Abstract
The germ cell lineage exhibits unique characteristics, which are essential towards generating totipotency. Among the distinctive events in this lineage is DNA demethylation and the erasure of parental imprints, which occur on embryonic day 11.5 (E11.5) after the primordial germ cells (PGCs) have entered into the developing gonads 1,2. Little is yet known about the mechanism involved, except that this appears to be an active process. Here we have examined the associated changes in the chromatin to gain further insights into this reprogramming event. We show that chromatin changes during this process occur in two-steps. The first changes observed in nascent PGCs at E8.5 establish a distinctive chromatin signature with some characteristics associated with pluripotency. Subsequently, at E11.5 when these PGCs are residing in the gonads, major changes occur in nuclear architecture with an extensive erasure of several histone modification marks along with exchange of histone variants. Furthermore, at this time, the histone chaperones, HIRA and NAP1, which are implicated in histone exchange, show accumulation in PGC nuclei undergoing reprogramming. We thus suggest that the mechanism of histone replacement is critical for these chromatin rearrangements to occur. The striking chromatin changes we show here are intimately linked with the process of genome-wide DNA demethylation. Based on the timing of the observed events, we propose, that if DNA demethylation entails DNA repair based mechanism, the evident histone replacement would rather than being a prerequisite, represent a repair-induced response event.
The specification of about 40 primordial germ cells (PGCs) from Blimp1 expressing PGCs precursors is accompanied by expression of stella on embryonic day 7.25 (E7.25) 3,4. PGCs then migrate into the developing gonads by E10.5. Between E11.5-E12.5 when there are approximately 2000 PGCs per gonad, PGCs exhibit a striking genome-wide DNA demethylation, including erasure of genomic imprints, which is supposedly an active process 1,2(Supplementary Figure 1). The mechanism of this DNA demethylation process is unknown but we reasoned that it might be linked with changes in chromatin and histone modifications. However, investigations of this fundamental event are technically difficult since only limited numbers of PGCs are available for analysis, and they are temporally asynchronous and difficult to culture in vitro.
We set out first to investigate the status of the chromatin in E8.5 PGCs (100 PGCs per embryo: Supplementary Figure 1), and found that the chromatin undergoes distinctive changes, including loss of dimethylation of lysine 9 of histone H3 (H3K9me2) despite the presence of G9a, a histone methyltransferase, which is responsible for this modification. PGC specification is also unaffected in G9a−/− embryos (5 K.A. and P.H. unpublished observation). The erasure of H3K9me2 in nascent PGCs could be due to competition with H3K9ac (Supplementary Figure 2), or due to down regulation of GLP, an essential regulatory component of the G9a/GLP complex 6 Early PGCs also show enhancement of trimethylation of lysine 27 of histone H3 (H3K27me3) concomitantly with Ezh2, a polycomb group enzyme. Additionally, there is enrichment of methylation of lysine 4 of histone H3 (H3K4me2 and H3K4me3), and of many histone acetylation marks, especially H3K9ac (Supplementary Figure 2), as well as of methylation of arginine 3 on histones H4 and H2A (H4/H2AR3me2s) attributed to the Blimp1/Prmt5 complex 5.
Notably, this germ cell chromatin signature is set up specifically in PGCs (not detected in the contemporary somatic cells) prior to their entry into the gonads, and is associated with the expression of pluripotency-specific genes; Sox2, Oct4, Nanog and Stella 7. Incidentally, this chromatin state is potentially crucial for the derivation of pluripotent embryonic germ cells (EG) from PGCs between E8.5-E11.5. It is against this background that subsequent events follow when PGCs enter into the developing gonads at E10.5.
The first sign of chromatin changes in PGCs that reside in gonads at E11.5 is a rapid loss of linker histone H1 (Fig. 1a), accompanied by “loosening” of the chromatin as revealed by the loss of detectable chromocenters (identified by intensively stained DAPI foci), and by the significant enlargement of nuclei (Fig. 1b, c). Notably, HP1s (α, β and γ), ATRX and M33, which are normally associated with constitutive and facultative heterochromatin also re-distribute or disappear during this time (Supplementary Figures 3a, b and data not shown). There is also concomitant loss of H3K9me3 and H3K27me3, which represent marks of constitutive and facultative heterochromatin, respectively (Fig. 2a, b and Supplementary Figure 3-7), and of the repressive H4/H2AR3me2s 5. While these chromatin changes could serve to remove heterochromatin structures and associated repressive histone modifications to make the chromatin more “permissive” for DNA demethylation, they may be indicative of more profound changes in nuclear structure. Indeed, we also found that modifications associated with the transcriptionally active chromatin, such as H3K9ac, are also lost (Supplementary Figure 7 and data not shown). Note that nuclear histones are still accessible to antibodies as the signal for H3 persists in PGCs during the reprogramming process (Supplementary Figure 6).
