Abstract
Background
Cells are exposed to multiple stressors that induce significant alterations in signaling pathways and in the cellular state. As obligate parasites, all viruses require host cell material and machinery for replication. Virus infection is a major stressor leading to numerous induced modifications. Previous gene array studies have measured infected cellular transcriptomes. More recently, mass spectrometry-based quantitative and comparative assays have been used to complement such studies by examining virus-induced alterations in the cellular proteome.
Methods
We used SILAC (stable isotope labeling with amino acids in cell culture), a non-biased quantitative proteomic labeling technique, combined with 2-D HPLC/mass spectrometry and reciprocal labeling to identify and measure relative quantitative differences in HeLa cell proteins in purified cytosolic and nuclear fractions after reovirus serotype 3 Dearing infection. Protein regulation was determined by z-score analysis of each protein’s label distribution.
Results
A total of 2856 cellular proteins were identified in cytosolic fractions by 2 or more peptides at >99% confidence and 884 proteins were identified in nuclear fractions. Gene ontology analyses indicated up-regulated host proteins were associated with defense responses, immune responses, macromolecular binding, regulation of immune effector processes, and responses to virus, whereas down-regulated proteins were involved in cell death, macromolecular catabolic processes, and tissue development.
Conclusions
These analyses identified numerous host proteins significantly affected by reovirus T3D infection. These proteins map to numerous inflammatory and innate immune pathways, and provide the starting point for more detailed kinetic studies and delineation of virus-modulated host signaling pathways.
Keywords: RNA virus, Virus infection, Host cell alterations, Proteomics, Mass spectrometry, Liquid chromatography, Bioinformatics
Background
The cellular proteome (the total protein repertoire, which includes how each protein may be co-translationally or post-translationally modified) is affected by numerous stresses, including infection by viruses. Numerous previous microarray studies have determined how cellular transcriptomes respond after virus infection (see for example: [1,2]). However, since mRNA levels cannot provide complete information about levels of protein synthesis or the types and degrees of post-translational modifications, there frequently is little concordance between microarray and protein data [3-5]. Therefore, quantitative and comparative proteomic analyses are also being used to provide additional information about host alterations to virus infection (reviewed in: [6,7]). Commonly used methods include 2-dimensional difference in gel electrophoresis (2D-DIGE (see for examples: [8,9]), isotope coded affinity tags (ICAT; [10,11]), isobaric tags for relative and absolute quantitation (iTRAQ; [12,13]), and stable isotope labeling by amino acids in cell culture (SILAC; [14-18]). The SILAC method involves labeling cells with “light” (normal; L) and “heavy” (H) isotopic forms of amino acids. Advantages of this particular technique include: experimental set-up is relatively simple, L and H samples are mixed together early in this process, thereby reducing sample-to-sample variability, and, if 12C6-Lys and 12C614N4-Arg (L), and 13C6-Lys and 13C615N4-Arg (H) amino acids are used, virtually every tryptic peptide should contain a labeled amino acid, thereby providing increased protein coverage. Indeed, several early comparative studies suggested SILAC provided more identifications than the other commonly used methods (reviewed in [7]). We have been using SILAC to measure comparative proteomic alterations induced by influenza virus in A549 [19] and in normal human bronchial epithelial (NHBE) airway cells [20]. We have also begun similar analyses with reovirus-infected cells, focusing initially upon reovirus serotype T1L-infected HEK-293 cells [21].
The mammalian reoviruses (MRV) are non-enveloped viruses with a genome that consists of 10 segments of double-stranded (ds)RNA. The dsRNA genome is enclosed in a double-layered concentric protein capsid composed of 8 viral structural proteins. For reviews, see [22-24]. MRV is the prototype member of the Orthoreovirus genus in the family Reoviridae. The Ortheoreoviruses include nonfusogenic MRV and fusogenic avian reovirus. The Reoviridae family also contains rotaviruses [25], orbiviruses [26], and at least 9 other genera, several of which can infect animals, insects and/or plants [23,24]. MRV infections are generally mild in humans but many of the other family members are highly pathogenic in their hosts. MRV currently consist of 4 identified serotypes, with each represented by a prototype strain: strain Lang (T1L) for serotype 1; strain Jones (T2J) for serotype 2, strain Dearing (T3D) for serotype 3 and strain Ndelle for serotype 4 [27]. The reoviruses have long served as models for understanding viral pathogenesis [22] and they have also been identified as potential oncolytic agents [28-30] because of their capacity to selectively kill cancer cells that contain activated Ras pathway and functional p53 [28,31].
MRV are capable of infecting a wide range of cells, including mouse L929 cells, often used for stock preparation and titration [32], and various human cells, including HEK-293 [21]. Numerous reovirus studies have also been performed in HeLa cells (see for examples: [33-36]). Global microarray analyses of MRV-infected cellular transcriptomes detected activation of numerous cellular genes, including many related to apoptosis [37-41]. These microarray assays have recently been complemented by quantitative and comparative proteomic analyses. For example, Li and colleagues recently demonstrated, using 2D-DIGE, that MRV-infected murine myocytes regulate several proteins, including heat shock proteins and interferon-response proteins [42]. We have also shown, using SILAC and 2D-HPLC/MS, that proteins involved in cell death, cell growth and proliferation, molecular transport, gene expression, and inflammatory response pathways are affected in MRV T1L-infected HEK-293 cells [21]. Similar pathways were also found regulated in a preliminary analysis of reovirus T3D-infected HeLa cells that concluded that inclusion of Proteominer® bead-based non-biased enrichment did not significantly improve proteomic coverage of unpurified cell extracts [43].
Thus, as part of an ongoing systematic delineation of reovirus-induced comparative host protein responses, we are examining how reoviruses T1L and T3D, two of the most commonly used MRV strains, affect various permissive cells. We have extended our previous T1L-infected HEK-293 cell study [21] to T3D-infected HEK-293 cells (Berard, in preparation). Our previous proteomic analysis of HeLa cells infected with T3D concluded that inclusion of Proteominer® bead-based non-biased enrichment did not significantly improve proteomic coverage of unpurified cell extracts [43]. Therefore, the current study extends these previous studies by examining proteomic alterations in purified cytosolic and nuclear fractions in order to globally assess sub-cellular protein distribution. Reciprocal labeling also was incorporated to identify probable contaminants which were removed from the analyses. We also extended previous protein identification by more detailed follow-up kinetic studies of some selected important proteins. This study identified and measured 2856 cytosolic proteins by 2 or more peptides at >99% confidence and 884 nuclear proteins. DAVID™ and IPA™ ontological analyses identified significantly up- and down-regulated proteins as well as significantly affected canonical pathways.
Materials and methods
Cells and viruses
Cell lines and media
Mouse L929 fibroblast cells (L929) were grown in suspension in Joklik’s modified minimal essential medium (J-MEM) (Gibco, Grand Island, NY) supplemented to contain 5% fetal bovine serum (FBS) (Invitrogen Canada Inc., Burlington, Ontario), and 2 mM L-glutamine as described [32]. Cells were sub-cultured daily.
Human HeLa cells were cultured as monolayers in Dulbecco’s modified MEM (D-MEM) supplemented with 0.2% (w/v) glucose, 10% FBS (Invitrogen), 2 mM l-glutamine, non-essential amino acids, and sodium pyruvate. Cells were sub-cultured 2 – 3 times each week.
Viruses
Reovirus strain Type 3 Dearing (T3D) is a laboratory stock. There are two commonly-used clones of T3D; the Cashdollar strain (T3DC) and the Fields strain (T3DF). We elected to use the T3DF strain because a large number of genetic reagents, including temperature-sensitive mutants (reviewed in [44,45]) and T1L/T3DF intertypic reassortants [46-48] were generated from this clone. Virus stocks were usually grown in L929 cell monolayers in J-MEM in the presence of 5% CO2 at 37°C as above, but with 3% FBS, 100 U/ml of penicillin, 100 μg/ml streptomycin sulfate and 100 μg/ml amphotericin-B as previously described [32].
Virus purification
Large quantities of reovirus T3D were grown in suspension L929 cell cultures and purified by routine procedures that make use of Vertrel-XF™ extraction and cesium chloride ultracentrifugation [49]. Purified virions were harvested and dialyzed against D-Buffer (150 mM NaCl, 15 mM MgCl2, 10 mM Tris, pH 7.4). Virus concentration was measured by optical density at 260 nm, using the relationship 1 ODU = 2.1 × 1012 particles per milliliter [34] and infectivity was titrated.
Infections
Sets of HeLa cells were infected with gradient-purified T3D at a multiplicity of infection (MOI) of 7 PFU per cell. For routine infections not destined for SILAC analysis (i.e. for photomicrography, virus growth kinetic determinations, or Western blot analyses – see below), cells were harvested at various time points (0 – 72 hours post infection; hpi), then fractionated as described below.
For SILAC labeling, HeLa cells were adapted through 3 passages (=6 doublings) into D-MEM media provided in a SILAC™ Phosphoprotein Identification and Quantification Kit (Invitrogen Canada Inc.; Burlington, Ontario), supplemented as above (except without non-essential amino acids), and with 10% dialyzed FBS (Invitrogen), plus 100mg each of normal (L; 12C6-lysine and 12C6-/14N4-arginine) or “heavy” (H; 13C6-lysine [6.0Da difference] and 13C6-/15N4-arginine [10.0Da] difference) per liter of D-MEM. Once HeLa cells had grown through six doublings in appropriate SILAC media, sets of cells were infected with gradient-purified T3D or were mock infected with diluent. Cells were overlaid with appropriate SILAC media and cultured for 24 hours. In one experiment (Run #1) the L cells were infected and the H cells were mock infected, whereas H cells were infected and L cells were mock infected in the reciprocal labeling experiment (Run #2).
Microscopy
HeLa cells were allowed to attach to 6-well culture dishes or onto sterile glass cover slips in 6-well culture dishes and incubated overnight at 37°C. Cells, at approximately 70% confluency, were washed twice with 1X PBS and T3D was added to each culture at an MOI of 7. Virus was adsorbed to cells on ice for one hour to ensure infection synchronization. Mock-infected cells received only diluent. Cells were overlaid with complete pre-warmed media and cells were then incubated for various periods of time from 0 to 72 hours at 37°C.
Infections were monitored at various times and aliquots taken for cell viability determinations, using trypan blue and ensuring > 200 cells were counted at each time point, and for virus titrations. Cell monolayers were also examined with a Nikon TE-2000 and cells were photographed with a Canon-A700 digital camera. Images were imported into Adobe and slight adjustments made in brightness and contrast, but which did not alter image context with respect to each other.
Cell fractionation
Infected and mock-infected cells were harvested at various times post-infection and counted. Aliquots of each culture were saved for virus titration to verify infection status. Non-SILAC-labeled cells were individually processed. For comparative SILAC assays, equivalent numbers of L and H cells in each experimental run were confirmed to contain equivalent amounts of protein by BCA™ Protein Assay (Pierce; Rockford, IL) and were mixed together 1:1. Harvested cells were washed 3× in >50 volumes of ice-cold Phosphate Buffered Saline (PBS). Washed cells were resuspended in 250 μl of ice-cold PBS supplemented with 1.5× complete™ Protease Inhibitor (Pierce) and lysed by the addition of 1/10th volume of 5% NP-40. Cells were incubated for 30 min with periodic mixing then centrifuged at 500×g for 10 min to pellet nuclei. The supernatants (cytosol and soluble membranes) were transferred to fresh microfuge tubes and the nuclear pellets were resuspended in 250 μl of PBS supplemented with 1× complete™ Protease Inhibitor + 10% sucrose + 0.44% NP-40 and nuclei re-pelleted, with the 2nd supernatants added to the first ones. The nuclei were then washed 4 times with 1ml of PBS + 0.25× complete™ Protease Inhibitor + 10% sucrose. Washed nuclei were extracted by a new 2-step MS-compatible procedure [20]. Briefly, nuclei were resuspended in 150 μl of High Salt Extraction Buffer (620 mM NaCl, 1 mM DTT, 1 mM MgSO4, 10 mM HEPES, pH 8.0), freeze-thawed, sonicated, insoluble material pelleted at 17,000×g for 10 min, and the supernatants transferred to fresh microfuge tubes. The insoluble pellets were resuspended in 50 μl of 8 M urea, freeze-thawed, sonicated, insoluble material pelleted at 17,000×g for 10 min, and the 2nd supernatants combined with the first.
