Dear Editor,
Insect genome engineering is crucial for gene functional analysis and the long-term development of novel pest control technologies. The recently developed engineered nucleases, zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), are capable of facilitating precise genome modifications1. However, constructing separate nucleases for each targeting site is time- and labor-consuming, especially for ZFNs. The recently identified CRISPR/Cas system, which is involved in an adaptive immune mechanism found in many bacterial and archaeal species, is a facile and efficient alternative to ZFNs and TALENs. Of the 4 known CRISPR/Cas systems, the type II bacterial CRISPR/Cas system is most suitable for genome engineering2. The Cas9 endonuclease from the Streptococcus pyogenes type II system cleaves DNA under the guidance of a synthetic single guide RNA (sgRNA)3. Indeed, the Cas9-sgRNA complex has been used in several organisms to introduce mutations at single or multiple sites4,5,6.
The domesticated silkworm, Bombyx mori (B. mori), is an economically valuable lepidopteran model insect. Genetic manipulation technologies such as germline transformation have been extensively applied in B. mori gene functional analysis and in the development of silkworms as bioreactors. In B. mori, ZFN- and TALEN-mediated gene mutagenesis targeting BmBLOS2 was reported7,8. BmBLOS2 is the homolog of the human biogenesis of lysosome-related organelles complex 1, subunit 2 gene, and is responsible for urate granule synthesis in B. mori larval epidermis9. When BmBLOS2 function is compromised, the larval integument is transformed from opaque to translucent, generating the oily skin phenotype (OSP). Thus, BmBLOS2 represents an excellent target gene for testing the development of novel genome engineering strategies.
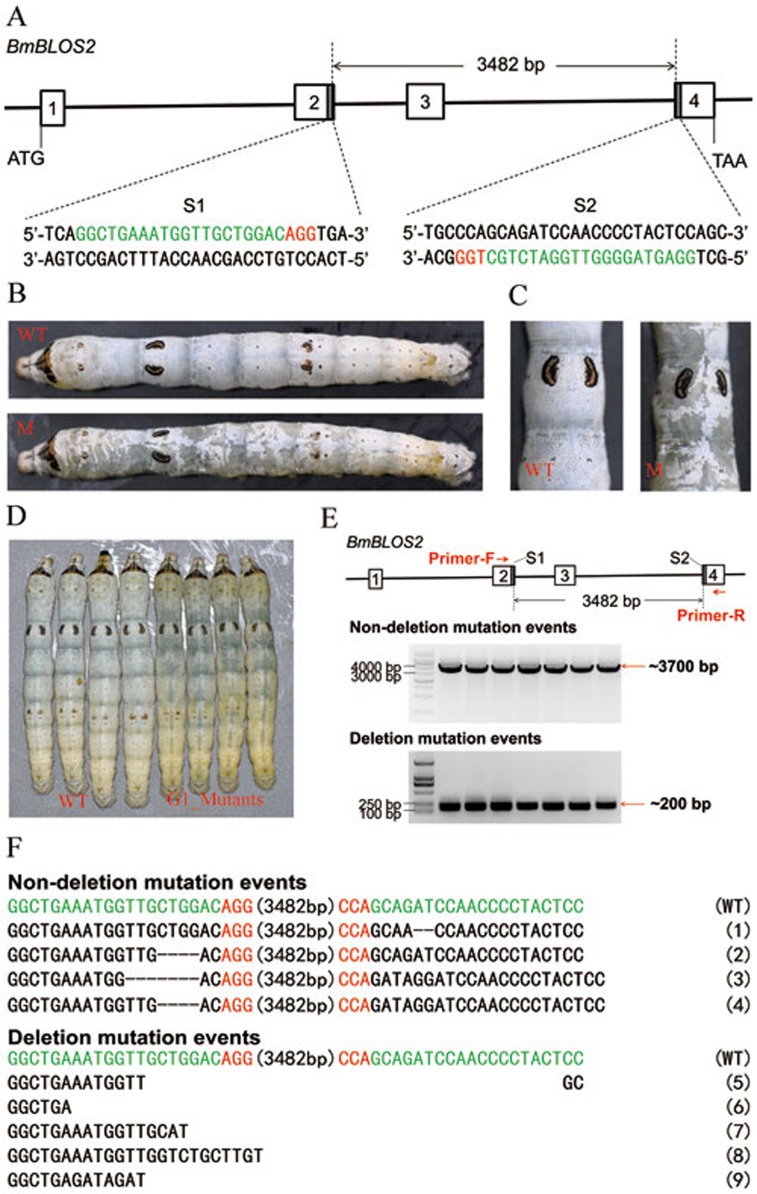
In the current study, we used the Cas9-sgRNA system to target BmBLOS2 in B. mori. We screened the BmBLOS2 open reading frame (ORF), and identified two 23-bp sgRNA-targeting sites, S1 and S2, by following the GGN19GG rule (Figure 1A). Each sgRNA was separately mixed with Cas9 mRNA and injected into the preblastoderm embryos. A mosaic translucent integument phenotype was detected in most larvae: 152 for sgRNA-1 (95.6%) and 94 for sgRNA-2 (94.0%). Mutants with severe OSP (> 95% of their integument area was translucent) were also recorded: 27 for sgRNA-1 (17.0%) and 3 for sgRNA-2 (3.0%) (Supplementary information, Figure S1 and Table S1). Genomic DNA was extracted from the second-instar larvae with severe OSP, and fragments spanning the S1 and S2 sites were separately amplified and sequenced. Similar to the results of the ZFNs and TALENs experiments7,8, Cas9-sgRNA induced small deletions or insertions at the targeting site, which resulted from non-homologous end joining (NHEJ) repair. Most of the mutations caused a frame-shift and abrogated BmBLOS2 function (Supplementary information, Figure S1).