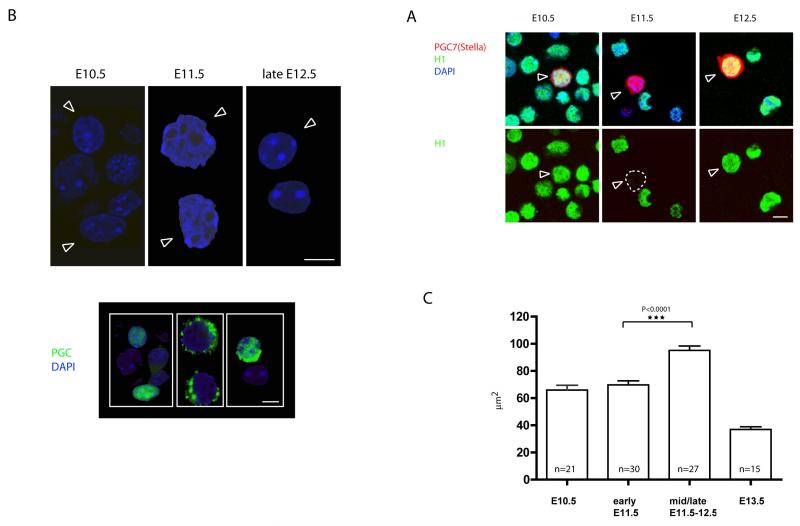
Figure 1. Following the entry of PGCs into the gonads, germ cell chromatin undergoes rapid conformational changes.
A) Dissapearance of linker histone H1 in PGCs at E11.5. PGC7/Stella (in red) was used as germ cell specific marker. PGCs are depicted by arrows. B) Increase in the nuclear size and disappearance of chromocenters in PGCs at E11.5 as observed by DAPI staining of cell suspension from genital ridges. PGCs are depicted by arrows. The lower panel shows identification of PGCs by staining with germ cell specific marker (SSEA1 or Oct4). Scale bars: 10μm. C) Measurement of the nuclear size on the cryosections from E10.5-E13.5 genital ridges shows transient change in the size of germ cell nuclei. The increase is highly statistically significant using t-test. The measured PGC nuclei were identified on the cryosections using Oct4 stainings (data not shown).
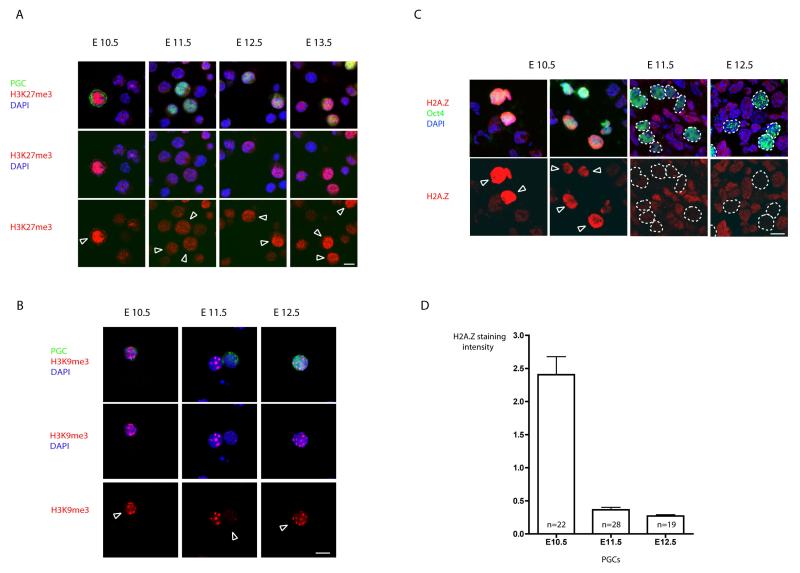
Figure 2. Chromatin changes observed in PGCs at E11.5.
