Abstract
Mouse primordial germ cells (PGC) undergo sequential epigenetic changes and genome-wide DNA demethylation to reset the epigenome for totipotency. Here, we demonstrate that erasure of CpG methylation (5mC) in PGCs occurs via conversion to 5-hydroxymethylcytosine (5hmC), driven by high levels of TET1 and TET2. Global conversion to 5hmC initiates asynchronously among PGCs at embryonic day (E) 9.5-E10.5 and accounts for the unique process of imprint erasure. Mechanistically, 5hmC enrichment is followed by its protracted decline thereafter at a rate consistent with replication-coupled dilution. The conversion to 5hmC is a significant component of parallel redundant systems that drive comprehensive reprogramming in PGCs. Nonetheless, we identify rare regulatory elements that escape systematic DNA demethylation in PGCs, providing a potential mechanistic basis for transgenerational epigenetic inheritance.
Specification of primordial germ cells (PGCs) from epiblast cells at ~E6.25 is linked with extensive epigenetic reprogramming, including global DNA demethylation, chromatin reorganisation and imprint erasure, that is vital for generating totipotency (1, 2). The erasure of CpG methylation (5mC) is a key component of this program, but the dynamics and underlying mechanisms of the process remain unclear (3). Here we report a comprehensive analysis of PGCs by combining immunofluorescence, genome-wide (h)meDIP-seq, single cell RNA-seq, bisulfite-seq and functional analyses to address the mechanistic basis of epigenetic reprogramming in PGCs.
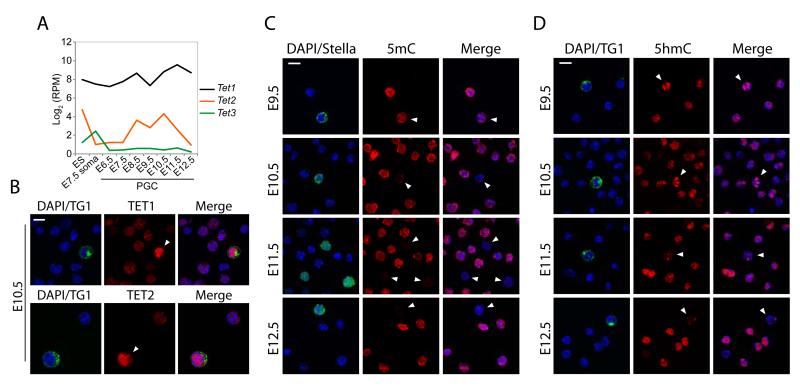
We investigated Tet expression using single cell RNA-seq, which revealed that Tet1 and Tet2 are expressed in PGCs and peak between E10.5-E11.5, but that Tet3 is undetectable (Fig. 1A). Immunofluorescence (IF) showed that TET1 and TET2 are nuclear and expressed at significantly higher levels in PGCs than neighbouring somatic cells between E9.5-E11.5 (Fig. 1B & S1-S2). This suggests that erasure of 5mC in PGCs could occur through conversion to 5-hydroxymethylcytosine (5hmC) by TET1/TET2 (4, 5).
Figure 1. Global dynamics of 5mC, 5hmC & TETs in PGCs.
(A) Single cell RNA-seq analysis of Tet1, Tet2 and Tet3 expression. Shown is Log2 reads per million (RPM). (B) Expression of TET1 and TET2 in E10.5 PGCs (arrowheads) and soma. (C) Dynamics of DNA methylation (5mC) in PGCs shows 5mC erasure between E9.5-E11.5. (D) 5-hydroxymethylcytosine (5hmC) kinetics in PGCs. TG1/STELLA mark PGCs. Scale bar = 10μm.