Protein content in each fraction was determined by BCA Protein Assay (Pierce) and bovine serum albumin standards. The cytosolic and nuclear fractions were stored at -80°C until further processing took place.
Immunoblotting
Equivalent cytosolic and nuclear fractions were resolved by either 10% linear mini sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE, (8.0 × 6.5 × 0.1 cm)) at 180 V for 60 min. or in 4-16% gradient SDS-PAGE (8.0 × 16.0 × 0.1 cm) at 20 mA per gel for 5.5 hours (or overnight for a cumulative total of 110 mAhr per gel). Proteins were transferred to 0.2 μm polyvinylidene difluoride (PVDF) membranes at 20 V for 40 min with a Semi-dry apparatus (BioRad), and protein transfer was confirmed by Ponceau-S staining. The membranes were blocked with 5% (w/v) skim milk in Tris-buffered saline with Tween-20 (TBST; 50 mM Tris, 150 mM NaCl, 0.05% Tween 20, pH 7.4) and probed with various primary antibodies. Primary antibodies were: in-house produced rabbit anti-reovirus, α- GAPDH (Cell Signaling, cat#2118), α-Mx1 (Ori-Gene # TA308496), α-PARP (Cell Signaling, cat#9541), α-ISG15 (Rockland, cat#200-401-438), α-IFIT (Abcam, cat#ab55837), and α-SAMD9 (Sigma, cat#HPA021318); and mouse α-Actin (Sigma, cat#A5441) and anti-STAT1 (Cell Signaling, cat#9176). Appropriate secondary horseradish peroxidase (HRP)-conjugated rabbit anti-mouse or goat anti-rabbit (Cell Signaling, cat#7076, cat#7074, respectively) were used to detect immune complexes. Bands were developed by enhanced chemiluminescence and imaged with an Alpha Innotech FluorChemQ MultiImage III instrument.
Comparative SILAC analyses
Protein digestion and peptide fractionation
After protein concentration determinations, SILAC-labeled samples were diluted with freshly made 100 mM ammonium bicarbonate to concentrations of ~1 mg/ml and pH ~8. Three hundred μl of each sample (~300 μg of protein) were reduced with dithiothreitol (DTT), alkylated with iodoacetic acid, quenched with additional DTT, and trypsin-digested overnight at 37°C with 6 μg of sequencing grade trypsin (Promega, Madison, WI) as previously described [19]. Digests were dried.
Tryptic peptides were fractionated by an orthogonal 2-dimensional reverse-phase (RP) high pH – RP low pH procedure [50,51]. Lyophilized tryptic digests were dissolved in 200 μl of 20 mM ammonium formate pH 10 (Buffer A), injected onto a 1×100 mm XTerra (Waters, Milford, MA) column and fractionated with a 0.67% acetonitrile per minute linear gradient (Agilent 1100 Series HPLC system, Agilent Technologies, Wilmington, DE) at a flow rate of 150 μL/min. Sixty 1-min fractions were collected (covering ~40% acetonitrile concentration range) and concatenated [51,52], with the last 30 fractions combined with the first 30 fractions in sequential order (i.e. #1 with #31; #2 with #32, etc.). Combined fractions were vacuum-dried and re-dissolved in buffer A for the second dimension RP separation (0.1% formic acid in water).
A splitless nano-flow Tempo LC system (Eksigent, Dublin, CA) with 20 μL sample injection via a 300 μm×5 mm PepMap100 pre-column (Dionex, Sunnyvale, CA) and a 100 μm×200 mm analytical column packed with 5 μm Luna C18(2) (Phenomenex, Torrance, CA) were used in the second dimension separation prior to MS analysis. Both eluents A (water) and B (acetonitrile) contained 0.1% formic acid as an ion-pairing modifier. A 0.33% acetonitrile per minute linear gradient (0-30% B) was used for peptide elution, providing a total 2-hour run time for each of the 30 concatenated samples.
Mass spectrometry, bioinformatics, and data mining
A QStar Elite mass spectrometer (Applied Biosystems, Foster City, CA) was used in a data-dependent MS/MS acquisition mode. One-second survey MS spectra were collected (m/z 400-1500) followed by MS/MS measurements on the 3 most intense parent ions (80 counts/sec threshold, +2 - +4 charge state, m/z 100-1500 mass range for MS/MS), using the manufacturer’s “smart exit” (spectral quality 5) settings. Previously targeted parent ions were excluded from repetitive MS/MS acquisition for 60 sec (50 mDa mass tolerance) and the bias correction option was used to correct for small pipetting errors. Raw data files (30 for each of the 4 experimental run samples) were submitted for simultaneous search using standard SILAC settings for QStar instruments and were analyzed by Protein Pilot®, version 4.0, using the non-redundant human gene database (NCBInr, released March 2011, downloaded from http://ftp.ncbi.nih.govrefseqH_sapiensmRNA_Prot, containing 37,391 entries). Proteins, their confidences, and their expression ratios, expressed as infected: mock (I:M), were returned with gi accession numbers. Only proteins for which at least 2 fully trypsin digested L and H peptides were detected at >99% confidence were used for subsequent comparative quantitative analyses. The false discovery rate (FDR), defined as the percentage of reverse proteins identified against the total protein identification, was determined to be < 0.8%.
Each of the 4 datasets were normalized, essentially as described [53] to allow dataset merging and comparison. Briefly, every I:M ratio was converted into log space to determine geometric means and standard deviations in each dataset. Every protein’s log2 I:M ratio was then converted into a z-score, using the formula:
where “b” represents an individual protein in a dataset population a….n, and z-score is the measure of how many standard deviation units (expressed as “σ”) that protein’s log2 I:M ratio is away from its population mean. Thus, a protein with a z-score > 1.960σ indicates that protein’s differential expression lies outside the 95% confidence level, > 2.576σ indicates outside the 99% confidence level, and 3.291σ indicates 99.9% confidence. Z-scores >1.960 were considered significant. Proteins that obtained significant positive z-scores in one labeling experiment, but that also obtained significant negative z-scores in the reciprocal labeling experiment were assumed to be contaminants, were removed from analysis, and z-scores iteratively re-calculated. Gi numbers of all significantly regulated proteins were converted into HGNC identifiers by Uniprot (http://www.uniprot.org/) and HGNC terms were submitted to and analyzed by STRING [54,55] and by the DAVID bioinformatic suite at the NIAID, version 6.7 [56,57] and gene ontologies examined with the “FAT” datasets. The gi numbers were also submitted to, and pathways constructed with, Ingenuity Pathway Analysis software (IPA™).
Results and discussion
T3D replicates in HeLa cells
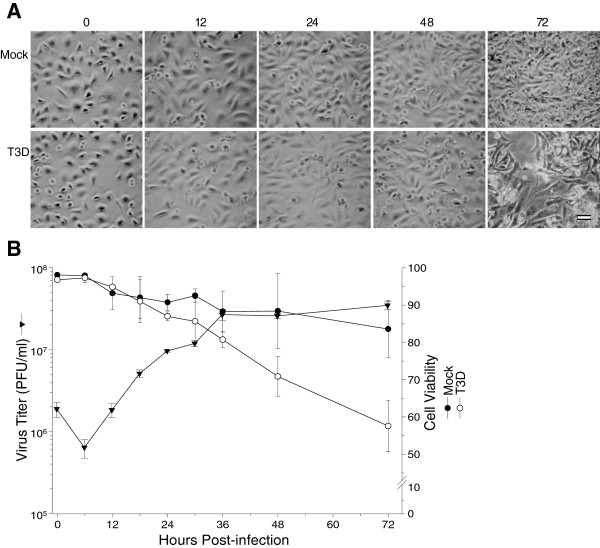
Reoviruses are routinely cultured and titrated in mouse L929 cells [22,23]. The virus also is capable of growing in HeLa cells [35]. We evaluated the time course and confirmed the capacity of T3D to enter and replicate in HeLa cells, and to cause cytopathic effect (CPE), under various infection conditions (Figure 1). Phase-contrast microscopy indicated mock-infected cells showed no evidence of virus infection and CPE was not noticeable until almost a week later, presumably due to nutrient deprivation. T3D-infected HeLa cells also showed little, if any, noticeable CPE until approximately 72 hpi (Figure 1A). Differences in CPE were detectable by trypan blue exclusion, with mock-infected cells showing gradual loss of cell viability from 0 to 72 hpi (Figure 1B). Differences in trypan blue-measured cell viability between the mock- and T3D-infected samples began to manifest at 24 hpi and became more apparent later (Figure 1B). Virus titers initially declined, corresponding to virus uncoating, and began to increase after 6 hpi, was exponential from 6 to 36 hpi, and plateaued after this point. Thus, we elected to use an initial MOI of 7, the same amount of virus used in our earlier quantitative proteomic analyses [19-21], and to examine comparative quantitative differences at the 24 hpi time point, prior to significant CPE development, to allow more meaningful comparison to these other studies.
Figure 1.
Confirmation of HeLa cell infectivity. A, HeLa cells were mock-infected (top), or infected with MRV strain T3D at an MOI of 7 (bottom). Cells were imaged at indicated times post-infection (labeled at top) with a Nikon TE-2000 inverted microscope and photographed with a Canon A700 digital camera. Images were imported into Adobe and slight adjustments made in brightness and contrast, that did not alter image context. Scale bar is 50 μm. B, Kinetics of virus production (▼) and cell viability after HeLa cells were infected (○) or mock-infected (●) with T3D. Error bars represent standard error of the mean from 2 or more replicates.
Identification of altered host proteins
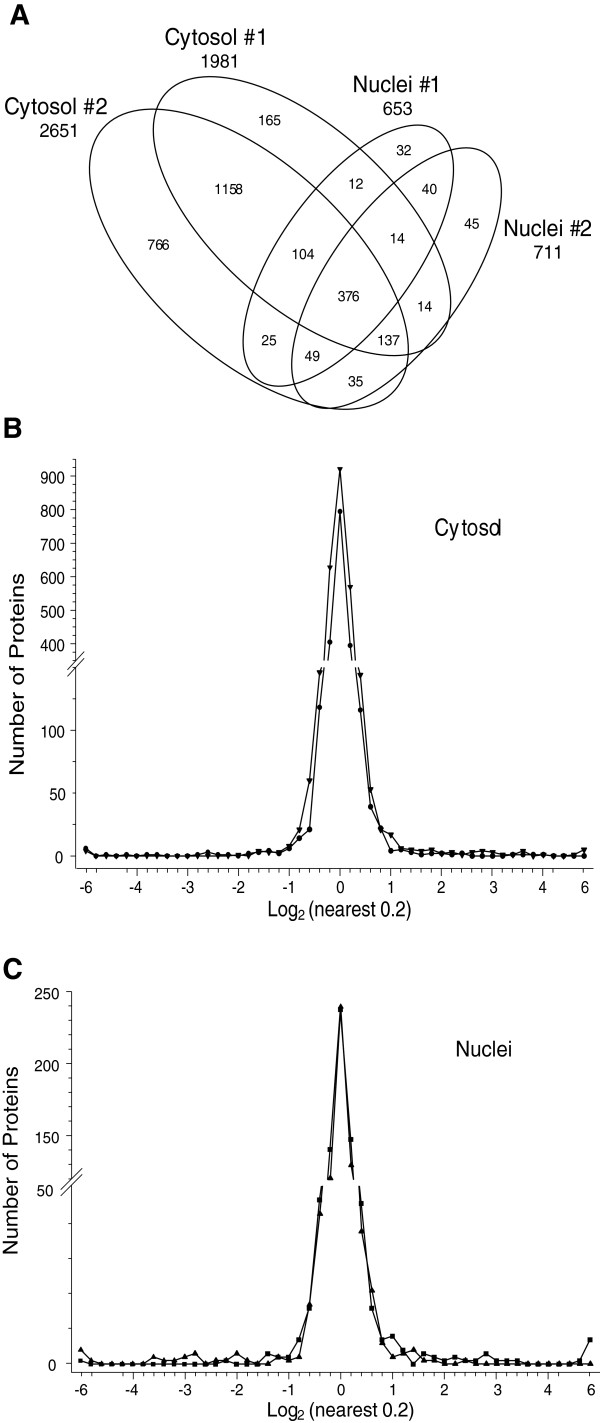
HeLa cells were infected (or mock) in duplicate and purified cytosolic and nuclear fractions obtained. Viruses generally induce profound alterations in infected cells. To rule out the possibility that infection might lead to profound alterations, including significant nuclear leakage by 24 hpi, which could affect our interpretations of protein localization, we determined the distribution of annotated nuclear proteins within each of our L-labeled cytoplasmic fractions (Additional file 1: Figure S1). 922 of the 2137 (43.1%) L-labeled proteins in the mock-infected cytoplasmic sample had GO nuclear annotations, whereas 43.6% of the 1583 L-labeled proteins in the infected cytoplasmic sample had GO nuclear annotations, suggesting no significant nuclear leakage by 24 hpi in the infected samples. 2D-HPLC/MS processing of the mixed samples identified 73,969 H:L peptide pairs, corresponding to 3230 proteins, at ≥99% confidence. 2067 proteins were found in the cytosolic fraction in experimental run #1 and 2787 were present in the run #2 cytosolic fraction (Table 1). Exclusion of those proteins identified by only a single peptide, as well as those that probably represent contaminants (see next paragraph), further reduced the number of identified proteins to 1981 in the run #1 cytosolic fraction (Table 1; Figure 2A). Using similar criteria, we analyzed 2651 proteins in the reciprocally-labeled cytosolic fraction (run #2) and 653 and 711 nuclear proteins (Table 1; Figure 2A).