A) Presence of H3K27me3 in PGCs. At E10.5 nuclei of PGCs contain high levels of H3K27me3 as part of their germline chromatin signature (for details see text). This chromatin mark is lost in PGCs at E11.5 and re-gained between E12.5 and E13.5. Notably the loss of H3K27me3 occurs despite the continuous presence of Ezh2 responsible for this modification in PGCs (data not shown). B) Loss of H3K9me3 in PGCs at E11.5. Stainings were performed on single cell suspensions from genital ridges between E10.5-E13.5. The shown epigenetic changes are detectable in 70-90% of PGCs depending on the developmental stage of the embryo. Germ cells were stained in green using the germ cell specific markers SSEA1 and Oct4, respectively, and are depicted by arrows. Scale bar: 10μm. C) The immunofluorescence stainings of cryosections of embryonic gonads (as described in Methods section). Oct4 positive germ cells (depicted by arrows) are stained in green. Note high levels of H2A.Z in early postmigratory PGCs at E10.5 and the disappearance of the H2A.Z staining in later developmental stages (E11.5 and E12.5), while the signal in surrounding somatic cells remains relatively constant. Scale bar: 10μm. D) Quantification of the H2A.Z signal in germ cells. In order to normalize between different samples, the intensity of H2A.Z staining was calculated as a mean pixel intensity in every PGC divided by the mean pixel intensity in surrounding somatic cells. The Y axis values thus represent the intensity of H2A.Z staining in PGCs in comparison with surrounding somatic cells.
Whereas the erasure of differential DNA demethylation on imprinted genes in E 11.5 PGCs 1,2 persists until new imprints are imposed later during gametogenesis 8, the chromatin decondensation and re-structuring are transient, with most PGCs re-gaining the typical appearance of bright DAPI stained chromocenters around E12.5 (Fig1b). Concomitantly, the H3K9me3 marks re-appear and the proteins associated with pericentromeric heterochromatin re-localize forming a pattern resembling that seen in the surrounding somatic cells (Fig. 2 and Supplementary Figure 3). Other chromatin changes also revert to the original state, though with diverse kinetics. For example, there is rapid re-appearance of linker histone H1, but that of H3K27me3 is slower. Notably, some histone modifications fail to re-appear altogether, including H3K9ac and H4/H2AR3me2s (Supplementary Figure 7). Hence, alongside permanent changes in DNA methylation, there are also persistent changes in chromatin modifications. The reprogramming process thus exerts the erasure of epigenetic memory at multiple and distinct levels. Importantly, none of these changes are seen in the surrounding somatic cells.
Three possible scenarios may explain the extensive loss of histone modifications in PGCs 9. First, site-specific de-modification enzymes, such as demethylases and deacetylases could remove histone tail modifications. Second, the N-terminal tails of histones may be clipped-off, and third, the existing histones could be replaced by new histones carrying different combinations of histone tail modifications; both of the latter would involve histone replacement. The concomitant disappearance of numerous histone modifications is suggestive of the involvement of histone replacement. Furthermore, while the rapid loss of H4/H2AR3me2s5 could be due to PADI4 enzymatic activity via citrulination 10,11, this enzyme acts predominantly on monomethylated arginines and is inhibited by arginine dimethylation 10. In any case, we could not detect PADI4 in our single cell cDNAs PGC libraries, or significant increase in citrulination of H4R3 (data not shown). These reasons prompted us to investigate histone replacement during this reprogramming event.
The existence of histone variants has recently been of considerable interest in the field of epigenetics. Histone variants H2A.Z and H3.3 12, as opposed to the canonical histones (H2A, H2B, H3.1, H3.2 and H4), are synthesized in a cell cycle independent manner and can be incorporated into the chromatin outside the S phase. We reasoned that if there is “large scale” histone replacement occurring, we might detect a shift in the contribution of certain histone variants in germ cell chromatin. Indeed, we detected significant levels of H2A.Z in early E10.5 PGCs, but most of the signal is lost by E11.5-E12.5, while the levels remain relatively constant in the neighbouring somatic cells (Fig. 2c,d and Supplementary Figure 8). This very substantial loss of H2A.Z occurs within 24hrs (Fig. 2c,d). Considering that the doubling time of PGCs is about 16hrs 7, active displacement of this histone variant is more likely than dilution through replication and cell division.