Figure 1.
Targeted mutagenesis induced by Cas9-sgRNAs in B. mori. (A) Schematic diagram of the sgRNA-targeting sites. The black line indicates the genome locus of BmBLOS2, and the 4 boxes represent the 4 exons of BmBLOS2. The sgRNA-targeting sites, S1 and S2, are located on the sense strand of exon-2 and the anti-sense strand of exon-4, respectively. The sgRNA-targeting sequence is in green, and the protospacer adjacent motif (PAM) sequence is in red. The interspace fragment between the S1 and S2 sites is 3 482 bp. (B, C) Mutants with mosaic translucent skin induced by Cas9-sgRNA injection. (C) is a partially enlarged image of (B). (D) G1 mutants with completely translucent integuments. The left 4 larvae are wild type at the final-instar stage, and the right 4 larvae are mutants at the final-instar stage. (E) Results of PCR analyses with primer-F and primer-R revealed non-deletion and deletion mutation events in G1 mutants that were derived from G0 B. mori injected with a mixture of Cas9 mRNA and 2 sgRNAs. Primer-F anneals upstream of the S1 site, and primer-R anneals downstream of the S2 site. For the non-deletion mutation event, fragments of ∼3.7 kb were detected, whereas for a deletion mutation event, fragments of ∼200 were detected, indicating a deletion of ∼3.5 kb. Forty-five non-deletion mutations and 7 deletion mutations were detected in the 52 mutants that yielded successful PCR results. (F) Various types of non-deletion or deletion mutations in the G1 mutants. In non-deletion mutation events, a small deletion or insertion was detected at a single targeting site (1 and 2) or both sites (3 and 4). For the deletion mutation events, a 3 482-bp interspace fragment and flanking sequences of different lengths were not detected (5-9). The sgRNA-targeting sequence is in green and the PAM sequence is in red. The number of 3482 bp in brackets refers to the 3 482-bp interspace fragment between the S1 and S2 sites.
Next, we coinjected sgRNA-1 and sgRNA-2 with Cas9 mRNA (double-targeting group) into preblastoderm embryos, which effectively induced loss-of-function mutations at the BmBLOS2 locus (Figure 1B and 1C): 317 individuals (95.5%) had mosaic OSP (Figure 1B and 1C) and 168 (50.6%) had severe OSP (data not shown). Genomic DNA was extracted from the individuals with severe OSP, and fragments spanning the S1 and S2 sites were amplified and sequenced. Three types of mutations were detected in somatic cells: a ∼3.5-kb large fragment deletion, double modifications at 2 separated sites, and modifications at a single site (Supplementary information, Figure S2).
