Abstract
Synthetic and natural peptide assemblies can possess transport or conductance activity across biomembranes through the formation of nanopores. The fundamental mechanisms of membrane insertion necessary for antimicrobial or synthetic pore formation are poorly understood. We observe a lipid-assisted mechanism for passive insertion into a model membrane from molecular dynamics simulations. The assembly used in the study, a generic nanotube functionalized with hydrophilic termini, is assisted in crossing the membrane core by transleaflet lipid flips. Lipid tails occlude a purely hydrophobic nanotube. The observed insertion mechanism requirements for hydrophobic-hydrophilic matching have implications for the design of synthetic channels and antibiotics.
The interaction between biological membranes and synthetic or natural macromolecules that have activity through the formation of transmembrane nanopores is intrinsic to the functioning of ion channels (1–8) and antimicrobial peptides (6, 9). It is surprising, therefore, that the insertion mechanism of amphiphilic molecules into (and even across) biomembranes is poorly understood (10–13). The use of fully atomistic computer simulations to probe the interactions of membrane proteins with lipid bilayers is of considerable current interest (8). However, despite many recent successes, such simulations are hampered by the accessible system size and timescale. To glean some insight into possible mechanisms associated with the membrane insertion process we have used molecular dynamics (MD) calculations to study the interaction of tubular macromolecules with a fully hydrated dimyristoylphosphatidylcholine (DMPC) lipid bilayer in its physiologically relevant liquid-crystalline phase. The calculations we report have been carried out by using a coarse-grain (CG) model, inspired by the model successfully used to study self-organizing surfactant systems (14). The tubular nanostructures we study are generic models for preassembled bundles of membrane-spanning proteins (3, 11), antimicrobial peptides (4), and cyclic peptides (1) and for a synthetic nanosyringe based on functionalizing a carbon nanotube (2, 15). Although many naturally existing ion channels and antimicrobial peptides form pores with a hydrophilic lumen, many systems exist, including viral ion channels, synthetic peptides, and nanodevices, for which the present model should be an adequate description.
The present simulations involve molecular systems that represent ≈70,000 atoms altogether. Simulations with larger systems have been reported (8, 16). The present CG approach, however, allows us to routinely calculate dynamics in the hundreds or thousands of nanoseconds and therefore allows us to study events that take place at longer scales than those currently accessible by all-atom calculations.
Methods
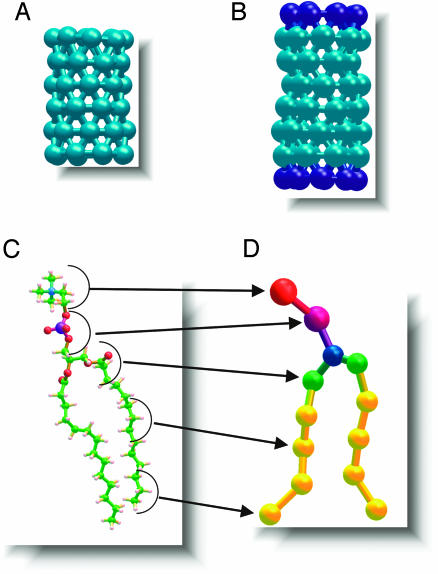
CG Model. In the CG model (17, 18), each DMPC lipid molecule consists of 13 interaction sites, eight of which are hydrophobic (four for each alkanoyl tail) and five of which are hydrophilic, three for the glycerol moiety and one each for the charged choline and phosphate (see Fig. 2). Groups of three water molecules are considered as a single unit (W) with no net charge. We have shown elsewhere that, with suitable parameterization, the CG-DMPC and W particles self-assemble spontaneously to form a stable hydrated bilayer, corresponding to the liquid crystalline Lα phase, in quantitative agreement with experimental data (18). The length of the purely hydrophobic cylindrical macromolecule (NT) was chosen to match the width of the hydrophobic core of a CG-DMPC membrane (20 Å) (18). The diameter (13 Å) was chosen to allow passage of CG W particles. In total the tube has 48 hydrophobic interaction sites uniformly distributed on its surface in a triangular lattice, i.e., six evenly separated rings with eight sites each (Fig. 1A). The hydrophobic macromolecule with hydrophilic caps (NT*) contains an additional ring of eight hydrophilic units at each end for an overall length of 30 Å (Fig. 1B). The macromolecular hydrophobic and hydrophilic sites share the same parameters as those determined for the alkanoyl tails of CG-DMPC and W, respectively (17).
