specialized sugars, such as the l-vancosamine in vancomycin and l-4-epi-vancosamine in chloroeremomycin, are synthesized from dTDP-d-glucose by an array of enzymes dedicated to deoxygenaton, amination, and methylation, which are also encoded in the same clusters (11, 20). The deoxy- and deoxyaminosugars can also be further decorated by acylation and methylation, which are important substituents for target recognition (3).
The glycopeptide antibiotics, vancomycin and teicoplanin, have been used as the last resort for the treatment of life-threatening Gram-positive bacterial infections such as methicillin-resistant Staphylococcus aureus, and Enterococcus faecium and faecalis. With the global emergence of VRE, tremendous efforts have been made to find new antibiotic therapeutics to fight life-threatening infections caused by these pathogens. These studies include alternation of the binding pocket of the heptapeptide scaffold, derivatization of the N or C terminus of scaffolds, and acylation and alkylation of sugar moieties appended to the heptapeptide scaffolds (3–6). To date, two semisynthetic compounds from these studies are in clinical trials. Oritavancin 4, developed from chloroeremomycin by reductively alkylating the epivancosaminyl group of the disaccharide moiety, exhibited promising activity against VanA-resistant phenotypes of enterococci (25). Dalbavancin, an amide version of teicoplanin type glycopeptide A40926, shows potent activity against a wide range of staphylococci, streptococci, and enterococci (5). Sugar modification has been shown to be an important element of pharmacophore recognition for the glycopeptides (4, 26, 27); however, variation of the aminosugar and sugar combinations have not been well studied.
To study the importance of sugar decoration on antibiotic activities of glycopeptides and to search for new generations of glycopeptide antibiotics by varying sugar identities, we have focused earlier studies both on the in vitro conversion of dTDP-β-l-4-epi-vancosamine from dTDP-d-glucose by five steps of enzymatic reaction (20) and on the characterization of a series of glycosyltransferases by using nucleotide diphosphate sugars to decorate the heptapeptide aglycone during vancomycin and chloroeremomycin biosynthesis (13, 18). In those studies, we have characterized the d-glucosyltransferases (GtfB and GtfE), the l-vancosaminyltransferase (GtfD), and the l-epi-vancosaminyltransferase (GtfC) that build the disaccharide moiety. In parallel, we have determined the high-resolution crystal structures of GtfB (28), GtfA (14), and GtfD (29) to evaluate how each Gtf catalyzes regioselective glycosylation. We have also devised a synthetic route for UDP-4-epi-vancosamine for the evaluation of GtfC and GtfD (13).
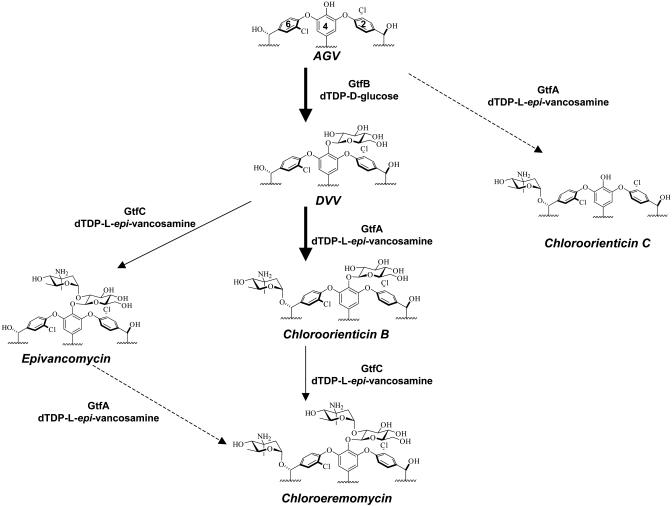
In this work, we report the heterologous expression and purification of GtfA as well as the chemical synthesis of dTDP-β-l-4-epi-vancosamine, which allowed us to study the timing and regioselectivity of GtfA action. GtfA strongly prefers DVV as its substrate rather than the heptapeptide aglycone AGV or epivancomycin, suggesting the likely glycosylation sequence of GtfB → GtfA → GtfC in the chloroeremomycin biosynthetic pathway (Fig. 5). This glycosylation sequence is reminiscent of glycosylation events in the balhimycin biosynthetic pathway. During balhimycin biosynthesis, bGtfA transfers the oxovancosaminyl moiety