Table 1.
Number of peptides, proteins, log2 Infected: Mock (I:M) ratio means and standard deviation, and Z-scores of SILAC-measured HeLa cell proteins after T3D infection
| |
Cytosol |
Nuclei |
||
|---|---|---|---|---|
| Run #1 | Run #2 | Run #1 | Run #2 | |
| Total number of peptide pairs1 |
22,591 |
34,393 |
8,011 |
8,974 |
| Total number of proteins2 |
2,067 |
2,787 |
670 |
728 |
| Number of proteins analysed3 |
1,987 |
2,684 |
635 |
686 |
| Mean Log2 I:M ratios |
0.0147 |
0.0126 |
0.0004 |
0.0187 |
| Standard deviation of Log2 I:M ratios |
0.3382 |
0.4211 |
0.4664 |
0.3423 |
| Number of proteins at Z-score cutoff of: ± 1.960σ (95%) |
33, 224 |
50, 20 |
7, 4 |
20, 12 |
| ± 2.576σ (99%) |
24, 11 |
29, 11 |
5, 2 |
14, 8 |
| ± 3.291σ (99.9%) | 14, 7 | 23, 8 | 2, 2 | 8, 4 |
1 Total number of H:L peptide pairs for all proteins identified at confidence level ≥ 99%.
2 Total number of proteins identified at confidence level ≥ 99%.
3 Number of proteins analyzed after those identified by only a single peptide, as well as possible contaminants, removed.
4 First value is number of up-regulated proteins outside the indicated confidence level; second number is number of down-regulated proteins outside the indicated confidence level.
Figure 2.
Distributions of proteins identified in various experiments. A, Venn diagram of numbers of identified proteins from various analyses. B, Frequency distributions of identified proteins in both cytosolic sample sets, with I:M ratios expressed as Log2 values. Positive values represent up-regulated host proteins in virus-infected cells; negative values represent down-regulated host proteins. C, Frequency distributions of identified proteins in both nuclear sample sets. Note that distributions are not identical, with different numbers of proteins differentially expressed away from the major central peak. Characteristics of all peptide and protein distributions, mean log2 I:M ratios and standard deviations of log2 I:M ratios are shown in Table 1.
Each protein’s infected-to-mock (I:M) H:L or L:H ratio was converted into a z-score to normalize the data and facilitate comparisons of each dataset as described in Materials and Methods (and in [19]). A number of proteins were found to have significantly high or low log2 values and corresponding z-scores in either the H:L or L:H labeling scheme, but z-scores that were significantly regulated in the opposite direction in the reciprocal labeling experiment. These included hornerin, keratins, some S100 calcium-binding proteins, and some other proteins (Additional file 2: Table S1; rows 3111-3243). These significant reciprocal values could arise if, for example, exogenous unlabeled (= L-labeled) proteins were introduced during processing. Thus, such proteins were assumed to represent possible contaminants and were removed from further analysis. Z-scores were iteratively recalculated until all probable contaminants had been removed.
Stratification of each protein’s I:M ratio and its corresponding z-score indicated that numerous proteins in each sample could be considered significantly regulated. For example, of the 1987 proteins identified in the cytosolic run #1 preparation, 33 were up-regulated at 95% confidence and 14 were also up-regulated at 99.9% confidence (Table 1). Twenty-two proteins in the same dataset were down-regulated at 95% confidence, and seven of these proteins were also down-regulated at 99.9% confidence. Inspection of protein I:M ratios and z-scores indicated that most proteins differentially regulated at >95% confidence had I:M ratios altered by > 1.6-fold. Thus, for proteins observed multiple times, we applied several filtering levels. We considered them significantly regulated if at least one of their observations had a z-score ≥ 1.960σ, if the other observation in the same type of fraction (cytosolic or nuclear) was no more than 0.75σ in the opposite direction, if most of the peptides used to determine the protein regulation were differentially regulated at >95% confidence, if peptide regulation variability, as measured by standard error of the mean, was relatively small, and if the average I:M ratio was > 1.6-fold. Application of these filters resulted in removal of several proteins which might otherwise have been considered significantly regulated. For example, mitochondria-associated granulocyte macrophage CSF signaling molecule (gi|27363461) had an I:M ratio of 1.63 (Additional file 2: Table S1; row 3133) but was excluded because two peptides had measured I:M ratios of ~1.22 and one peptide had a measured ratio of 100 (STD = 12.64). For proteins observed a single time, they were considered significantly regulated if the z-score was ≥ 1.960σ and if the I:M ratios were altered > 1.6-fold. Using the above criteria, we identified and measured 45 proteins that were significantly up-regulated and 19 proteins that were significantly down-regulated in the cytosolic fraction and 16 proteins that were significantly up-regulated and 9 proteins that were significantly down-regulated in the nuclear fraction (Table 2).
Table 2.
Significantly-regulated HeLa cell proteins after T3D infection
| |
|
|
|
|
|
Run # 1 (H:L) |
Run # 2 (L:H) |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession | HGID | Name | Inf / Mock | (±S.E.M.) | # Pep | % Cov | Z-Score | # Pep | % Cov | Z-score | |
|
Cytoplasmic |
|
|
|
|
|
|
|
|
|
|
|
|
Up-regulated |
|
|
|
|
|
|
|
|
|
|
|
|
Measured more than once |
|
|
|
|
|
|
|
|
|||
| |
gi|222136619 |
MX1 |
myxovirus resistance protein 1 |
9.76 |
0.52 |
8 |
55.9 |
10.925 |
23 |
51.7 |
7.427 |
| |
gi|94536771 |
CCD56 |
coiled-coil domain containing 56 |
5.48 |
1.62 |
5 |
56.6 |
0.223 |
3 |
20.7 |
15.154 |
| |
gi|27881482 |
DDX58 |
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide RIG-I |
4.76 |
0.83 |
3 |
27.7 |
7.191 |
12 |
23.2 |
5.203 |
| |
gi|72534658 |
IFIT3 |
interferon-induced protein with tetratricopeptide repeats 3 |
4.65 |
0.39 |
4 |
25.3 |
7.260 |
14 |
46.9 |
5.067 |
| |
gi|4826774 |
ISG15 |
ISG15 ubiquitin-like modifier |
4.60 |
0.51 |
14 |
47.3 |
6.164 |
14 |
53.9 |
5.442 |
| |
gi|116534937 |
IFIT1 |
interferon-induced protein with tetratricopeptide repeats 1 isoform 2 |
3.80 |
1.36 |
2 |
40.4 |
11.782 |
8 |
25.1 |
3.308 |
| |
gi|156105693 |
PR285 |
PPAR-alpha interacting complex protein 285 isoform 1 |
3.45 |
1.53 |
2 |
22.0 |
5.564 |
5 |
10.6 |
4.109 |
| |
gi|153082755 |
IFIT2 |
interferon-induced protein with tetratricopeptide repeats 2 |
3.37 |
0.81 |
3 |
33.7 |
1.291 |
11 |
33.1 |
4.978 |
| |
gi|6274552 |
STAT1 |
signal transducer and activator of transcription 1 isoform alpha |
3.23 |
0.39 |
13 |
44.4 |
4.386 |
34 |
50.4 |
4.170 |
| |
gi|45007007 |
OAS3 |
2'-5'oligoadenylate synthetase 3 |
3.00 |
0.32 |
4 |
19.5 |
4.560 |
9 |
17.3 |
3.761 |
| |
gi|208973246 |
DHPR |
quinoid dihydropteridine reductase |
2.40 |
0.48 |
2 |
27.9 |
0.693 |
4 |
20.5 |
4.171 |
| |
gi|38201706 |
SAMD9 |
sterile alpha motif domain containing 9 |
2.15 |
0.48 |
1 |
18.8 |
2.560 |
10 |
15.4 |
2.648 |
| |
gi|4506103 |
E2AK2 |
eukaryotic translation initiation factor 2-alpha kinase 2 |
1.97 |
0.72 |
3 |
45.7 |
2.503 |
10 |
16.2 |
2.374 |
| |
gi|24307901 |
IFI35 |
interferon-induced protein 35 |
1.92 |
0.44 |
3 |
29.2 |
2.119 |
5 |
18.4 |
2.508 |
| |
gi|166706903 |
GBP1 |
guanylate binding protein 1, interferon-inducible, 67 kDa |
1.85 |
0.88 |
5 |
25.7 |
1.682 |
11 |
27.2 |
2.393 |
| |
gi|5031863 |
LG3BP |
galectin 3 binding protein |
1.81 |
0.86 |
6 |
19.5 |
1.731 |
12 |
29.2 |
2.303 |
| |
gi|22035653 |
APOL2 |
apolipoprotein L2 |
1.80 |
0.53 |
4 |
30.3 |
1.501 |
6 |
32.6 |
2.507 |
| |
gi|27477136 |
ZCCHV |
zinc finger antiviral protein isoform 1 |
1.80 |
0.69 |
2 |
26.2 |
2.245 |
16 |
25.5 |
2.007 |
| |
gi|20270303 |
MIRO2 |
ras homolog gene family, member T2 |
1.79 |
0.92 |
2 |
15.7 |
9.955 |
6 |
15.7 |
−0.055 |
| |
gi|4757876 |
BST2 |
bone marrow stromal cell antigen 2 |