Fig. 2.
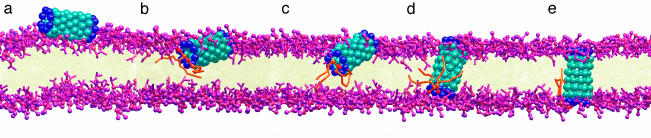
The NT* molecule spontaneously adsorbs onto the membrane (a). Partial immersion subsequently takes place with the NT* long axis remaining essentially parallel to the membrane plane (b). During this process a few lipids form salt bridges with the hydrophilic termini of the tube. Next, random thermal fluctuations drive one end of the NT* toward the core of the membrane (c). During this latter process two lipids remain attached to the immersed hydrophilic end of NT*. These lipids occlude the tube terminus and prevent lipid tails from entering the interior of the nanotube. After this metastable structure passes through the core, the flanking lipids and the hydrophilic terminus interact with the opposite side of the membrane (d). At this point, the long axis of the nanotube aligns perpendicular to the bilayer plane. The escort lipids detach from the hydrophilic terminus to join the bulk lipids in the far leaflet, thereby allowing the formation of a transmembrane pore, which can now conduct water (e). (Color key: turquoise, hydrophobic sites; dark blue, hydrophilic sites; pink, head groups; orange, chaperone lipids.)
Fig. 1.
Stick-and-ball renditions of the hydrophobic (turquoise) nanotube composed of hydrophobic sites (A), the nanotube functionalized with hydrophilic caps (blue) at its termini (B), and a schematic representation showing the all-atom DMPC molecule (C) and its CG counterpart (D). The uncapped tube measures 20 Å in length, whereas the capped tube measures 30 Å in length. Both tubes measure 13 Å in diameter.
MD Simulations. All simulations reported herein use the cm3d program (www.cmm.upenn.edu/∼moore) and the npt ensemble. All the simulations were carried out at 303.15 K and 1 atm as has been described (19). The van der Waals interactions were truncated at 15 Å, whereas the real part of the electrostatic interactions was truncated at 18.7 Å, and a multiple time-step integrator scheme was used to optimize the simulations. The molecular species in each simulation were distributed as follows: The simulation of a NT* above a lipid bilayer was composed of the tube, 256 CG DMPC lipids, and 4,263 W sites (representing 12,789 water molecules). The simulations of NT and NT* embedded in a bilayer consisted of 256 CG DMPC lipids, 2,192 W sites (representing 6,576 water molecules), and a single NT* or NT as necessary. All other simulations mentioned in the article are geometrical variations of these basic simulations. The interaction potential parameters for the CG model were fit iteratively to the results of all-atom MD simulations on corresponding systems. From our previous all-atom and CG simulation results (18) we concluded that the CG systems evolve several orders of magnitude faster than their all-atom counterparts, thereby providing access to physical phenomena on larger spatial and temporal scales (18) (see Movies 1–3, which are computer animations of the insertion process published as supporting information on the PNAS web site). The longest simulation in the present work, that of NT* penetrating the lipid bilayer as shown in Fig. 2, was run for 180 ns. The remaining simulations were all run for ≈40 ns.
Results and Discussion
Insertion Mechanism. Our MD simulations demonstrate that a generic hydrophobic nanotube (NT, Fig. 1), with hydrophilic functionality at its termini (NT*, Fig. 1), spontaneously inserts into, aligns, and conducts molecules across a lipid bilayer (Fig. 2). Pore formation occurs in a two-stage process whereby the nanotube is first adsorbed onto and accommodated into the membrane surface with its long axis parallel to the lipid–water interface. Subsequently, the functionalized nanotube spontaneously reorients to become a transmembrane pore. Reorientation, which involves transport of one of the hydrophilic ends across the hydrophobic core of the membrane, is facilitated by chaperone lipids whose head groups form salt bridges to the tube caps. These chaperone lipids detach from the nanotube once reorientation is complete and become incorporated into the far membrane leaflet. Although the insertion process is difficult to study experimentally, it has been shown that transmembrane peptides can induce lipid flips (20). The CG model for the tubes and lipid molecules (17, 18) is shown in Fig. 1.