after OH-PheGly4 has been glucosylated by bGtfB, because the bGtfB-deficient mutant accumulates only heptapeptide intermediate (30). In contrast, the glycosyltransferases (tGtfA and tGtfB) from a teicoplainin-producing Actinoplanes teichomyceticus ATCC31131 displayed no preferential glycosylation sequence in decorating the aglycone backbone (31, 32). The glycosylation sequence of chloroeremomycin is also supported by the crystal structures of GtfA and GtfD. The GtfA structure suggests the enzyme prefers DVV rather than vancomycin as acceptor (14). The GtfD structure reveals the existence of a crevice mouth not only providing room but also contributing binding for the 4-epi-vancosamine unit attached at residue 6, suggesting that the related GtfC may prefer chloroorienticin B as the optimal acceptor.
Fig. 5.
Glycosylation pathway for the maturation of chloroeremomycin. The kinetically favored glycosylation steps are labeled by thick arrows. The glycosylation steps labeled by dash arrows are enzymatically disfavored. The detailed kinetics of the GtfC-mediated steps, marked by thin arrows, remain to be determined.
The postsource-decay mass analysis of chloroeremomycin obtained from the tandem actions of GtfA and GtfC on DVV as well as the differential actions of GtfA and GtfC on 2-aminoglucosyl-AGV further confirm that GtfA transfers epivancosamine to residue 6, proving that it has distinct regioselectivity from GtfC. These data are also consistent with the crystal structure of GtfA complexed with DVV and dTDP, where the β-OH of β-OH-Tyr6 is oriented toward bound dTDP presented by the C-terminal domain of the enzyme (14).
In addition to the different regioselectivity toward DVV between GtfA and GtfC, these two enzymes displayed distinct preference toward NDP-sugars. GtfA was active only when dTDP-l-epi-vancosamine was used, whereas GtfC and GtfD could use both UDP- and dTDP-4-epi-vancosamine as substrates. Steady-state analysis of GtfA indicated that this enzyme is a slow enzyme (kcat = 2.3 min–1) compared to other glycopeptide Gtfs where the maximal rates vary from 10 to 70 per min (13).
The availability of synthetic dTDP-l-4-epi-vancosamine, active GtfA, and the other glycopeptide Gtfs (18) allows us to reconstitute chloroeremomycin from the heptapeptide aglycone, dTDP-glucose and dTDP-4-epi-vancosamine by using the tandem action of GtfB, GtfA, and GtfC. Because we have previously shown the conversion of DVV from heptapeptide AGV by the action of GtfB, in this paper, we demonstrated the in vitro reconstitution of chloroeremomycin from DVV by the sequential reaction catalyzed by GtfA and GtfC. The success of chloroeremomycin reconstitution sets the stage for variant glycosyl tailoring of the crosslinked heptapeptide scaffold.
The importance of carbohydrate modification on antibiotic activity and the recent development of in vitro studies on glycopeptide Gtfs have made it attractive to use these enzymes tandemly to attach various sugars to the aglycone efficiently and regiospecifically. Because reductive alkylation and variation of sugar modification have led to the discovery of new antibiotics with enhanced efficacy, it will be of interest to scale up the in vitro reconstitutions of both chloroeremomycin and its analogs followed by reductive alkylation of terminal l-aminohexose to evaluate new lipoglycopeptides for VRE and methicillin-resistant Staphylococcus aureus activity.