We pursued this possibility by IF and found a progressive reduction of 5mC in PGCs between E9.5-E10.5, until it became undetectable by E11.5 (Fig. 1C). The loss of 5mC occurs concurrently with a global enrichment of 5hmC in PGCs between E9.5-E10.5, suggesting a genome-scale conversion of 5mC to 5hmC (Fig. 1D). The global conversion to 5hmC initiates asynchronously among PGCs from E9.5, perhaps reflecting developmental heterogeneity (Fig. S3-S5). Indeed, TET1 upregulation also initially occurs in a subset of PGCs from E9.5, which apparently exhibit lower 5mC signal (Fig. S6). In contrast to soma and ES cells (6), we observed that 5hmC exhibited a distinct localisation in PGCs that coincided with DAPI-dense chromocentres, indicating that the conversion of 5mC to 5hmC includes heterochromatic satellite regions (Fig. S7). The enrichment of 5hmC in PGCs at E10.5 is followed thereafter by its progressive reduction, suggesting that 5hmC is an intermediate towards demethylation to unmodified cytosine (C) (Fig. 1D). We checked if 5hmC is subsequently converted to 5-formylcytosine (5fC) or 5-carboxycytosine (5caC) but found no detectable enrichment of these derivatives in PGCs (Fig. S8) (7).
To gain further insight into the dynamics of 5mC to 5hmC conversion, we performed meDIP-seq and hmeDIP-seq in E10.5-E13.5 PGCs (Fig. S9). As prior to E10.5 PGCs were highly limiting we also profiled epiblast stem cells (EpiSC), which are derived from the same post-implantation epiblast as nascent PGCs, and embryonic soma (E10.5) as references (Fig. S10). Unlike bisulfite sequencing, our approach distinguishes between 5mC and 5hmC but generates a relative rather than a quantitative measure of modifications (6). We therefore initially examined exonic sequences, which are highly methylated and thus exhibit an informative dynamic range of relative (h)meDIP signal when they become demethylated. We found significantly reduced 5mC in E10.5 PGCs relative to EpiSC and soma, and erasure by E11.5 (Fig. 2A, S11-S13). The loss of 5mC in PGCs is paralleled by a strong exonic enrichment of 5hmC, indicating 5mC to 5hmC conversion (Fig. 2A & S11). Once 5mC is converted to 5hmC, it is set on a pathway towards demethylation, as there are no 5hmC maintenance mechanisms (6). Consistent with this, 5hmC undergoes a progressive depletion during PGC development, which is delayed relative to loss of 5mC (Fig. 2A & 2B).
Figure 2. 5mC erasure is coupled to 5hmC conversion.
(A) Enrichment of 5mC and 5hmC in E10.5-E13.5 PGCs and Epiblast stem cells (EpiSC) over internal exons. (B) 5mC and 5hmC distribution relative to a metagene. (C) Profiles of 5mC (blue) and 5hmC (red) at the Dazl promoter. (D) Glucosyltransferase-qPCR showing quantitative levels of 5mC and 5hmC at a CCGG site in the Dazl promoter. Error bars represent S.E.M. (E & F) DNA methylation (%) by bisulfite sequencing of uninduced (−DOX) or induced (+DOX) Tet1/Tet2 miR or non-targeting (NT) miR PGCLCs at (E) gene promoters and (F) repeat elements. Open and closed circles represent unmethylated and methylated CpGs, respectively. (G) DNA methylation in PGCLCs stably expressing catalytically active (WT) or mutant (Mut) TET1 and TET2. Error bars represent S.E.M of allelic methylation.
Next we examined methylation-dependent genes such as Dazl, which are activated by promoter demethylation in PGCs (8, 9), and observed strong 5hmC enrichment coincident with loss of 5mC at their promoters (Fig. 2C & S14). We confirmed that 5mC erasure is coupled to 5hmC enrichment at the Dazl promoter quantitatively, using the glucosyltransferase-qPCR (Glu-qPCR) assay (Fig. 2D). RNA-seq revealed that transcriptional activation of Dazl and other methylation-dependent germline genes initiates at E9.5 and increases progressively until ~E11.5 (Fig. S15). This represents an important functional readout of the timing of DNA demethylation in PGCs.