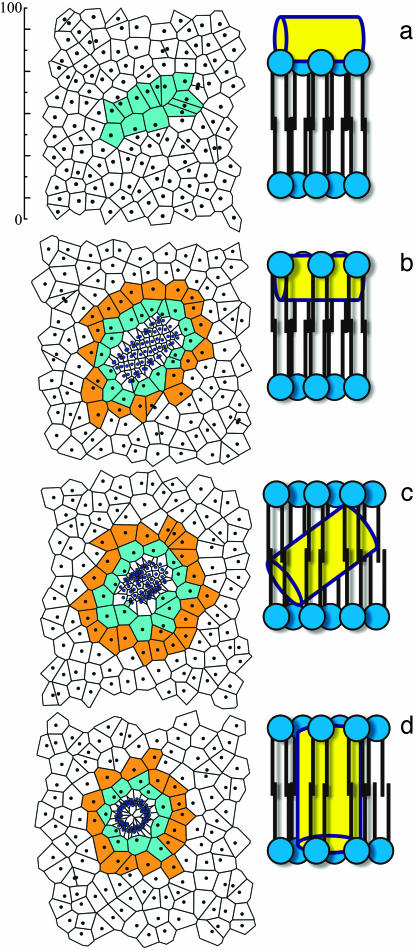
Lipid Behavior. A direct relation exists between the surface tension of a leaflet and the area per head group of its constituent lipids, which is typically expressed by surface pressure/area per lipid isotherms (21). We use 2D tessellations with Voronoi polyhedra (22) to study local instantaneous changes in the area per head group of the lipids in the vicinity of the NT* tube. Fig. 3a shows the centers of mass of the lipid molecules as the tube begins to penetrate the membrane. During the insertion process, the area per head group of the CG lipids in the neighborhood of NT* (Fig. 3 Left, cyan) decreases from its equilibrium value (19) of ≈70 Å2:49 Å2 during immersion (Fig. 3b), 54 Å2 after the tube has submerged into the membrane (Fig. 3c), 60 Å2 as the tube rotates, and finally 56 Å2 on completion of the rotation (Fig. 3d). The lipids in the second shell follow a similar, albeit less drastic, trend in their area per head group changes (within 5 Å2 of their equilibrium value). Thus, only the lipids in the first shell around the tube display a significant response to the stress induced by tube immersion. The lower area per head group value of those lipids immediately surrounding the tube is attributed, at least in part, to strongly attractive membrane–tube interactions.
Fig. 3.
(Left) Voronoi tessellations for the centers of mass of each lipid and the NT* tube sites, as the tube penetrates the model membrane. (Right) The corresponding cartoon rendition is shown. (a) At 0 ns the tube is adsorbed onto the surface, but penetration has not started. (b) At 20 ns the tube has penetrated laterally onto the bilayer, and two shells of different areas per head group surround the tube. (c) At 60 ns the tube is halfway through its rotation, and the area of the lipid atoms in contact with the tube changes slowly. (d) At 80 ns the tube has completely inserted into the membrane. (Left, color key: black dots, lipids center of mass; blue dots, tube sites; cyan, lipids in contact with the tube; and orange, lipids in the second shell. Right, color key: yellow, tube; blue, lipids.) The scale depicted on the upper left is in angstroms.
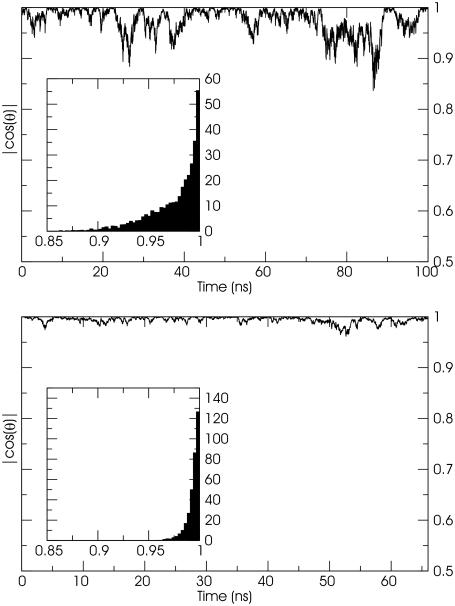
Tube Behavior and Water Transport. To further characterize the interaction of cylindrical macromolecules with a biomembrane, MD simulations were performed with NT and NT* molecules initially embedded in the DMPC bilayer, each with their long axes perpendicular to the membrane surface. Water is observed to populate the pore during the initial stages of the NT simulation, but, subsequently, the tube tilts and lipid tails enter the tube ends from both leaflets and block the transport of water. The problem of the hydrophobic lipid tails occluding the lumen of the NT pore is alleviated in the embedded NT*. In this case, the tilting of the tube is reduced by the presence of hydrophilic termini that can make form-specific interactions with the lipid head groups. In our simulations the long axis of each tube remains approximately perpendicular to the membrane plane (Fig. 4), but the tilting oscillations of the NT are large enough for lipid tails to enter and occlude the pore as discussed above (Fig. 4 Upper).