The lack of a reliable antibody for histone variant H3.3, which differs from the canonical H3 histones (H3.1 and H3.2) by only 5 and 4 amino acids substitutions, respectively 13, precludes immunofluorescence studies. Biochemical approaches to separate the histone variants (e.g. by AUT gels) 14, were, despite our efforts, also technically unfeasible due to the limited number and asynchronous nature of PGCs.
However, we discovered an alternative route for our investigations: FACS sorted PGCs from Oct4-GFP transgenic mice 1, revealed a change in GFP intensity at E11.5. Under appropriate conditions we could detect two major populations of E11.5 PGCs: those with lower (population A), and higher GFP signal (population B). By contrast, the GFP signal of PGCs from E10.5 and E12.5 embryos always appeared homogeneous (Fig. 3a). The two populations of E11.5 PGCs also differed in their side scatter values (SSC), suggesting changes in their intracellular physical properties (data not shown). Strikingly, population B is highly transient and is detectable only within the time window of 3-4 hours.
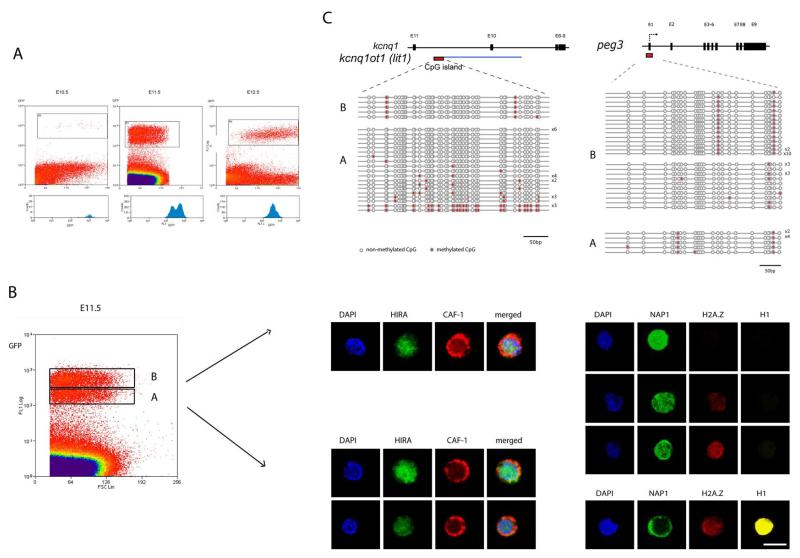
Figure 3. Separation and analysis of PGCs undergoing distinct phases of the reprogramming process.
A) Transient appearance of two populations of GFP expressing PGCs at E11.5. The X and Y axis represent the FSC (forward scatter) and GFP intensity values, respectively. The lower panels show the profiles of the GFP (germ cell specific) signal. Note the appearance of two distinct peaks at E11.5. B) Separation and analysis of chromatin configuration of the two populations of PGC, denoted as A and B, at E11.5. The population B is more advanced and shows loss of histone H1 and H2A.Z. Note the cytoplasmic localisation of CAF-1 in these populations and relocalisation of NAP1 from the cytoplasm to the nucleus in population B (for details see the text). Cells shown here are representative of the populations. Between 50 and 100 PGCs were examined for each population. Scale bar: 10μm. C) Analysis of DNA methylation of peg3 and lit1 (kcnqtot1) DMRs in dinstict populations of E11.5 PGCs using bisulphite genomic sequencing. Each line represents an independent clone. Open and closed circles represent non-methylated and methylated CpGs, respectively.
We found distinct differences between populations A and B of E11.5 PGCs, Population A stains for linker histone H1 and H2A.Z, while population B is devoid of both, which confirms that population B is at a more advanced stage (Fig. 3b). Interestingly, the loss of linker histone H1 precedes the loss of nucleosomal histone H2A.Z (Fig. 3b). Furthermore, the loss of H2A.Z in population B correlates with the loss of other modifications, such as H3K9ac, H3K9me3 and H3K27me3, as well as H2A/H4R3me2s 5. Notably, the modifications on histone H3 are lost prior to the disappearance of H2A.Z.