To functionally link 5hmC to DNA demethylation, we used in vitro PGC-like cells (PGCLC). PGCLCs are specified from epiblast-like cells (EpiLC) and exhibit the fundamental properties of migratory PGCs in vivo, including global DNA demethylation and chromatin reorganisation (Fig.S10) (10). As TET1 and TET2 are both active in PGCs, we generated PGCLCs carrying a doxycycline(DOX)-inducible compound microRNA (miR) knockdown of Tet1 and Tet2 (T-KD). We found that genes known to be demethylated in PGCs in vivo (8), also underwent DNA demethylation upon specification of control uninduced (−DOX) T-KD PGCLCs and in non-targeting (NT) miR PGCLCs (+/−DOX). In contrast, induction of Tet1/Tet2 miR (+DOX) resulted in a substantial inhibition of DNA demethylation in PGCLCs, but not did not reduce the efficiency of their specification (Fig. 2E & S16). Knockdown of Tet1/Tet2 also inhibited DNA demethylation at LINE-1 elements and prevented the limited erasure of 5mC that occurs at intracisternal-A-particles (IAP) (Fig. 2F). These findings are significant considering that both maintenance and de novo DNA methylation systems are repressed in PGC/PGCLCs (10), which likely accounts for some direct passive demethylation. Moreover, constitutive overexpression of catalytically-active, but not catalytic-mutant, TET1 and TET2 in PGCLCs promoted 5mC erasure to a greater extent (Fig. 2G). Thus, TET-mediated 5hmC conversion is a key event towards DNA demethylation in PGCs.
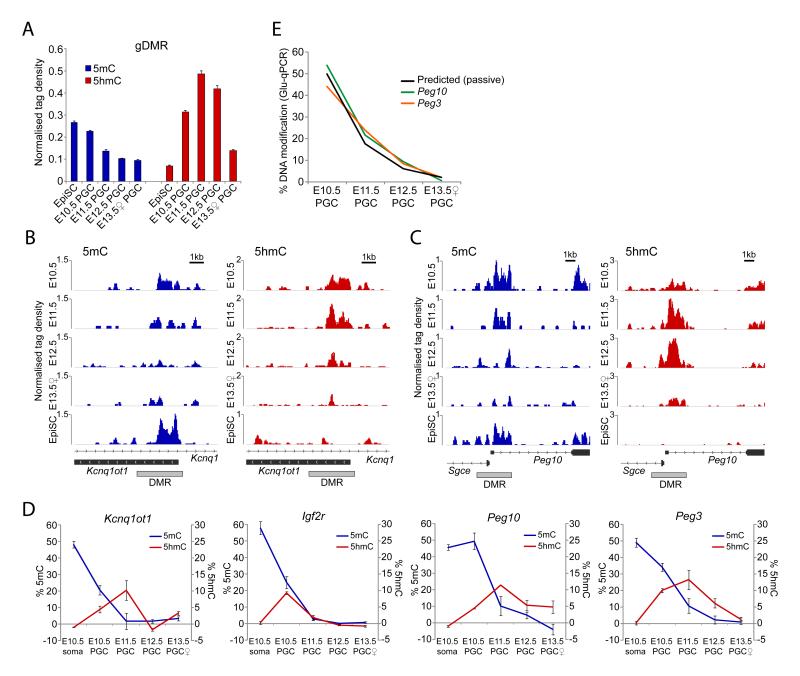
The reprogramming of gonadal PGCs in vivo uniquely entails the complete erasure of genomic imprints (11). Analysis of imprinted gametic differentially methylated regions (gDMRs) (n=21) in PGCs revealed that erasure of 5mC is coupled to a significant increase of 5hmC enrichment (Fig. 3A). However, the precise timing of 5mC erasure is imprinted locus-specific. For example, the DMRs at Kcnq1ot1 and Igf2r exhibit significant loss of 5mC by E10.5 relative to EpiSC (which represent ~50% allelic 5mC), and erasure by E11.5 (Fig. 3B), while Peg10 and Peg3 remain methylated until E11.5 (Fig. 3C & S17). Moreover, Kcnq1ot1 and Igf2r are enriched in 5hmC by E10.5, whereas 5hmC enrichment at Peg10 and Peg3 is delayed until E11.5, suggesting that conversion to 5hmC follows a defined temporal order at imprinted DMRs, which dictates the timing of demethylation in PGCs. Indeed, we observed that other genomic regions also exhibited differential onset of 5mC erasure (compare Peg10 DMR versus exon, Fig. 3C). Glu-qPCR analysis confirmed that the Peg10 and Peg3 DMRs maintained 5mC levels of 50% and 34%, respectively, in E10.5 PGCs, while Kcnq1ot1 and Igf2r DMRs were already reduced to 21% and 25%, respectively (Fig. 3D). Glu-qPCR also established the quantitative enrichment of 5hmC at imprinted DMRs in PGCs. The cumulative data suggest that conversion of 5mC to 5hmC by TET1/TET2 is a general mechanism for the erasure of imprints in PGCs.