Fig. 4.
The NT and NT* tube are approximately perpendicular to the bilayer plane throughout the simulation as discussed in the text. The oscillations in the positioning of the NT tube (Upper), however, allow for lipid tails to penetrate the lumen of the tube and occlude the pore. Simulations with longer purely hydrophobic tubes resulted in tubes with larger tilt angles and pore occlusion. The oscillations are reduced by the addition of hydrophilic caps to the NT* tube (Lower). This tube can conduct water sites across the membrane. The embedded graphs represent normalized histograms of the tilt angles collected from the simulations.
In a separate MD simulation, a NT* tube initially perpendicular to the bilayer plane rotates, becoming parallel to the membrane before its immersion, and then follows the same insertion mechanism as described in Fig. 1. MD simulations starting with a purely hydrophobic NT placed in the water subphase results in a slow insertion process and a tube that becomes solvated and occluded by membrane lipid tails.
It is essential to select suitable matching lengths for the hydrophobic and hydrophilic nanotube subdomains to anchor the nanopore in the membrane (23). Shorter tubes result in oscillations and occlusion of the pore because of lipid tail insertions. Longer tubes will tilt to maximize the contact of the hydrophobic portion of the tube with the hydrophobic membrane core. The hydrophilic functionality at either tube terminus also enhances the accessibility of water to the channel and, as discussed above, is vital for the spontaneous transport of the tube end across the membrane core. Although the NT* molecule conducts CG water throughout the simulation without occlusion by the lipids, such events are too scarce in our simulation for meaningful statistics to be acquired about this process. However, because the CG water units used in this study represent three loosely packed water molecules, a fully atomistic simulation of the present system should accommodate a continuous water column or at least a wire across the pore as has been shown (8, 11).
Note that it is feasible to perform a simulation with atomistic detail of the present system. It would be difficult, however, to access the necessary scales for insertion without using biasing forces or other approaches to accelerate the insertion of the nanotube into the membrane. The results suggest a mechanism of insertion which should be now probed with experimental approaches and simulations including more detail.
Conclusion
We conclude that the insertion of a functionalized hydrophobic macromolecule into a biomembrane can occur in a spontaneous fashion. The mechanism involves a two-step process where the molecule is first accommodated onto the membrane surface and then reorients to adopt a transmembrane configuration. The observed transport of a hydrophilic tube end across the membrane is assisted by chaperone lipids which undergo transbilayer flips. By analogy, we postulate that positively charged residues in antibiotic molecules adhere to negatively charged lipids, which subsequently chaperone the transmembrane insertion process and could perhaps also play a role in peptide oligomerization. This implies that transport of hydrophilic groups across a membrane can be possible when the appropriate lipid–molecule interaction can be established. Hydrophilic functionality at the termini of a tubular macromolecule and close matching of its subdomains with the hydrophobic and hydrophilic domains of a biomembrane seem to be necessary and sufficient requirements for the transport of molecules across the membrane without occlusion of the pore.
Supplementary Material
Acknowledgments
We thank Dr. Srinivas Goundla for suggestions that enhanced this article. This work was supported by generous grants from the National Institutes of Health (Grant GM40712), the National Science Foundation, and the Natural Sciences and Engineering Research Council of Canada.
Abbreviations: NT, hydrophobic cylindrical macromolecule; NT*, hydrophobic macromolecule with hydrophilic caps; MD, molecular dynamics; DMPC, dimyristoylphosphatidylcholine; CG, coarse-grain.
See Commentary on page 4337.