1.68 |
0.38 |
5 |
19.4 |
1.610 |
5 |
18.3 |
2.201 |
| |
gi|188528628 |
PNPT1 |
polyribonucleotide nucleotidyltransferase 1 |
1.63 |
0.20 |
13 |
47.6 |
2.221 |
24 |
36.8 |
1.575 |
|
Measured once |
|
|
|
|
|
|
|
|
|
||
| |
gi|156415992 |
ATP8* |
transcription factor B1, mitochondrial |
84.09 |
0.71 |
|
|
|
2 |
9.2 |
15.154 |
| |
gi|4557321 |
APOA1 |
apolipoprotein A-I preproprotein |
7.99 |
0.97 |
|
|
|
2 |
23.2 |
7.089 |
| |
gi|11342664 |
MX2 |
myxovirus resistance protein 2 |
7.72 |
0.57 |
|
|
|
6 |
11.3 |
6.972 |
| |
gi|4502511 |
CO9 |
complement component 9 precursor |
7.26 |
4.88 |
|
|
|
2 |
5.0 |
6.760 |
| |
gi|166706909 |
IF44L |
interferon-induced protein 44-like |
6.18 |
3.89 |
|
|
|
3 |
15.9 |
6.211 |
| |
gi|74229015 |
OAS1 |
2',5'-oligoadenylate synthetase 1 isoform 3 |
5.85 |
0.86 |
|
|
|
3 |
11.6 |
6.024 |
| |
gi|19923717 |
DTX3L |
deltex 3-like |
3.68 |
3.31 |
|
|
|
4 |
9.7 |
4.438 |
| |
gi|74229019 |
OAS2 |
2'-5'-oligoadenylate synthetase 2 isoform 1 |
3.50 |
2.14 |
|
|
|
2 |
4.9 |
4.263 |
| |
gi|41350201 |
EPN1 |
epsin 1 isoform c |
3.20 |
2.53 |
|
|
|
2 |
5.5 |
3.953 |
| |
gi|5174751 |
YAP1 |
Yes-associated protein 1, 65 kD |
3.12 |
1.32 |
|
|
|
3 |
7.9 |
3.867 |
| |
gi|222831595 |
DDX60 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
2.92 |
1.56 |
|
|
|
5 |
7.6 |
3.636 |
| |
gi|15208660 |
RO52 |
tripartite motif protein 21 |
2.69 |
0.63 |
|
|
|
8 |
22.7 |
3.364 |
| |
gi|110825982 |
HERC5 |
hect domain and RLD 5 |
2.50 |
0.86 |
|
|
|
2 |
4.7 |
3.104 |
| |
gi|4557499 |
CTBP2 |
C-terminal binding protein 2 isoform 1 |
2.49 |
1.36 |
|
|
|
7 |
25.8 |
3.093 |
| |
gi|4506191 |
PSMB10 |
proteasome beta 10 subunit proprotein |
2.21 |
0.71 |
|
|
|
2 |
7.3 |
2.688 |
| |
gi|73747915 |
TAP2 |
transporter 2, ATP-binding cassette, sub-family B isoform 1 |
2.16 |
0.70 |
|
|
|
3 |
10.8 |
2.615 |
| |
gi|38016914 |
SAMH1 |
SAM domain- and HD domain-containing protein 1 |
2.16 |
0.48 |
|
|
|
6 |
21.7 |
2.610 |
| |
gi|126012562 |
LRP1 |
low density lipoprotein-related protein 1 |
2.11 |
0.60 |
|
|
|
3 |
2.9 |
2.529 |
| |
gi|4503445 |
TYPH |
endothelial cell growth factor 1 (platelet-derived) precursor |
2.10 |
0.85 |
|
|
|
2 |
18.1 |
2.504 |
| |
gi|6912630 |
IFIT5 |
interferon-induced protein with tetratricopeptide repeats 5 |
2.05 |
0.63 |
|
|
|
3 |
13.9 |
2.435 |
| |
gi|112789562 |
IF16 |
interferon, gamma-inducible protein 16 |
1.95 |
0.64 |
|
|
|
6 |
21.8 |
2.263 |
| |
gi|45580709 |
UN93B |
unc-93 homolog B1 |
1.93 |
0.77 |
|
|
|
2 |
4.2 |
2.231 |
| |
gi|9665248 |
TAP1 |
transporter 1, ATP-binding cassette, sub-family B |
1.88 |
0.78 |
|
|
|
2 |
11.8 |
2.135 |
| |
gi|52630342 |
HLA-C |
major histocompatibility complex, class I, C precursor |
1.80 |
0.34 |
|
|
|
20 |
45.4 |
1.982 |
|
Down-regulated |
|
|
|
|
|
|
|
|
|
|
|
|
Measured more than once |
|
|
|
|
|
|
|
|
|||
| |
gi|134288865 |
S4A7 |
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
0.064 |
0.21 |
2 |
21.7 |
−3.832 |
2 |
3.8 |
−15.807 |
| |
gi|29294627 |
PPFIBP1 |
PTPRF interacting protein binding protein 1 isoform 1 |
0.30 |
0.21 |
3 |
26.5 |
−10.860 |
3 |
11.0 |
0.328 |
| |
gi|4506431 |
RASA1 |
RAS p21 protein activator 1 isoform 1 |
0.36 |
0.06 |
1 |
16.5 |
−1.418 |
2 |
1.8 |
−4.688 |
| |
gi|5453704 |
PRAF3 |
ADP-ribosylation-like factor 6 interacting protein 5 |
0.56 |
0.18 |
4 |
40.4 |
−3.941 |
2 |
29.8 |
0.235 |
| |
gi|21956645 |
MTPN |
myotrophin |
0.56 |
0.08 |
9 |
49.1 |
−4.823 |
8 |
39.0 |
0.091 |
|
Measured once |
|
|
|
|
|
|
|
|
|
||
| |
gi|167466177 |
CDC27 |
cell division cycle protein 27 isoform 1 |
0.010 |
0.36 |
|
|
|
2 |
4.0 |
−15.807 |
| |
gi|8922549 |
ASF1B |
anti-silencing function 1B |
0.33 |
0.000 |
|
|
|
2 |
23.8 |
−3.834 |
| |
gi|33598948 |
PLCG1 |
phospholipase C gamma 1 isoform a |
0.33 |
0.34 |
|
|
|
2 |
6.9 |
−3.803 |
| |
gi|46049063 |
XPO6 |
exportin 6 |
0.34 |
0.05 |
|
|
|
2 |
5.2 |
−3.706 |
| |
gi|116734706 |
I2BP2 |
interferon regulatory factor 2 binding protein 2 isoform B |
0.36 |
0.38 |
|
|
|
3 |
13.7 |
−3.517 |
| |
gi|116686114 |
FERM1 |
kindlin-1 |
0.38 |
0.004 |
|
|
|
3 |
12.3 |
−3.348 |
| |
gi|5453543 |
AK1C1 |
aldo-keto reductase family 1, member C1 |
0.39 |
0.05 |
14 |
72.8 |
−4.099 |
|
|
|
| |
gi|63025188 |
HERC4 |
hect domain and RLD 4 isoform a |
0.41 |
0.000 |
|
|
|
2 |
2.5 |
−3.077 |
| |
gi|14589951 |
RPAB1 |
DNA directed RNA polymerase II polypeptide E |
0.46 |
0.14 |
|
|
|
6 |
21.4 |
−2.683 |
| |
gi|51317370 |
NDUA6 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6, 14 kDa |
0.47 |
0.000 |
|
|
|
2 |
14.9 |
−2.637 |
| |
gi|10190746 |
RDH14 |
retinol dehydrogenase 14 (all-trans and 9-cis) |
0.51 |
0.38 |
|
|
|
2 |
8.6 |
−2.306 |
| |
gi|13236559 |
MMTA2 |
hypothetical protein LOC79169 |
0.54 |
0.01 |
|
|
|
2 |
35.0 |
−2.138 |
| |
gi|115430235 |
UHRF1 |
ubiquitin-like with PHD and ring finger domains 1 isoform 1 |
0.55 |
0.10 |
|
|
|
2 |
11.1 |
−2.105 |
| |
gi|71773010 |
AP1G1 |
adaptor-related protein complex 1, gamma 1 subunit isoform a |
0.55 |
0.25 |
4 |
22.3 |
−2.570 |
|
|
|
|
Nuclear |
|
|
|
|
|
|
|
|
|
|
|
|
Up-regulated |
|
|
|
|
|
|
|
|
|
|
|
|
Measured more than once |
|
|
|
|
|
|
|
|
|||
| |
gi|4826774 |
ISG15 |
ISG15 ubiquitin-like modifier |
4.05 |
0.40 |
5 |
42.4 |
4.065 |
4 |
23.0 |
6.275 |
| |
gi|19923236 |
SP100 |
nuclear antigen Sp100 isoform 2 |
2.96 |
0.93 |
4 |
18.1 |
2.812 |
8 |
26.2 |
4.880 |
| |
gi|112789562 |
IF16 |
interferon, gamma-inducible protein 16 |
2.92 |
0.40 |
2 |
11.8 |
2.384 |
6 |
19.3 |
4.879 |
| |
gi|67089149 |
PML |
promyelocytic leukemia protein isoform 1 |
2.34 |
0.60 |
2 |
8.4 |
2.758 |
9 |
21.7 |
3.496 |
| |
gi|4757876 |
BST2 |
bone marrow stromal cell antigen 2 |
2.03 |
0.56 |
3 |
18.3 |
2.512 |
2 |
13.9 |
2.290 |
|
Measured once |
|
|
|
|
|
|
|
|
|
||
| |
gi|4503253 |
DAD1 |
defender against cell death 1 |
5.22 |
0.62 |
3 |
17.7 |
5.110 |
|
|
|
| |
gi|188219599 |
NMI |
N-myc and STAT interactor |
4.33 |
1.17 |
|
|
|
2 |
16.9 |
6.125 |
| |
gi|24307901 |
IFI35 |
interferon-induced protein 35 |
3.60 |
0.93 |
|
|
|
3 |
15.6 |
5.347 |
| |
gi|4758024 |
COIL |
coilin |
2.46 |
0.83 |
2 |
12.8 |
2.788 |
|
|
|
| |
gi|5453690 |
DNJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
2.32 |
0.000 |
|
|
|
2 |
11.8 |
3.499 |
| |
gi|58530840 |
DESP |
desmoplakin isoform I |
2.09 |
0.91 |
|
|
|
3 |
9.3 |
3.054 |
| |
gi|156104878 |
GLSK |
glutaminase |
2.03 |
0.75 |
|
|
|
3 |
4.8 |
2.931 |
| |
gi|27477136 |
ZCCHV |
zinc finger antiviral protein isoform 1 |
1.95 |
0.74 |
|
|
|
2 |
2.2 |
2.754 |
| |
gi|4507951 |
1433F |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
1.89 |
1.06 |
|
|
|
2 |
22.8 |
2.633 |
| |
gi|5901998 |
LSM6 |
Sm protein F |
1.85 |
0.60 |
|
|
|
3 |
56.3 |
2.538 |
| |
gi|4506003 |
PP1A |
protein phosphatase 1, catalytic subunit, alpha isoform 1 |
1.70 |
0.95 |
|
|
|
4 |
13.9 |
2.184 |
|
Down-regulated |
|
|
|
|
|
|
|
|
|
|
|
|
Measured more than once |
|
|
|
|
|
|
|
|
|||
| |
gi|55953087 |
NOG1 |
G protein-binding protein CRFG |
0.10 |
0.21 |
2 |
4.4 |
−13.385 |
2 |
6.3 |
−1.353 |
| |
gi|4503453 |
EDF1 |
endothelial differentiation-related factor 1 isoform alpha |
0.63 |
0.17 |
4 |
28.4 |
−0.672 |
4 |
41.2 |
−2.991 |
| |
gi|4507555 |
LAP2A |
thymopoietin isoform alpha |
0.64 |
0.03 |
10 |
15.3 |
−2.464 |
10 |
12.5 |
−0.479 |
|
Measured once |
|
|
|
|
|
|
|
|
|
||
| |
gi|88853069 |
VTNC |
vitronectin precursor |
0.013 |
0.04 |
2 |
4.8 |
−13.385 |
|
|
|
| |
gi|25777615 |
PSD7 |
proteasome 26S non-ATPase subunit 7 |
0.39 |
0.30 |
|
|
|
3 |
12.3 |
−4.046 |
| |
gi|4757732 |
AIFM1 |
programmed cell death 8 isoform 1 |
0.40 |
0.18 |
|
|
|
2 |
18.4 |
−3.870 |
| |
gi|57863257 |
TCPA |
T-complex protein 1 isoform a |
0.50 |
0.04 |
|
|
|
2 |
5.2 |
−2.951 |
| |
gi|4506701 |
RS23 |
ribosomal protein S23 |
0.54 |
0.19 |
|
|
|
4 |
32.9 |
−2.647 |
| gi|151108473 | FIS1 | tetratricopeptide repeat domain 11 | 0.63 | 0.000 | 2 | 7.2 | −1.990 | ||||
*: Obsolete record; removed from NCBI database.