Our findings are supported by two additional observations: First, PGCs of both “A” and “B” populations are in the G2 phase of the cell cycle (Supplementary Figure 9), which suggests that the loss of histone modifications is not linked to replication-coupled dilution. Second, prior to the disappearance of H1 and H2A.Z, we transiently detected the signal for these histones, as well as for macroH2A and H4K5ac in the cytoplasm of some PGCs of population B (Supplementary Figures 10 and 11). This suggests that the at least some of the nucleosomal histones may be transferred to the cytoplasm, possibly for degradation.
The mobilisation of histones, and their apparent eviction and replacement in PGCs would require the participation of histone chaperones. While a complex containing Chromatin Assembly Factor 1 (CAF-1) can deposit the canonical histone H3.1 onto chromatin during DNA replication, the complex containing HIRA can deposit the non-replicative H3.3 histone variant 15. It is striking therefore that the largest subunit of CAF-1, p150, is predominantly cytoplasmic, while HIRA is enriched in the nucleus in PGCs in both A and B populations (Fig. 3b and Supplementary Figure 12a). This unusual localisation of p150 suggests that the process at work in this context is likely to involve complexes other than CAF-1.
Next we examined the histone chaperone with affinity for H1, H2A and H2B, namely Nucleosome Assembly Protein 1 (NAP1) 16,17,18,19. By forming complexes with H2A/H2B, NAP1 enhances the release of H3 and H4 during transcription 17. Moreover, NAP1 is also capable of extracting linker histones from chromatin of HeLa cells, which results in extended chromatin fibers 16. Indeed, we detected much higher levels of NAP1 in PGCs compared to the signal in neighbouring somatic cells where the protein was predominantly in the cytoplasm (Supplementary Figure 12b). Detailed analysis showed that the disappearance of H1 from PGCs correlates with the re-localisation of NAP1 from the cytoplasm to the nucleus (compare populations A and B), suggesting that NAP1 may be involved in the removal of H1 in PGCs (Fig. 4b). Since NAP1 can also interact with H2A.Z as well as direct nucleosomal disassembly in vitro 17,18,19, it is likely that NAP1 plays a significant role in the reprogramming process. Indeed, a recent study in fission yeast shows that NAP1 may be involved in nucleosomal disassembly also in vivo 20. These to our knowledge are the first indications of the potential role of NAP1 in chromatin disassembly processes in vivo.
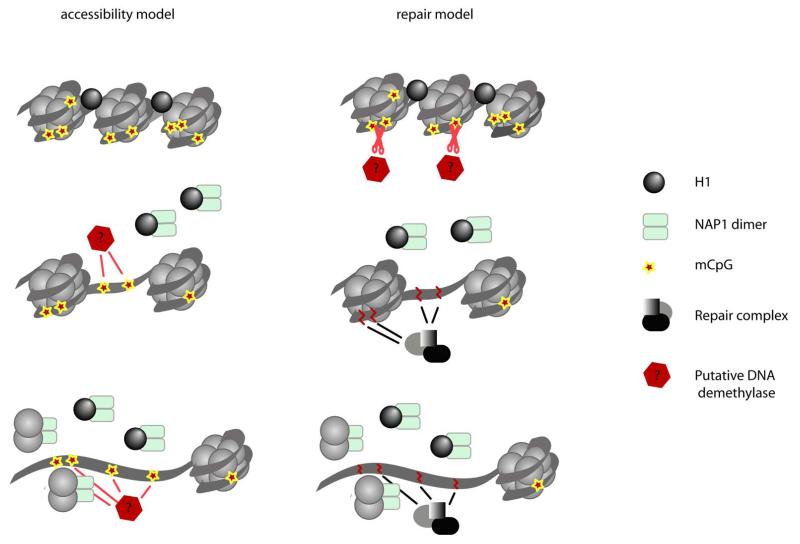
Figure 4. Possible connections between chromatin changes and DNA demethylation process.
Changes in chromatin structure may be seen as prerequisite for the DNA demethylation process (accessibility model.) Alternatively, DNA demethylation in PGCs can occur via DNA repair pathway as described in plants 24, 25, 23. The resulting DNA damage and repair could potentially induce chromatin remodelling, which would occur as a consequence of DNA demethylation (for details see the text).