References
- 1.Fernandez-Lopez, S., Kim, H. S., Choi, E. C., Delgado, M., Granja, J. R., Khasanov, A., Kraehenbuehl, K., Long, G., Weinberger, D. A., Wilcoxen, K. M., et. al. (2001) Nature 412, 452–455. [DOI] [PubMed] [Google Scholar]
- 2.Hummer, G., Rasaiah, J. C. & Noworyta, J. P. (2001) Nature 414, 188–190. [DOI] [PubMed] [Google Scholar]
- 3.Tew, G. N., Liu, D., Chen, B., Doerksen, R. J., Kaplan, J., Carroll, P. J., Klein, M. L. & DeGrado, W. F. (2002) Proc. Natl. Acad. Sci. USA 99, 5110–5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang, H. W. (2000) Biochemistry 39, 8347–8352. [DOI] [PubMed] [Google Scholar]
- 5.Wang, H. & Branton, D. (2001) Nat. Biotechnol. 19, 622–623. [DOI] [PubMed] [Google Scholar]
- 6.Yang, L., Harroun, T. A., Weiss, T. M., Ding, L. & Huang, H. W. (2001) Biophys. J. 81, 1475–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dieckmann, G. R., Lear, J. D., Zhong, Q. F., Klein, M. L., DeGrado, W. F. & Sharp, K. A. (1999) Biophys. J. 76, 618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tajkhorshid, E., Nollert, P., Jensen, M. O., Miercke, L. J. W., O'Connell, J., Stroud, R. M. & Schulten, K. (2002) Science 296, 525–530. [DOI] [PubMed] [Google Scholar]
- 9.Zasloff, M. (2002) Nature 415, 389–395. [DOI] [PubMed] [Google Scholar]
- 10.Wimley, W. C. & White, S. H. (2000) Biochemistry 39, 4432–4442. [DOI] [PubMed] [Google Scholar]
- 11.Beckstein, O., Biggin, P. C. & Sansom, M. S. P. (2001) J. Phys. Chem. B 105, 12902–12905. [Google Scholar]
- 12.Epand, R. F., Umezawa, N., Porter, E. A., Gellman, S. H. & Epand, R. M. (2003) Eur. J. Biochem. 270, 1240–1248. [DOI] [PubMed] [Google Scholar]
- 13.Schleiff, E. & Klosgen, R. B. (2001) Biochim. Biophys. Acta 1541, 22–33. [DOI] [PubMed] [Google Scholar]
- 14.Karaborni, S., Esselink, K., Hilbers, P. A. J., Smit, B., Karthauser, J., Vanos, N. M. & Zana, R. (1994) Science 266, 254–256. [DOI] [PubMed] [Google Scholar]
- 15.Berezhkovskii, A. & Hummer, G. (2002) Phys. Rev. Lett. 89, 64503-1–64503-4. [DOI] [PubMed] [Google Scholar]
- 16.Jensen, M. Ø., Tajkhorshid, E. & Schulten, K. (2001) Structure (London) 9, 1083–1093. [DOI] [PubMed] [Google Scholar]
- 17.Shelley, J. C., Shelley, M. Y., Reeder, R. C., Bandyopadhyay, S. & Klein, M. L. (2001) J. Phys. Chem. B 105, 4464–4470. [Google Scholar]
- 18.Lopez, C. F., Moore, P. B., Shelley, J. C., Shelley, M. Y. & Klein, M. L. (2002) Comput. Phys. Commun. 147, 1–6. [Google Scholar]
- 19.Lopez, C. F., Nielsen, S. O., Moore, P. B., Shelley, J. C. & Klein, M. L. (2002) J. Phys. Condens. Matter 14, 9431–9444. [Google Scholar]
- 20.Kol, M. A., van Laak, A. N. C., Rijkers, D. T. S., Killian, J. A., de Kroon, A. I. P. M. & de Kruijff, B. (2003) Biochemistry 42, 231–237. [DOI] [PubMed] [Google Scholar]
- 21.Ybert, C., Lu, W., Moller, G. & Knobler, C. M. (2002) J. Phys. Chem. B 106, 2004–2008. [Google Scholar]
- 22.Okabe, A., Boots, B. & Sugihara, K. (1992) Spatial Tessellations: Concepts and Applications of Voronoi Diagrams (Wiley, New York).
- 23.Ridder, A. N. J. A., van de Hoef, W., Stam, J., Kuhn, A., de Kruijff, B. & Killian, J. A. (2002) Biochemistry 41, 4946–4952. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.