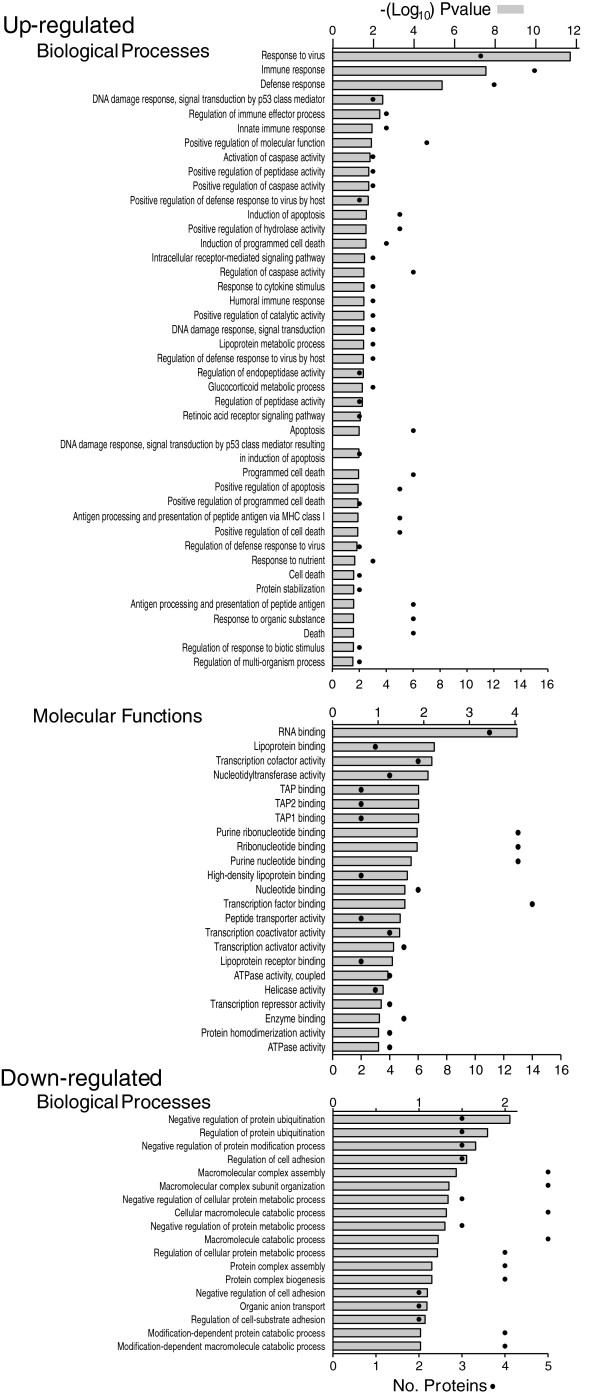
HeLa cell proteins up-regulated by T3D infection are associated with defense responses, immune responses, macromolecular binding, regulation of immune effector processes, and responses to virus
Proteins, and their levels of regulation, were analyzed several ways. Protein gi numbers were imported into Uniprot (http://www.uniprot.org/) and converted into HUGO nomenclature committee (HGNC) identifiers. Significantly up-regulated and down-regulated proteins (95% confidence interval) were then imported into DAVID [56,58] and gene ontologies identified (Figure 3).
Figure 3.
Gene ontology analyses of up-regulated and down-regulated proteins. Sets of up- and down-regulated proteins identified in Table 2 were separately imported into the DAVID gene ontology suite of programs at the NIAID, gene identifications converted by that program, and ontological functions determined by GOTERM.
Up-regulated proteins were assigned to 42 GOTERM biological processes at 95% confidence (Figure 3, upper), that included responses to viruses, immune responses, defense responses, regulation of immune effector processes and numerous related processes. Up-regulated proteins were also assigned to 23 functional groups (Figure 3, middle) including primarily macromolecular binding and transcription cofactor and nucleotidyltransferase activities.
Protein HGNC identifiers were also uploaded into STRING [54,55] and interactions depicted (Figure 4B, left). Many immunomodulatory proteins (ie. MX1, MX2, ISG15, various IFITs and various OAS molecules) form a major interacting cluster and interact either directly or indirectly with numerous other regulated proteins.
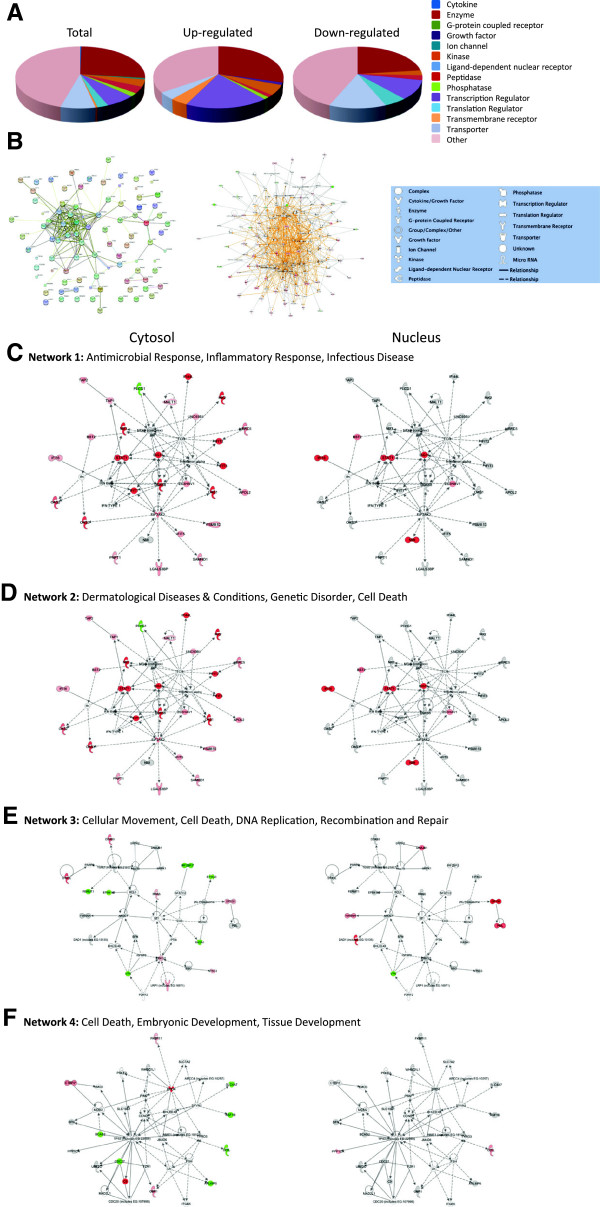
Figure 4.
Molecular pathways of regulated proteins. Proteins and their levels of regulation were imported into the Ingenuity Pathways Analysis (IPA™) tool and interacting pathways were constructed. A, Ontological classifications of all measured proteins (Total) as well as those significantly up- and down-regulated. B, Merged networks, determined by uploading the regulated proteins only into STRING [54,55] (left), or by uploading all molecules and their degree of regulation into IPA™ (middle). C-F, The top 4 IPA networks, identified at 95% confidence and each of which contained 12 or more “focus” molecules (molecules significantly up- or down-regulated), with pathway names indicated. Solid lines: direct known interactions; dashed lines: suspected or indirect interactions. Significantly regulated proteins identified in either the cytosolic or nuclear fractions were overlaid onto each network; red: significantly up-regulated proteins; pink: moderately up-regulated proteins; grey: proteins identified but not significantly regulated; light green: moderately down-regulated proteins; dark green: significantly down-regulated proteins; white: proteins known to be in network, but not identified in our study.
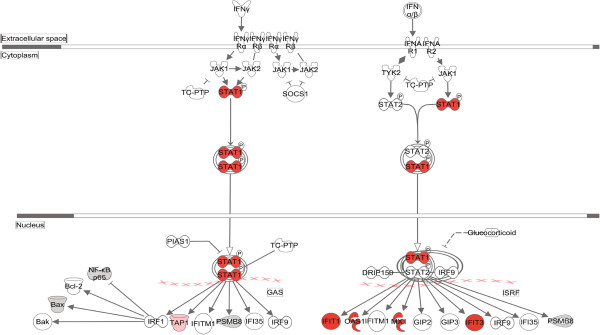
Protein gi numbers and levels of regulation were also imported into the Ingenuity Pathways Analysis (IPA™) tool to build interacting pathways. IPA indicated up-regulated proteins were enriched in cytokines, growth factors, transcription regulators and transmembrane receptors. IPA identified 8 interacting pathways at a confidence level of 95% or greater. Four of these pathways, each with 12 or more “focus” members (significantly up- or down-regulated proteins), shared common members and it was possible to build a single, merged pathway (Figure 4B, middle). Other pathways contained 2 or fewer focus molecules. The four major pathways were those involved in antimicrobial response, inflammatory response, infectious diseases, dermatological diseases and conditions, cell-to-cell signaling and interaction, cell cycle, and cell death. Regulated proteins identified in each of the purified cytosolic and nuclear fractions were overlaid onto each network (Figure 4C–F). Proteins identified in our analyses as up-regulated and present in the pathways are depicted in shades of red. Some proteins, such as IFI16, ISG15, SP100, and STAT1, were highly regulated in both the cytosol and the nucleus, whereas some, such as IFIT1, Mx1, and TRIM21, were up-regulated in one fraction but not the other. Many of the identified regulated proteins also are involved in several known canonical pathways, including interferon signaling (Figure 5), glycogen degradation, pattern recognition receptors, PDGF signaling, antigen presentation, protein ubiquitination, and others (Additional file 3: Figure S2). Many of the highly up-regulated proteins, such as STAT, ISG15 and Mx1 also represent interaction nodes within various pathways, interacting with numerous other proteins. Many of these proteins play important roles in innate immunity. In addition, the Mx proteins, which are large GTPase dynamin-like interferon-induced molecules, are important anti-viral proteins, particularly against RNA viruses [59,60]. These have been identified as up-regulated by various virus infections, including by influenza [4,7,61] and by reoviruses [62-64].
Figure 5.
Significantly affected canonical pathway “Interferon signaling”, as determined by IPA™ analysis. Red indicates highly up-regulated proteins, light green indicates down-regulated, grey represents not significantly affected, and white represents molecules known to be part of the pathway but not identified by SILAC.