We previously showed that PGCs undergo genome-wide DNA demethylation around E11.5 1. In light of this work, we chose to investigate the kinetics of DNA demethylation of Peg3 and lit1 (kcnq1ot1) loci as representatives of imprinted genes that undergo DNA demethylation between E11.5-E12.5 in PGCs. Using bisulphite analysis under optimal conditions 1,21,22, we repeatedly detected only the demethylated Peg3 and lit1 alleles in PGCs of B population, while PGCs in population A revealed the presence of some methylated alleles of lit1 but none from Peg3 locus. Surprisingly and unexpectedly, bisulphite analysis of population A proved technically challenging requiring a larger number of cells. Furthermore, the PCR products often showed signs of clonality, suggesting that only a limited number of amplifiable DNA molecules are present in the PGCs in population A. Additional analysis of global levels of 5methylcytosine (5mC) revealed variable DNA methylation within population A, while population B appeared to be devoid of 5mC (Supplementary Figure 13). Taken together, DNA demethylation occurs in population A and is followed by chromatin remodelling in population B.
Our study adds to our so far limited knowledge concerning the erasure of epigenetic modifications, which is pivotal for genomic reprogramming generally. We suggest two alternative models to explain the striking temporal and spatial relationship between DNA demethylation and chromatin changes. First, the ‘accessibility model’ predicts that decondensation of chromatin may precede DNA demethylation to allow a putative DNA demethylase access to DNA (Fig. 4a). Alternatively, DNA demethylation may occur via the DNA repair pathway, as exemplified by the methylcytosine specific DNA glycosylases in plants 23,24,25. While no similar enzymes have yet been identified in mammals, it is possible that DNA demethylation here may also be connected with DNA repair 26. Since it has been shown that DNA repair may be intimately connected with the structural chromatin changes and histone replacement 27,28 (as recently demonstrated for nucleotide excision repair and ds-break repair processes 29,30), DNA repair driven demethylation could directly induce chromatin changes and histone replacement (Fig. 4b “repair model”). Our evidence suggests that the loss of DNA methylation occurs prior to histone replacement, which supports the repair model. Notably, our evidence also shows that the erasure of epigenetic memory in PGCs occurs not only at the level of DNA methylation, but also at the level of chromatin. This broad erasure of epigenetic information in PGCs may be crucial for resetting the genome for the eventual acquisition of totipotency.
Methods Summary
PGCs were isolated by FACS sorting of GFP positive cells from OCT4-GFP transgenic mice. Immunofluorescence staining of PGCs and neighbouring somatic cells was carried out on trypsinised single cell suspensions that were fixed and processed for confocal microscopy. Alternatively, the urogenital ridges were cryosectioned and prepared for staining and confocal microscopy. The FACS-sorted PGCs (Populations A and B) were also prepared for 5 methylcytosine staining using monoclonal 5mC antibody as described in Supplementary Information. DNA methylation analysis of Peg3 and lit1 imprinted genes was carried out by bisulphite sequencing 1. All the antibodies used in this study have previously been used in other studies and verified to generate authentic signals. Further details are provided in the Supplementary Information.
Acknowledgments
The authors are very grateful to N. Miller for technical assistance and expertise with FACS sorting and to S. Jackson for stimulating discussions and critical reading of the manuscript. Additionally, we would like to thank D. Tremethick, P. Adams and T. Jenuwein for sharing their antibodies. K.A. was a recipient of a Marie Curie (PRZ/OO4/RG32856) and a Newton Trust (PRZ/008/RG38647) fellowships, F.C. was a holder of a Marie Curie Fellowship (MEIF-CT-2005-514427) and U.C.L was supported by a Wellcome Trust PhD studentship. This work was funded by grants from the Wellcome Trust to M.A.S.