Mutated individuals in the double-targeting group were crossed with wild-type B. mori, and genetically transformed mutants were easily detected based on their completely translucent skin from the second-instar stage in generation 1 (G1) (Figure 1D). To calculate the germline transmission efficiency, we surveyed 104 G1 egg batches and recovered 37 positive egg batches (35.6%), in which one or more individuals showed completely translucent skin (Supplementary information, Table S1). We randomly selected 100 OSP individuals (i.e., 10 larvae from each of 10 positive egg batches) for genotyping. Among these, successful PCR results were obtained from 52 individuals, in which 7 mutants contained a ∼3.5-kb large fragment deletion, and 45 mutants did not (Figure 1E and 1F). In the 45 mutants without the ∼3.5-kb large fragment deletion, small deletions or insertion were detected at a single site (S1 or S2) or both sites. Sequencing analysis was also performed in the G2 mutants to confirm the germline transmission of the genotype that appeared in the G1 mutants, and the results demonstrated the stable inheritance of the mutations (Supplementary information, Figure S3). We also tested a higher dosage of Cas9 and 2 sgRNAs (injection of twice amount of Cas9 and sgRNAs). In this case, G0 individuals showed completely translucent skin, which was normally detected only in G1 mutants in lower-dose experiments (Supplementary information, Figure S2 and Table S1).
These results demonstrate that the Cas9-sgRNA system effectively induced targeted mutagenesis in B. mori. Based on these results, we conclude that the CRISPR/Cas system is a powerful tool for genome manipulation in B. mori and, most likely other lepidoptera, particularly the pest species. However, some potential problems with this system need to be resolved. Mutations in many loci do not cause phenotypic effects that are easy to assess, posing a difficulty in mutation screening. Furthermore, the targeting sequences of the CRISPR/Cas system are limited to 23 bp. Considering the tolerance for mismatches at the 5′-ends of the targeting site, this limitation may lead to serious off-target effects. Nonetheless, we report here a proof-of-principle study that exemplifies targeted mutagenesis of BmBLOS2 by direct microinjection of Cas9 mRNA and sgRNAs. The ensuing highly efficient mutagenesis demonstrates a new, facile genome engineering technology in B. mori.
Acknowledgments
We thank Dr David Stanley (USDA-ARS, Columbia, MO, USA) for comments on earlier drafts of this paper. We are grateful to Ms Lin Cao for technical assistance. This work was supported by grants from the National Science Foundation of China (31030060) and the National Basic Research Program of China (2012CB114101 and 2012CB721103).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Single-targeting induced by Cas9-sgRNA injection.
Double-targeting induced three types of mutations in somatic cells.
Genotypes in G2 mutants.
Mutagenesis induced by four different combinations (Com).
Materials and Methods
Sequences
References
- Gaj T, Gersbach CA, Barbas CF., 3rd Trends in Biotechnol. 2013. pp. 397–405. [DOI] [PMC free article] [PubMed]
- Koonin EV, Makarova KS. RNA Biol. 2013. pp. 679–686. [DOI] [PMC free article] [PubMed]
- Jinek M, Chylinski K, Fonfara I, et al. Science. 2012. pp. 816–821. [DOI] [PMC free article] [PubMed]
- Wang H, Yang H, Shivalila CS, et al. Cell. 2013. pp. 910–918. [DOI] [PMC free article] [PubMed]
- Hwang WY, Fu Y, Reyon D, et al. Nat Biotechnol. 2013. pp. 227–229. [DOI] [PMC free article] [PubMed]
- Gratz SJ, Cummings AM, Nguyen JN, et al. Genetics. 2013. pp. 1029–1035. [DOI] [PMC free article] [PubMed]
- Takasu Y, Kobayashi I, Beumer K, et al. Insect Biochem Mol Biol. 2010. pp. 759–765. [DOI] [PubMed]
- Ma S, Zhang S, Wang F, et al. PLoS One. 2012. p. e 45035. [DOI] [PMC free article] [PubMed]
- Fujii T, Daimon T, Uchino K, et al. Insect Mol Biol. 2010. pp. 659–667. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Single-targeting induced by Cas9-sgRNA injection.
Double-targeting induced three types of mutations in somatic cells.
Genotypes in G2 mutants.
Mutagenesis induced by four different combinations (Com).
Materials and Methods
Sequences