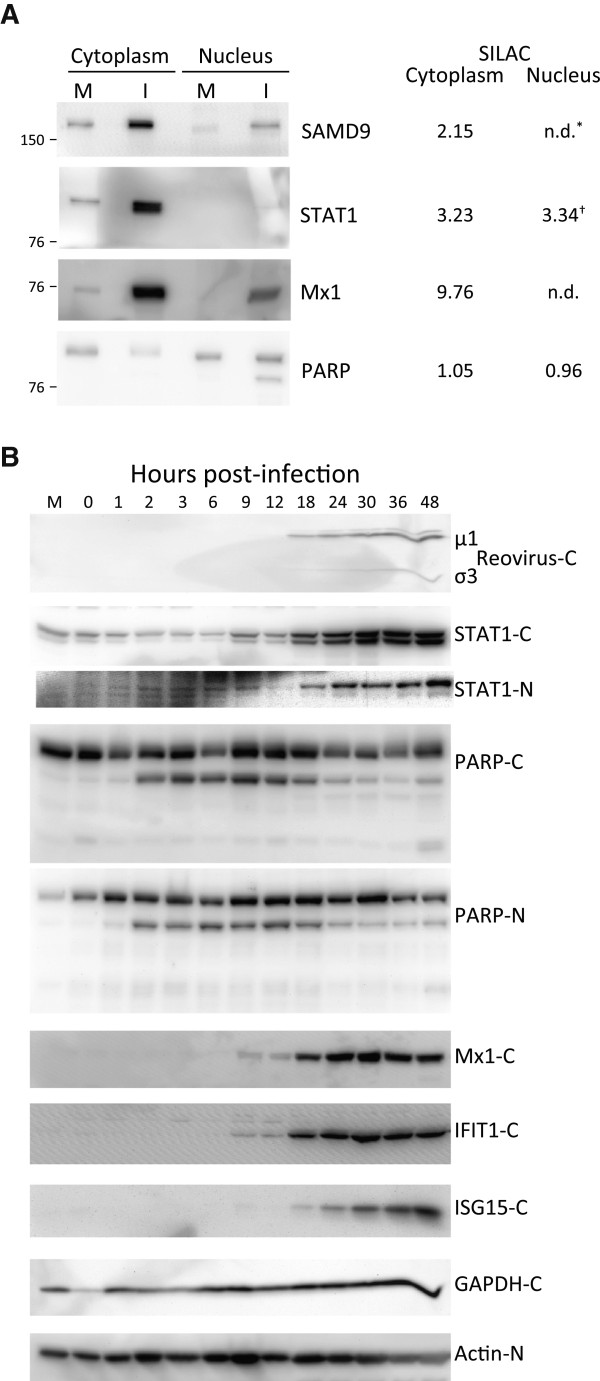
To both validate some of the SILAC data, as well as determine temporal expression of various immune-modulated proteins, we harvested T3D-infected HeLa cells at various times, extracted the cytosolic and nuclear fractions, and immuno-probed for various molecules (Figure 6). Most Western blot results confirmed the SILAC-determined results (Figure 6A). For example, STAT1 was measured as up-regulated in both the cytoplasmic as well as nuclear fractions by SILAC and Western blot of both fractions confirmed these results. A few differences in the specific measured ratios (for example, SILAC indicated STAT1 was up-regulated 3.2–fold in the cytoplasm, but Western blot densitometry suggested the increase was approximately 5–fold) probably reflect different levels of sensitivity of the two assays. In addition, SILAC suggested the relative levels of PARP were essentially unchanged, although Western blot suggested a small decrease in cytoplasmic levels and nuclear fractions demonstrated PARP cleavage (Figure 6A). Kinetic analyses revealed more details. Reovirus proteins were not detected at early time points but became detectable by 18 hpi. STAT-1, an important effector protein that plays a key role in numerous canonical pathways (Figure 5; Additional file 3: Figure S2), and that was found up-regulated in both the cytoplasmic as well as nuclear fractions by SILAC (Figure 6A; Table 2), was detected in mock-infected cells as well as in infected cells. STAT-1 expression appeared to remain constant until 9 hpi, and then increased from 9 hpi onward, reaching levels about 3-fold higher at 24 hpi in the cytoplasm compared to mock and about 6-fold higher at later time points. Significant STAT1 increases were seen at later times, 18 hpi, in the nucleus (Figure 6B). Similarly, expression of Mx1 was detected in mock and at early times in infected cells and began to increase at 9 hpi, reaching ~8-fold higher levels by 24 hpi (Figure 6B). Mx1, IFIT1, and ISG15 were detected in cytoplasmic fractions and showed clear up-regulation by 18 hpi (by 9 hpi in the case of Mx1 and IFIT1) (Figure 6B), but none of these three proteins were detected in nuclear fractions at any time point up to 48 hpi (data not shown). PARP, cleavage of which serves as an apoptosis marker, demonstrated increased cleavage in both the cytoplasm and the nucleus as early as 2 hpi and this cleavage appeared to wane by about 18-24 hpi, shortly after reovirus proteins became detectable and shortly after the other discussed proteins started to increase, consistent with known interactions between these and other innate immune and inflammatory response molecules [65-67].
Figure 6.
Western blot validation of host protein regulation. A, HeLa cells were mock-infected or infected for 24 h, or B, for indicated periods of time, harvested and lysed with 0.5% NP-40 detergent. The cytosolic and nuclear fractions were separately purified, dissolved in SDS electrophoresis sample buffer, and proteins resolved in 10% (A), or in 4-16% gradient (B) SDS-PAGE, transferred to PVDF, and probed with indicated antibodies. Antibody binding was detected with HRP-conjugated secondary antibodies and ECL, and visualized with an Alpha Innotech FluorChemQ MultiImage III instrument. Molecular weight standards are indicated at left and SILAC-measured ratios are indicated on right in A. *: not detected in indicated fraction; †: based on single peptide only.
HeLa cell proteins down-regulated by T3D infection are associated with cell death, macromolecular catabolic processes, and tissue development
DAVID assigned down-regulated proteins to 18 biological processes at 95% confidence (Figure 3, lower), that included ubiquitination regulation, cell adhesion regulation, macromolecular complex assembly and catabolic processes. IPA indicated down-regulated proteins were enriched in translation regulation and transporter factors (Figure 4A). Many down-regulated proteins also are related to mitochondrial dysfunction (Additional file 3: Figure S2). Many other proteins, such as EIF4G3, FERMT1, CDC27 and COIL were mapped to the cytoplasmic fractions (Figure 4E, F).
An apparent down-regulation in protein levels in either of the sub-cellular fractions examined could represent reduced protein biosynthesis, protein degradation, redistribution of proteins from one fraction to another, or various combinations of the above. Protein redistribution could be manifested by the same protein apparently being up-regulated in one compartment at the same time it appeared down-regulated in the other compartment. While there were numerous proteins that were detected as either up-regulated or down-regulated in one compartment and unaffected in the other compartment (i.e. IFIT3, MX2, PML, RASA1, XPO6) there were very few proteins detected that were up-regulated in one compartment and simultaneously down-regulated in the other. For example coilin (COIL) was found up-regulated about 2.5-fold in purified nuclear fractions, based upon 2 peptides; however, it appeared down-regulated by about 2-fold, but only based upon a single peptide (Additional file 2: Table S1). This molecule is an integral component of nuclear suborganelles called Cajal bodies that play roles in small RNA post-transcriptional modifications [68,69]. More detailed analyses of fates of various proteins, with regards to the possible altered biosynthesis, degradation and redistribution as a result of virus infection, are warranted.
Similarities and differences between HeLa cell and HEK293 cell protein responses to reovirus infection
Transcriptomic responses to T3D infection have been previously reported [40]. That study identified more than 100 interferon- and NF-κB-responsive genes that were either positively or negatively regulated by T3D infection. We identified and measured 30 of these genes’ proteins and assessed correlation between the mRNA and protein levels. There was good correlation for some genes and proteins. For example, OAS1 and PML mRNA and protein levels were highly up-regulated, 9 other proteins we identified as up-regulated also had up-regulated mRNA levels, although the degree of up-regulation differed, and some proteins and genes (i.e. UBC and POLD1) were similarly non-regulated or only slightly down-regulated. Analysis of the 30 proteins after assigning proteins and genes to highly-up, slightly-up, non-regulated, slightly-down and highly-down regulated classes resulted in an r2 correlation of 0.62 (data not shown), slightly higher than what has been found in other studies that correlate mRNA levels to protein levels [3,4]. Unfortunately, most mRNAs and proteins could not be compared because the other ~70 interferon- and NF-κB-responsive genes reported by O’Donnell et al. [40] were not found by us.
We previously determined HEK293 responses to reovirus T1L infection [21]. That study identified and measured 2992 proteins at 24 hpi, 104 of which were up-regulated and 49 of which were down-regulated. Only 194 (~ 6.5%) of these same proteins were identified in the current study (Additional file 1: Figure S3). A small number of proteins were similarly regulated by both viruses in both cell types. Most proteins (159-187, depending upon how cutoffs were set) were non-regulated in both cell types, and no proteins were up-regulated in one cell but down-regulated in the other (Additional file 1: Figure S3C). Although there was only ~6.5% overlap between the 2 protein datasets, which could represent differences in the virus used and/or in the type of cell analyzed, many of the highly regulated pathways and processes were similar between the two experimental conditions. For example, regulation of interferon signaling, immunomodulation and responses to virus were highly up-regulated in both studies. Only 5 of the 194 host proteins identified and measured in both the T1L-infected HEK-293 and T3D-infected HeLa cells were significantly regulated. Of these, only 1 (STAT1), discussed in more detail earlier, was significantly regulated by both cell/virus conditions, being up-regulated 2.1-fold in T1L-infected HEK293 cells and up-regulated 3.2-fold in T3D-infected HeLa cells. One other protein (SCO1) was up-regulated 8.4-fold in T1L-infected HEK-293 cells but only moderately up-regulated (1.4-fold, Z-score = 1.088σ) in T3D-infected HeLa cells. SCO1 is a metallochaperone involved in copper homeostasis and found in the mitochondrial intermembrane space [70]. The SCO genes appear evolutionarily conserved [71] and have not yet been reported to have roles in virus replication. Two proteins were significantly up-regulated in one cell type but not regulated in the other. PRKCDBP, a potential tumor suppressor gene [72], and COMT, a degradative methyltransferase [73] that has been associated with cognitive deficits in herpes simplex type 1 virus infections [74], were up-regulated 2.3-, and 3.1-fold, respectively, in T1L-infected HEK-293 cells. However, the COMT ratio is based upon a single measured peptide so less reliable. One protein was significantly down-regulated in one cell type but not regulated in the other. PLCG1, involved in immune regulated signal transduction [75], was down-regulated 3-fold (I:M ratio of 0.33) in T3D-infected HeLa cells. Twenty six other proteins were moderately regulated (Z score 1.000 – 1.959σ, corresponding to ~ 0.9- – 2.2-fold up-regulation; or -1.000 – -1.959σ, corresponding to ~ 0.9- – 1.6-fold down-regulation [I:M ratios of 0.63 – 1.13) (Additional file 3: Table S2). Although measured values were not significant, one additional protein (C2orf43), a gene that has been associated with prostate cancer [76], was moderately down-regulated in both the T1L and T3D infections, with infected:mock values of .66 and 0.61, respectively. A few of these determined values are based upon 1 or 2 peptides. Therefore, additional work is needed to determine whether these apparent similarities and differences are real and virus- and/or cell-specific. We are currently analyzing T1L- and T3D-infected HeLa cells labeled by the iTRAQ reagent to simultaneously compare multiple virus types to mock and to use a complementary approach that may identify additional regulated proteins.
In summary, this non-biased global analysis has identified numerous host proteins significantly affected by reovirus T3D infection. These proteins complement others determined in other studies, fit within numerous inflammatory and innate immune pathways, and will provide the starting point for more detailed kinetic studies, such as initiated herein, as well as studies aimed at delineation of virus-specific pathways.
Competing interests
The author declares no conflicts of interest.
Supplementary Material
Gene ontology distribution of light-labeled proteins in indicated samples collected at 24 hpi from Mock- and T3D-infected HeLa cells;  , nuclear-annotated proteins;
, nuclear-annotated proteins;  , non-nuclear-annotated proteins. Total numbers of proteins and percentages indicated within each pie section. Figure S2. Additional top-ranked canonical pathways identified by IPA™. Red indicates highly up-regulated proteins, light green indicates down-regulated, grey represents not significantly affected, and white represents molecules known to be part of the pathway but not identified by SILAC. Figure S3. Comparison of reovirus-induced protein identifications and regulation in HEK293 cells [21] and in HeLa cells (this study). A, Venn diagram of numbers of identified proteins, and their overlap, from the 2 studies. B, Dot plot distributions of the Z-scores of each of the 194 proteins identified in both studies. C, Comparative distributions of regulated proteins, using Z-score category cutoffs of ± 1.960 (upper value in each cell) and ± 1.000 (lower italicized value in each cell). For example, 1 protein (STAT1) had a Z-score > 1.960 in both the HEK293 and HeLa cell lists and 2 proteins (STAT1 and cytochrome oxidase deficient homolog 1) had Z-scores > 1.000 in both the HEK293 and HeLa cell lists.