Figure 3. Imprint erasure in PGCs.
(A) Average (h)meDIP-seq enrichment of 5mC and 5hmC across all imprinted gDMRs in PGCs. EpiSC represent monoallelic methylation. (B & C) 5mC and 5hmC profiles of DMRs that undergo (B) early or (C) late 5mC erasure and corresponding delay in 5hmC enrichment. (D) Glu-qPCR analysis of 5mC and 5hmC levels at early (Kcnq1ot1 and Igf2r) and late (Peg10 and Peg3) reprogrammed DMRs. Error bars represent S.E.M. (E) Rate of demethylation to unmodified cytosine. Shown is the predicted rate of passive demethylation and observed rates for Peg10 and Peg3.
Conversion of 5mC to 5hmC at exons, promoters and gDMRs in PGCs was followed by a protracted period of progressive 5hmC depletion between E11.5-E13.5 (Fig. 2A-D, Fig. 3), suggesting a replication-coupled process (12). This prompted us to examine the rate of DNA demethylation between E10.5-E13.5 quantitatively using Glu-qPCR. Because demethylation commences asynchronously in PGCs it is necessary to examine loci that have not initiated significant 5mC erasure by E10.5, such as Peg10 and Peg3. As PGCs have an estimated cell cycle of ~16hrs between E10.5-E13.5 (13), we would predict a reduction of DNA modification of ~3-fold per 24hrs (1.5 population doublings) if the process is coupled to DNA replication. We observed that the rate of demethylation at Peg10 (p=0.0022) and Peg3 (p=0.0019) fits highly significantly with the predicted rate (Fig. 3E), suggesting that 5hmC may be removed from these loci by replication-coupled dilution. We obtained similar results for the Dazl promoter (p=0.0014).
We next asked whether any promoters or regulatory elements can escape the comprehensive 5mC reprogramming in PGCs. We screened for CpG islands (CGI) that remain methylated in female PGCs at E13.5, as these cells represent the lowest point of global demethylation (Fig. S18) (14). We identified 11 CGIs with significant 5mC enrichment in E13.5 PGCs (Fig. S19 & S20). Validation by bisulfite sequencing showed that the promoter CGIs of Vmn2r29 and Sfi1 and the exonic CGI of Srrm2 were all methylated in PGCs at E10.5 and maintained significant CpG methylation throughout reprogramming (Fig. 4A).
Figure 4. Inheritance of 5mC through reprogramming.
(A) The Vmn2r29, Sfi1 and Srrm2 CGIs escape reprogramming in PGCs. Open and closed circles represent unmethylated and methylated CpGs, respectively. Dazl is representative of demethylation at most loci. (B) Model for the mechanisms and dynamics of DNA demethylation in PGCs.
To define the extent of 5mC erasure at single-base resolution we performed whole genome bisulfite sequencing (WGBS), which revealed that global CpG methylation is reduced to 2.2% in female E13.5 PGCs (Fig. S21). However, we identified 4730 loci that escape demethylation (>40% 5mC) in PGCs, which are predominately repeat associated (>95%). Resistant loci predominately correspond to IAP elements but interestingly, the IAPLTR1 subclass is significantly more methylated than any other (Fig. S22). IAPLTR1 is the most active and hence hazardous IAP subclass to genomic integrity, suggesting specific systems are mobilised to maintain 5mC at IAPLTR1 during PGCs reprogramming to protect genome stability (15). We were unable to determine any unique sequence characteristics of the 233 single-copy loci with >40% 5mC, suggesting positional context or chromatin structure may contribute to their escape from reprogramming. Indeed, ‘escapees’ were often adjacent to IAP elements or telomeric regions. Considered with the recent observations that many regulatory elements can evade zygotic 5mC erasure (16, 17), our data suggests that rare but potentially functionally relevant 5mC epialleles could be inherited over multiple generations by evading erasure during both zygotic and PGC reprogramming.