References
- 1.Hajkova P, et al. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 2.Lee J, et al. Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development. 2002;129:1807–17. doi: 10.1242/dev.129.8.1807. [DOI] [PubMed] [Google Scholar]
- 3.Saitou M, et al. Specification of germ cell fate in mice. Philos Trans R Soc Lond B Biol Sci. 2003;358:1363–70. doi: 10.1098/rstb.2003.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohinata Y, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–13. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- 5.Ancelin K, et al. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8:623–30. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- 6.Seki Y, et al. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development. 2007;134:2627–38. doi: 10.1242/dev.005611. [DOI] [PubMed] [Google Scholar]
- 7.Surani MA, et al. Mechanism of mouse germ cell specification: a genetic program regulating epigenetic reprogramming. Cold Spring Harb Symp Quant Biol. 2004;69:1–9. doi: 10.1101/sqb.2004.69.1. [DOI] [PubMed] [Google Scholar]
- 8.Allegrucci C, Thurston A, Lucas E, Young L. Epigenetics and the germline. Reproduction. 2005;129:137–49. doi: 10.1530/rep.1.00360. [DOI] [PubMed] [Google Scholar]
- 9.Bannister AJ, Schneider R, Kouzarides T. Histone methylation: dynamic or static? Cell. 2002;109:801–6. doi: 10.1016/s0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- 10.Cuthbert GL, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–53. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–83. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 12.Henikoff S, Furuyama T, Ahmad K. Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet. 2004;20:320–6. doi: 10.1016/j.tig.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Kaufman PD, Almouzni G. In: DNA Replication and Human Disease. DePamphilis ML, editor. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New york: 2006. pp. 121–40. [Google Scholar]
- 14.Zweidler A. Resolution of histones by polyacrylamide gel electrophoresis in presence of nonionic detergents. Methods Cell Biol. 1978;17:223–33. [PubMed] [Google Scholar]
- 15.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 16.Kepert JF, Mazurkiewicz J, Heuvelman GL, Toth KF, Rippe K. NAP1 modulates binding of linker histone H1 to chromatin and induces an extended chromatin fiber conformation. J Biol Chem. 2005;280:34063–72. doi: 10.1074/jbc.M507322200. [DOI] [PubMed] [Google Scholar]
- 17.Levchenko V, Jackson V. Histone release during transcription: NAP1 forms a complex with H2A and H2B and facilitates a topologically dependent release of H3 and H4 from the nucleosome. Biochemistry. 2004;43:2359–72. doi: 10.1021/bi035737q. [DOI] [PubMed] [Google Scholar]
- 18.Park YJ, Chodaparambil JV, Bao Y, McBryant SJ, Luger K. Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J Biol Chem. 2005;280:1817–25. doi: 10.1074/jbc.M411347200. [DOI] [PubMed] [Google Scholar]
- 19.Lorch Y, Maier-Davis B, Kornberg RD. Chromatin remodeling by nucleosome disassembly in vitro. Proc Natl Acad Sci U S A. 2006;103:3090–3. doi: 10.1073/pnas.0511050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walfridsson J, Khorosjutina O, Matikainen P, Gustafsson CM, Ekwall K. A genome-wide role for CHD remodelling factors and Nap1 in nucleosome disassembly. Embo J. 2007;26:2868–79. doi: 10.1038/sj.emboj.7601728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durcova-Hills G, et al. Influence of sex chromosome constitution on the genomic imprinting of germ cells. Proc Natl Acad Sci U S A. 2006;103:11184–8. doi: 10.1073/pnas.0602621103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulsen M, et al. Sequence conservation and variability of imprinting in the Beckwith-Wiedemann syndrome gene cluster in human and mouse. Hum Mol Genet. 2000;9:1829–41. doi: 10.1093/hmg/9.12.1829. [DOI] [PubMed] [Google Scholar]
- 23.Choi Y, et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 24.Gehring M, et al. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong Z, et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–14. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 26.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14(1):R47–58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 27.Green CM, Almouzni G. When repair meets chromatin. First in series on chromatin dynamics. EMBO Rep. 2002;3:28–33. doi: 10.1093/embo-reports/kvf005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nilsen H, Lindahl T, Verreault A. DNA base excision repair of uracil residues in reconstituted nucleosome core particles. Embo J. 2002;21:5943–52. doi: 10.1093/emboj/cdf581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polo SE, Roche D, Almouzni G. New histone incorporation marks sites of UV repair in human cells. Cell. 2006;127:481–93. doi: 10.1016/j.cell.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 30.Linger J, Tyler JK. The yeast histone chaperone chromatin assembly factor 1 protects against double-strand DNA-damaging agents. Genetics. 2005;171:1513–22. doi: 10.1534/genetics.105.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]