, non-nuclear-annotated proteins. Total numbers of proteins and percentages indicated within each pie section. Figure S2. Additional top-ranked canonical pathways identified by IPA™. Red indicates highly up-regulated proteins, light green indicates down-regulated, grey represents not significantly affected, and white represents molecules known to be part of the pathway but not identified by SILAC. Figure S3. Comparison of reovirus-induced protein identifications and regulation in HEK293 cells [21] and in HeLa cells (this study). A, Venn diagram of numbers of identified proteins, and their overlap, from the 2 studies. B, Dot plot distributions of the Z-scores of each of the 194 proteins identified in both studies. C, Comparative distributions of regulated proteins, using Z-score category cutoffs of ± 1.960 (upper value in each cell) and ± 1.000 (lower italicized value in each cell). For example, 1 protein (STAT1) had a Z-score > 1.960 in both the HEK293 and HeLa cell lists and 2 proteins (STAT1 and cytochrome oxidase deficient homolog 1) had Z-scores > 1.000 in both the HEK293 and HeLa cell lists.
All identified proteins and their associated # peptides, % coverage, Infected: Mock ratio, and calculated Z-scores a.
Common proteins regulated in either T1L-infected HEK-293 or T3D-infected HeLa cells.
Acknowledgements
This work was supported by grant MT-11630 from the Canadian Institutes of Health Research to KMC and is MIAPE-compliant. The author thanks Kolawole Opanubi for cell maintenance, Peyman Ezatti for mass spectrometry, John Cortens for database assistance, Matthew Stuart-Edwards for developing scripts for some bioinformatics analysis, and members of the laboratory for reviewing the manuscript.
References
- Geiss GK, Salvatore M, Tumpey TM, Carter VS, Wang XY, Basler CF, Taubenberger JK, Bumgarner RE, Palese P, Katze MG, Garcia-Sastre A. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: The role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc Natl Acad Sci USA. 2002;99:10736–10741. doi: 10.1073/pnas.112338099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobasa D, Jones SM, Shinya K, Kash JC, Copps J, Ebihara H, Hatta Y, Kim JH, Halfmann P, Hatta M. et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature. 2007;445:319–323. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]
- Tian Q, Stepaniants SB, Mao M, Weng L, Feetham MC, Doyle MJ, Yi EC, Dai H, Thorsson V, Eng J. et al. Integrated genomic and proteomic analyses of gene expression in mammalian cells. Mol Cell Proteomics. 2004;3:960–969. doi: 10.1074/mcp.M400055-MCP200. [DOI] [PubMed] [Google Scholar]
- Baas T, Baskin CR, Diamond DL, Garcia-Sastre A, Bielefeldt-Ohmann H, Tumpey TM, Thomas MJ, Carter VS, Teal TH, Van Hoeven N. et al. Integrated molecular signature of disease: analysis of influenza virus-infected macaques through functional genomics and proteomics. J Virol. 2006;80:10813–10828. doi: 10.1128/JVI.00851-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradet-Balade B, Boulme F, Beug H, Mullner EW, Garcia-Sanz JA. Translation control: bridging the gap between genomics and proteomics? Trends Biochem Sci. 2001;26:225–229. doi: 10.1016/S0968-0004(00)01776-X. [DOI] [PubMed] [Google Scholar]
- Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annu Rev Biomed Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- Coombs KM. Quantitative proteomics of complex mixtures. Expert Rev Proteomics. 2011;8:659–677. doi: 10.1586/epr.11.55. [DOI] [PubMed] [Google Scholar]
- Lucitt MB, Price TS, Pizarro A, Wu W, Yocum AK, Seiler C, Pack MA, Blair IA, Fitzgerald GA, Grosser T. Analysis of the zebrafish proteome during embryonic development. Mol Cell Proteomics. 2008;7:981–994. doi: 10.1074/mcp.M700382-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgener A, Boutilier J, Wachihi C, Kimani J, Carpenter M, Westmacott G, Cheng K, Ball TB, Plummer F. Identification of differentially expressed proteins in the cervical mucosa of HIV-1-resistant sex workers. J Proteome Res. 2008;7:4446–4454. doi: 10.1021/pr800406r. [DOI] [PubMed] [Google Scholar]
- Stewart JJ, White JT, Yan X, Collins S, Drescher CW, Urban ND, Hood L, Lin B. Proteins associated with Cisplatin resistance in ovarian cancer cells identified by quantitative proteomic technology and integrated with mRNA expression levels. Mol Cell Proteomics. 2006;5:433–443. doi: 10.1074/mcp.M500140-MCP200. [DOI] [PubMed] [Google Scholar]
- Booy AT, Haddow JD, Ohlund LB, Hardie DB, Olafson RW. Application of isotope coded affinity tag (ICAT) analysis for the identification of differentially expressed proteins following infection of Atlantic salmon (Salmo salar) with infectious hematopoietic necrosis virus (IHNV) or Renibacterium salmoninarum (BKD) J Proteome Res. 2005;4:325–334. doi: 10.1021/pr049840t. [DOI] [PubMed] [Google Scholar]
- Dwivedi RC, Dhindsa N, Krokhin OV, Cortens J, Wilkins JA, El Gabalawy HS. The effects of infliximab therapy on the serum proteome of rheumatoid arthritis patients. Arthritis ResTherapy. 2009;11:32. doi: 10.1186/ar2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Niu D, Sui J, Ching CB, Chen WN. Protein profile in hepatitis B virus replicating rat primary hepatocytes and HepG2 cells by iTRAQ-coupled 2-D LC-MS/MS analysis: Insights on liver angiogenesis. Proteomics. 2009;9:2836–2845. doi: 10.1002/pmic.200800911. [DOI] [PubMed] [Google Scholar]
- de Hoog CL, Foster LJ, Mann M. RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell. 2004;117:649–662. doi: 10.1016/S0092-8674(04)00456-8. [DOI] [PubMed] [Google Scholar]
- Everley PA, Krijgsveld J, Zetter BR, Gygi SP. Quantitative cancer proteomics: stable isotope labeling with amino acids in cell culture (SILAC) as a tool for prostate cancer research. Mol Cell Proteomics. 2004;3:729–735. doi: 10.1074/mcp.M400021-MCP200. [DOI] [PubMed] [Google Scholar]
- Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.M200025-MCP200. [DOI] [PubMed] [Google Scholar]
- Yan W, Lee H, Yi EC, Reiss D, Shannon P, Kwieciszewski BK, Coito C, Li XJ, Keller A, Eng J. et al. System-based proteomic analysis of the interferon response in human liver cells. Genome Biol. 2004;5:R54. doi: 10.1186/gb-2004-5-8-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs KM, Berard A, Xu W, Krokhin O, Meng X, Cortens JP, Kobasa D, Wilkins J, Brown EG. Quantitative proteomic analyses of influenza virus-infected cultured human lung cells. J Virol. 2010;84:10888–10906. doi: 10.1128/JVI.00431-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeker AL, Ezzati P, Halayko AJ, Coombs KM. Response of primary human airway epithelial cells to Influenza infection – A quantitative proteomic study. J Proteome Res. 2012;11:4132–4136. doi: 10.1021/pr300239r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berard AR, Cortens JP, Krokhin O, Wilkins JA, Severini A, Coombs KM. Characterization of the host response proteome after mammalian T1L reovirus infection. PLoS One. 2012;7:e51939. doi: 10.1371/journal.pone.0051939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff LA, Nibert ML, Tyler KL. In: Fields Virology, Volume 5. Knipe DM, Howley PM, editor. Philadelphia: Lippincott Williams & Wilkins; 2007. Orthoreoviruses and their replication; pp. 1853–1915. [Google Scholar]
- Coombs KM. Reoviruses. In Encyclopedia of Life Sciences . London, U.K.: Macmillan; 2011. [DOI] [Google Scholar]
- Mertens PPC, Attoui H, Duncan R, Dermody TS. In: Virus Taxonomy Eighth Report of the International Committee on Taxonomy of Viruses. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editor. London: Elsevier/Academic Press; 2005. Reoviridae; pp. 447–454. [Google Scholar]
- Estes MK, Kapikian AZ. In: Fields Virology, Volume 5. Knipe DM, Howley PM, editor. Philadelphia: Lippincott Williams & Wilkins; 2007. Rotaviruses; pp. 1917–1974. [Google Scholar]
- Roy P. In: Fields Virology, Volume 5. Knipe DM, Howley PM, editor. Philadelphia: Lippencott Williams & Wilkins; 2007. Orbiviruses; pp. 1975–1997. [Google Scholar]
- Attoui H, Biagini P, Stirling J, Mertens PPC, Cantaloube JF, Meyer A, de Micco P, de Lamballerie X. Sequence characterization of Ndelle virus genome segments 1, 5, 7, 8, and 10: Evidence for reassignment to the genus Orthoreovirus, family Reoviridae. Biochem Biophys Res Commun. 2001;287:583–588. doi: 10.1006/bbrc.2001.5612. [DOI] [PubMed] [Google Scholar]
- Coffey MC, Strong JE, Forsyth PA, Lee PW. Reovirus therapy of tumors with activated Ras pathway. Science. 1998;282:1332–1334. doi: 10.1126/science.282.5392.1332. [DOI] [PubMed] [Google Scholar]
- Forsyth P, Roldan G, George D, Wallace C, Palmer CA, Morris D, Cairncross G, Matthews MV, Markert J, Gillespie Y. et al. A phase I trial of intratumoral administration of reovirus in patients with histologically confirmed recurrent malignant gliomas. Mol Ther. 2008;16:627–632. doi: 10.1038/sj.mt.6300403. [DOI] [PubMed] [Google Scholar]
- Thirukkumaran CM, Nodwell MJ, Hirasawa K, Shi ZQ, Diaz R, Luider J, Johnston RN, Forsyth PA, Magliocco AM, Lee P. et al. Oncolytic viral therapy for prostate cancer: efficacy of reovirus as a biological therapeutic. Cancer Res. 2010;70:2435–2444. doi: 10.1158/0008-5472.CAN-09-2408. [DOI] [PubMed] [Google Scholar]
- Pan D, Pan LZ, Hill R, Marcato P, Shmulevitz M, Vassilev LT, Lee PWK. Stabilisation of p53 enhances reovirus-induced apoptosis and virus spread through p53-dependent NF-kappa B activation. Br J Cancer. 2011;105:1012–1022. doi: 10.1038/bjc.2011.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berard A, Coombs KM. Mammalian reoviruses: propagation, quantification, and storage. Curr Protoc Microbiol. 2009. Chapter 15:Unit15C 11. [DOI] [PubMed]
- Loh PC, Soergel M. Growth characteristics of reovirus type 2: actinomycin D and the synthesis of viral RNA. Proc Natl Acad Sci USA. 1965;54:857–863. doi: 10.1073/pnas.54.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RE, Zweerink HJ, Joklik WK. Polypeptide components of virions, top component and cores of reovirus type 3. Virology. 1969;39:791–810. doi: 10.1016/0042-6822(69)90017-8. [DOI] [PubMed] [Google Scholar]