We demonstrate here that comprehensive DNA demethylation in PGCs, including imprint erasure, entails conversion of 5mC to 5hmC, likely redundantly by TET1 and TET2. In vivo 5hmC conversion initiates asynchronously in PGCs between E9.5-E10.5, and is largely complete by E11.5. The rate of progressive decline of 5hmC thereafter, both globally and at single-copy loci, is consistent with a replication-dependent mechanism of demethylation towards unmodified cytosines (Fig. 4B). In parallel to 5hmC conversion, repression of the de novo (Dnmt3a/b) and maintenance (Uhrf1) DNA methylation systems in PGCs prevents cyclical re-methylation and simultaneously renders PGCs permissive for direct passive 5mC depletion (Fig. S23) (18), which may contribute to the partial demethylation observed in Tet1/Tet2 knockdown PGCLCs. Thus, while in zygotes 5mC reprogramming is mechanistically compartmentalised into TET3-mediated 5hmC conversion of the paternal genome, and direct passive 5mC depletion on the maternal genome (12, 19-21), both of these mechanisms operate together in PGCs (Fig. 4B). In addition, upregulation of the base excision repair (BER) pathway in PGCs may both protect against cumulative genetic damage, and act as an auxiliary active demethylation mechanism, perhaps for specific loci (22, 23). Reprogramming in PGCs therefore involves multiple redundant mechanisms to reset the epigenome for totipotency, which accounts for the apparent fertility (albeit subfertile) of mice lacking individual components, such as Tet1 (24). The existence of multiple mechanisms may also underpin the comprehensive nature of DNA demethylation in PGCs (3). Nonetheless, some rare single-copy sites of CpG methylation escape from 5mC erasure, which may provide mechanistic avenues for investigations into transgenerational epigenetic inheritance.
Supplementary Material
Acknowledgments
We thank Nigel Miller for FACS analysis, Fuchou Tang and Walfred Tang for experimental support and Guo-Liang Xu for reagents. This work was funded by the Wellcome Trust (RG49135, RG44593 & 083563). Sequencing data has been deposited in SRA (SRA60914).
References and Notes
- 1.Surani MA, Hayashi K, Hajkova P. Cell. 2007;128:747. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 2.Hajkova P, et al. Nature. 2008;452:877. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hackett JA, Zylicz JJ, Surani MA. Trends Genet. 2012;28:164. doi: 10.1016/j.tig.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Tahiliani M, et al. Science. 2009;324:930. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito S, et al. Nature. 2010;466:1129. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ficz G, et al. Nature. 2011;473:398. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 7.Ito S, et al. Science. 2011;333:1300. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maatouk DM, et al. Development. 2006;133:3411. doi: 10.1242/dev.02500. [DOI] [PubMed] [Google Scholar]
- 9.Hackett JA, et al. Development. 2012;139:3623. doi: 10.1242/dev.081661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M. Cell. 2011;146:519. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 11.Hajkova P, et al. Mechanisms of Development. 2002;117:15. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 12.Inoue A, Zhang Y. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tam PP, Snow MH. J Embryol Exp Morphol. 1981;64:133. [PubMed] [Google Scholar]
- 14.Popp C, et al. Nature. 2010;463:1101. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin C, et al. Mol Carcinog. 2010;49:54. doi: 10.1002/mc.20576. [DOI] [PubMed] [Google Scholar]
- 16.Smallwood SA, et al. Nat Genet. 2011;43:811. doi: 10.1038/ng.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borgel J, et al. Nat Genet. 2010;42:1093. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 18.Kurimoto K, et al. Genes & Development. 2008;22:1617. doi: 10.1101/gad.1649908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu TP, et al. Nature. 2011;477:606. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 20.Wossidlo M, et al. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 21.Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Proc Natl Acad Sci U S A. 2011;108:3642. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hajkova P, et al. Science. 2010;329:78. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cortellino S, et al. Cell. 2011;146:67. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dawlaty MM, et al. Cell Stem Cell. 2011;9:166. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.