Supplementary Material
Acknowledgments
We thank Dr. Heather C. Losey for her initial work on GtfA. J.T. thanks Kim Albizati and Van Martin for generous support of secondment (to Harvard Medical School). We thank Dr. Ryan Kruger for critical reading of the manuscript. This work was supported in part by National Institute of Health Grants 49338 (to C.T.W.) and 66174 (to D.K.).
Abbreviations: VRE, vancomycin-resistant Enterococci; Gtfs, glycosyltransferases; AGV, vancomycin aglycone; DVV, desvancosaminyl-vancomycin; MALDI-TOF, matrix-assisted laser desorption ionization–time-of-flight; dTDP, 2′-deoxy-thymidine 5′-diphosphate.
References
- 1.Chang, S., Sievert, D. M., Hageman, J. C., Boulton, M. L., Tenover, F. C., Downes, F. P., Shah, S., Rudrik, J. T., Pupp, G. R., Brown, W. J., et al. (2003) N. Engl. J. Med. 348, 1342–1347. [DOI] [PubMed] [Google Scholar]
- 2.Weigel, L. M., Clewell, D. B., Gill, S. R., Clark, N. C., McDougal, L. K., Flannagan, S. E., Kolonay, J. F., Shetty, J., Killgore, G. E. & Tenover, F. C. (2003) Science 302, 1569–1571. [DOI] [PubMed] [Google Scholar]
- 3.Malabarba, A. & Ciabatti, R. (2001) Curr. Med. Chem. 8, 1759–1773. [DOI] [PubMed] [Google Scholar]
- 4.Ge, M., Chen, Z., Onishi, H. R., Kohler, J., Silver, L. L., Kerns, R., Fukuzawa, S., Thompson, C. & Kahne, D. (1999) Science 284, 507–511. [DOI] [PubMed] [Google Scholar]
- 5.Malabarba, A., Nicas, T. I. & Thompson, R. C. (1997) Med. Res. Rev. 17, 69–137. [DOI] [PubMed] [Google Scholar]
- 6.Zweifel, M. J., Snyder, N. J., Cooper, R. D., Nicas, T. I., Mullen, D. L., Butler, T. F. & Rodriguez, M. J. (2003) J. Antibiot. 56, 289–295. [DOI] [PubMed] [Google Scholar]
- 7.Marahiel, M. A., Stachelhaus, T. & Mootz, H. D. (1997) Chem. Rev. 97, 2651–2674. [DOI] [PubMed] [Google Scholar]
- 8.Walsh, C. T., Chen, H., Keating, T. A., Hubbard, B. K., Losey, H. C., Luo, L., Marshall, C. G., Miller, D. A. & Patel, H. M. (2001) Curr. Opin. Chem. Biol. 5, 525–534. [DOI] [PubMed] [Google Scholar]
- 9.Williams, D. H. & Bardsley, B. (1999) Angew. Chem. Int. Ed. 38, 1172–1193. [DOI] [PubMed] [Google Scholar]
- 10.Nicas, T. I., Mullen, D. L., Flokowitsch, J. E., Preston, D. A., Snyder, N. J., Zweifel, M. J., Wilkie, S. C., Rodriguez, M. J., Thompson, R. C. & Cooper, R. D. (1996) Antimicrob. Agents Chemother. 40, 2194–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Wageningen, A. M., Kirkpatrick, P. N., Williams, D. H., Harris, B. R., Kershaw, J. K., Lennard, N. J., Jones, M., Jones, S. J. & Solenberg, P. J. (1998) Chem. Biol. 5, 155–162. [DOI] [PubMed] [Google Scholar]
- 12.Solenberg, P. J., Matsushima, P., Stack, D. R., Wilkie, S. C., Thompson, R. C. & Baltz, R. H. (1997) Chem. Biol. 4, 195–202. [DOI] [PubMed] [Google Scholar]