- Golden JW, Linke J, Schmechel S, Thoemke K, Schiff LA. Addition of exogenous protease facilitates reovirus infection in many restrictive cells. J Virol. 2002;76:7430–7443. doi: 10.1128/JVI.76.15.7430-7443.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokiej M, Ogden KM, Ikizler M, Reiter DM, Stehle T, Dermody TS. Optimum Length and Flexibility of Reovirus Attachment Protein sigma 1 Are Required for Efficient Viral Infection. J Virol. 2012;86:10270–10280. doi: 10.1128/JVI.01338-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggioli GJ, DeBiasi RL, Bickel R, Jotte R, Spalding A, Johnson GL, Tyler KL. Reovirus-induced alterations in gene expression related to cell cycle regulation. J Virol. 2002;76:2585–2594. doi: 10.1128/JVI.76.6.2585-2594.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBiasi RL, Clarke P, Meintzer S, Jotte R, Kleinschmidt-Demasters BK, Johnson GL, Tyler KL. Reovirus-induced alteration in expression of apoptosis and DNA repair genes with potential roles in viral pathogenesis. J Virol. 2003;77:8934–8947. doi: 10.1128/JVI.77.16.8934-8947.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler KL, Leser JS, Phang TL, Clarke P. Gene expression in the brain during reovirus encephalitis. J Neurovirol. 2010;16:56–71. doi: 10.3109/13550280903586394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell SM, Holm GH, Pierce JM, Tian B, Watson MJ, Chari RS, Ballard DW, Brasier AR, Dermody TS. Identification of an NF-kappaB-dependent gene network in cells infected by mammalian reovirus. J Virol. 2006;80:1077–1086. doi: 10.1128/JVI.80.3.1077-1086.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, Schmechel SC, Raghavan A, Abelson M, Reilly C, Katze MG, Kaufman RJ, Bohjanen PR, Schiff LA. Reovirus induces and benefits from an integrated cellular stress response. J Virol. 2006;80:2019–2033. doi: 10.1128/JVI.80.4.2019-2033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Sevinsky JR, Rowland MD, Bundy JL, Stephenson JL, Sherry B. Proteomic analysis reveals virus-specific Hsp25 modulation in cardiac myocytes. J Proteome Res. 2010;9:2460–2471. doi: 10.1021/pr901151k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Opanubi KJ, Coombs KM. Non-biased enrichment does not improve quantitative proteomic delineation of reovirus T3D-infected HeLa cell protein alterations. Front Microbiol. 2012;3:310. doi: 10.3389/fmicb.2012.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs KM. Temperature-sensitive mutants of reovirus. Curr Top Microbiol Immunol. 1998;233:69–107. doi: 10.1007/978-3-642-72092-5_4. [DOI] [PubMed] [Google Scholar]
- Ramig RF, Fields BN. In: The reoviridae. Joklik WK, editor. New York: Plenum; 1983. Genetics of reovirus; pp. 197–228. [Google Scholar]
- Weiner HL, Powers ML, Fields BN. Absolute linkage of virulence and central nervous system cell tropism of reoviruses to viral hemagglutinin. J Infect Dis. 1980;141:609–616. doi: 10.1093/infdis/141.5.609. [DOI] [PubMed] [Google Scholar]
- Nibert ML, Margraf RL, Coombs KM. Nonrandom segregation of parental alleles in reovirus reassortants. J Virol. 1996;70:7295–7300. doi: 10.1128/jvi.70.10.7295-7300.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Cheang M, Coombs KM. The M1 gene is associated with differences in the temperature optimum of the transcriptase activity in reovirus core particles. J Virol. 1996;70:1223–1227. doi: 10.1128/jvi.70.2.1223-1227.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez II, Hermann LL, Hazelton PR, Coombs KM. A comparative analysis of freon substitutes in the purification of reovirus and calicivirus. J Virol Methods. 2000;90:59–67. doi: 10.1016/S0166-0934(00)00217-2. [DOI] [PubMed] [Google Scholar]
- Gilar M, Olivova P, Daly AE, Gebler JC. Orthogonality of separation in two-dimensional liquid chromatography. Anal Chem. 2005;77:6426–6434. doi: 10.1021/ac050923i. [DOI] [PubMed] [Google Scholar]
- Spicer V, Yamchuk A, Cortens J, Sousa S, Ens W, Standing KG, Wilkins JA, Krokhin O. Sequence-specific retention calculator. A family of peptide retention time prediction algorithms in reversed-phase HPLC: applicability to various chromatographic conditions and columns. Anal Chem. 2007;79:8762–8768. doi: 10.1021/ac071474k. [DOI] [PubMed] [Google Scholar]
- Dwivedi RC, Spicer V, Harder M, Antonovici M, Ens W, Standing KG, Wilkins JA, Krokhin OV. Practical implementation of 2D HPLC scheme with accurate peptide retention prediction in both dimensions for high-throughput bottom-up proteomics. Anal Chem. 2008;80:7036–7042. doi: 10.1021/ac800984n. [DOI] [PubMed] [Google Scholar]
- Keshamouni VG, Jagtap P, Michailidis G, Strahler JR, Kuick R, Reka AK, Papoulias P, Krishnapuram R, Srirangam A, Standiford TJ. et al. Temporal quantitative proteomics by iTRAQ 2D-LC-MS/MS and corresponding mRNA expression analysis identify post-transcriptional modulation of actin-cytoskeleton regulators during TGF-beta-Induced epithelial-mesenchymal transition. J Proteome Res. 2009;8:35–47. doi: 10.1021/pr8006478. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P. et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Kruger B, Snel B, Bork P. STRING 7 - recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:60. doi: 10.1186/gb-2003-4-9-r60. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang QL, Wang L, Bai SY, Lin WS, Chen WN, Lin JY, Lin X. Global Proteome Analysis of Hepatitis B Virus Expressing Human Hepatoblastoma Cell Line HepG2. J Med Virol. 2009;81:1539–1550. doi: 10.1002/jmv.21593. [DOI] [PubMed] [Google Scholar]
- Haller O, Kochs G. Interferon-induced mx proteins: Dynamin-like GTPases with antiviral activity. Traffic. 2002;3:710–717. doi: 10.1034/j.1600-0854.2002.31003.x. [DOI] [PubMed] [Google Scholar]
- Haller O, Staeheli P, Kochs G. Protective role of interferon-induced Mx GTPases against influenza viruses. Rev Sci Tech. 2009;28:219–231. doi: 10.20506/rst.28.1.1867. [DOI] [PubMed] [Google Scholar]
- Vester D, Rapp E, Gade D, Genzel Y, Reichl U. Quantitative analysis of cellular proteome alterations in human influenza A virus-infected mammalian cell lines. Proteomics. 2009;9:3316–3327. doi: 10.1002/pmic.200800893. [DOI] [PubMed] [Google Scholar]
- Goody RJ, Beckham JD, Rubtsova K, Tyler KL. JAK-STAT signaling pathways are activated in the brain following reovirus infection. J Neurovirol. 2007;13:373–383. doi: 10.1080/13550280701344983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurney J, Kobayashi T, Holm GH, Dermody TS, Sherry B. Reovirus mu2 protein inhibits interferon signaling through a novel mechanism involving nuclear accumulation of interferon regulatory factor 9. J Virol. 2009;83:2178–2187. doi: 10.1128/JVI.01787-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry B. Rotavirus and reovirus modulation of the interferon response. J Interferon Cytokine Res. 2009;29:559–567. doi: 10.1089/jir.2009.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghavi N, Samuel CE. Protein kinase PKR catalytic activity is required for the PKR-dependent activation of mitogen-activated protein kinases and amplification of interferon beta induction following virus infection. Virology. 2012;427:208–216. doi: 10.1016/j.virol.2012.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ba XQ, Garg NJ. Signaling Mechanism of Poly(ADP-Ribose) Polymerase-1 (PARP-1) in Inflammatory Diseases. Am J Pathol. 2011;178:946–955. doi: 10.1016/j.ajpath.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG. Cajal bodies: The first 100 years. Annu Rev Cell Dev Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- Nizami Z, Deryusheva S, Gall JG. The Cajal Body and Histone Locus Body. Cold Spring Harb Perspect Biol. 2010;2:a000653. doi: 10.1101/cshperspect.a000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary SC, Cobine PA, Nishimura T, Verdijk RM, de Krijger R, de Coo R, Tarnopolsky MA, Winge DR, Shoubridge EA. COX19 mediates the transduction of a mitochondrial redox signal from SCO1 that regulates ATP7A-mediated cellular copper efflux. Mol Biol Cell. 2013;24:683–691. doi: 10.1091/mbc.E12-09-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcelli D, Oliva M, Duchi S, Latorre D, Cavaliere V, Barsanti P, Villani G, Gargiulo G, Caggese C. Genetic, functional and evolutionary characterization of scox, the Drosophila melanogaster ortholog of the human SCO1 gene. Mitochondrion. 2010;10:433–448. doi: 10.1016/j.mito.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Tong SY, Lee JM, Ki KD, Seol HJ, Choi YJ, Lee SK. Genetic Polymorphism of PRKCDBP is Associated with an Increased Risk of Endometrial Cancer. Cancer Invest. 2012;30:642–645. doi: 10.3109/07357907.2012.727054. [DOI] [PubMed] [Google Scholar]
- Naaz H, Pandey VP, Singh S, Dwivedi UN. Structure-function analyses and molecular modeling of caffeic acid-O-methyltransferase and caffeoyl-CoA-O-methyltransferase: Revisiting the basis of alternate methylation pathways during monolignol biosynthesis. Biotechnol Appl Biochem. 2013;60:170–189. doi: 10.1002/bab.1075. [DOI] [PubMed] [Google Scholar]
- Dickerson F, Stallings C, Sullens A, Origoni A, Leister F, Krivogorsky B, Yolken R. Association between cognitive functioning, exposure to Herpes Simplex Virus type 1, and the COMT Val158Met genetic polymorphism in adults without a psychiatric disorder. Brain Behav Immun. 2008;22:1103–1107. doi: 10.1016/j.bbi.2008.04.156. [DOI] [PubMed] [Google Scholar]
- Suh PG, Park JI, Manzoli L, Cocco L, Peak JC, Katan M, Fukami K, Kataoka T, Yun S, Ryu SH. Multiple roles of phosphoinositide-specific phospholipase C isozymes. BMB Rep. 2008;41:415–434. doi: 10.5483/BMBRep.2008.41.6.415. [DOI] [PubMed] [Google Scholar]
- Long QZ, Du YF, Ding XY, Li X, Song WB, Yang Y, Zhang P, Zhou JP, Liu XG. Replication and Fine Mapping for Association of the C2orf43, FOXP4, GPRC6A and RFX6 Genes with Prostate Cancer in the Chinese Population. PLoS One. 2012;7:e37866. doi: 10.1371/journal.pone.0037866. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene ontology distribution of light-labeled proteins in indicated samples collected at 24 hpi from Mock- and T3D-infected HeLa cells;  , nuclear-annotated proteins;
, nuclear-annotated proteins;  , non-nuclear-annotated proteins. Total numbers of proteins and percentages indicated within each pie section. Figure S2. Additional top-ranked canonical pathways identified by IPA™. Red indicates highly up-regulated proteins, light green indicates down-regulated, grey represents not significantly affected, and white represents molecules known to be part of the pathway but not identified by SILAC. Figure S3. Comparison of reovirus-induced protein identifications and regulation in HEK293 cells [21] and in HeLa cells (this study). A, Venn diagram of numbers of identified proteins, and their overlap, from the 2 studies. B, Dot plot distributions of the Z-scores of each of the 194 proteins identified in both studies. C, Comparative distributions of regulated proteins, using Z-score category cutoffs of ± 1.960 (upper value in each cell) and ± 1.000 (lower italicized value in each cell). For example, 1 protein (STAT1) had a Z-score > 1.960 in both the HEK293 and HeLa cell lists and 2 proteins (STAT1 and cytochrome oxidase deficient homolog 1) had Z-scores > 1.000 in both the HEK293 and HeLa cell lists.
, non-nuclear-annotated proteins. Total numbers of proteins and percentages indicated within each pie section. Figure S2. Additional top-ranked canonical pathways identified by IPA™. Red indicates highly up-regulated proteins, light green indicates down-regulated, grey represents not significantly affected, and white represents molecules known to be part of the pathway but not identified by SILAC. Figure S3. Comparison of reovirus-induced protein identifications and regulation in HEK293 cells [21] and in HeLa cells (this study). A, Venn diagram of numbers of identified proteins, and their overlap, from the 2 studies. B, Dot plot distributions of the Z-scores of each of the 194 proteins identified in both studies. C, Comparative distributions of regulated proteins, using Z-score category cutoffs of ± 1.960 (upper value in each cell) and ± 1.000 (lower italicized value in each cell). For example, 1 protein (STAT1) had a Z-score > 1.960 in both the HEK293 and HeLa cell lists and 2 proteins (STAT1 and cytochrome oxidase deficient homolog 1) had Z-scores > 1.000 in both the HEK293 and HeLa cell lists.
All identified proteins and their associated # peptides, % coverage, Infected: Mock ratio, and calculated Z-scores a.
Common proteins regulated in either T1L-infected HEK-293 or T3D-infected HeLa cells.