- 13.Losey, H. C., Peczuh, M. W., Chen, Z., Eggert, U. S., Dong, S. D., Pelczer, I., Kahne, D. & Walsh, C. T. (2001) Biochemistry 40, 4745–4755. [DOI] [PubMed] [Google Scholar]
- 14.Mulichak, A. M., Losey, H. C., Lu, W., Wawrzak, Z., Walsh, C. T. & Garavito, R. M. (2003) Proc. Natl. Acad. Sci. USA 100, 9238–9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Busca, P. & Martin, O. R. (1998) Tetrahedron Lett. 39, 8101–8104. [Google Scholar]
- 16.Silva, D. J., Wang, H., Allanson, N. M. & Sofia, M. J. (1999) J. Org. Chem. 64, 5926–5929. [Google Scholar]
- 17.Tai, V. W., O'Reilly, M. K. & Imperiali, B. (2001) Bioorg. Med. Chem. 9, 1133–1140. [DOI] [PubMed] [Google Scholar]
- 18.Losey, H. C., Jiang, J., Biggins, J. B., Oberthur, M., Ye, X. Y., Dong, S. D., Kahne, D., Thorson, J. S. & Walsh, C. T. (2002) Chem. Biol. 9, 1305–1314. [DOI] [PubMed] [Google Scholar]
- 19.Losey, H. C. (2003) Ph.D. thesis (Harvard University, Cambridge, MA).
- 20.Chen, H., Thomas, M. G., Hubbard, B. K., Losey, H. C., Walsh, C. T. & Burkart, M. D. (2000) Proc. Natl. Acad. Sci. USA 97, 11942–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, J., Albermann, C. & Thorson, J. S. (2003) ChemBioChem 4, 443–446. [DOI] [PubMed] [Google Scholar]
- 22.Sheldrick, G. M., Jones, P. G., Kennard, O., Williams, D. H. & Smith, G. A. (1978) Nature 271, 223–225. [DOI] [PubMed] [Google Scholar]
- 23.Thorson, J. S., Hosted, T. J., Jiang, J., Biggins, J. B. & Ahlert, J. (2001) Curr. Org. Chem. 5, 139–167. [Google Scholar]
- 24.Walsh, C., Freel Meyers, C. L. & Losey, H. C. (2003) J. Med. Chem. 46, 3425–3436. [DOI] [PubMed] [Google Scholar]
- 25.Malabarba, A., Ciabatti, R., Scotti, R., Goldstein, B. P., Ferrari, P., Kurz, M., Andreini, B. P. & Denaro, M. (1995) J. Antibiot. 48, 869–883. [DOI] [PubMed] [Google Scholar]
- 26.Kerns, R., Dong, S. D., Fukuzawa, S., Carbeck, J., Kohler, J., Silver, L. L. & Kahne, D. (2000) J. Am. Chem. Soc. 122, 12608–12609. [Google Scholar]
- 27.Fu, X., Albermann, C., Jiang, J., Liao, J., Zhang, C. & Thorson, J. S. (2003) Nat. Biotechnol. 21, 1467–1469. [DOI] [PubMed] [Google Scholar]
- 28.Mulichak, A. M., Losey, H. C., Walsh, C. T. & Garavito, R. M. (2001) Structure (Cambridge, Mass.) 9, 547–557. [DOI] [PubMed] [Google Scholar]
- 29.Mulichak, A. M., Lu, W., Losey, H. C., Walsh, C. T. & Garavito, R. M. (2004) Biochemistry, in press. [DOI] [PubMed]
- 30.Pelzer, S., Sussmuth, R., Heckmann, D., Recktenwald, J., Huber, P., Jung, G. & Wohlleben, W. (1999) Antimicrob. Agents Chemother. 43, 1565–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sosio, M., Kloosterman, H., Bianchi, A., Dijkhuizen, L. & Donadio, S. (2004) Microbiology 150, 95–102. [DOI] [PubMed] [Google Scholar]
- 32.Li, T. L., Huang, F., Haydock, S. F., Mironenko, T., Leadlay, P. F. & Spencer, J. B. (2004) Chem. Biol. 11, 107–119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.