Abstract
Potassium channels are widely distributed. To serve their physiological functions, such as neuronal signaling, control of insulin release, and regulation of heart rate and blood flow, it is essential that K+ channels allow K+ but not the smaller and more abundant Na+ ions to go through. The narrowest part of the channel pore, the selectivity filter formed by backbone carbonyls of the GYG-containing K+ channel signature sequence, approximates the hydration shell of K+ ions. However, the K+ channel signature sequence is not sufficient for K+ selectivity. To identify structural elements important for K+ selectivity, we randomly mutagenized the G protein-coupled inwardly rectifying potassium channel 3.2 (GIRK2) bearing the S177W mutation on the second transmembrane segment. This mutation confers constitutive channel activity but abolishes K+ selectivity and hence the channel's ability to complement the K+ transport deficiency of Δtrk1Δtrk2 mutant yeast. S177W-containing GIRK2 mutants that support yeast growth in low-K+ medium contain multiple suppressors, each partially restoring K+ selectivity to S177W-containing double mutants. These suppressors include mutations in the first transmembrane segment and the pore helix, likely exerting long-range actions to restore K+ selectivity, as well as a mutation of a second transmembrane segment residue facing the cytoplasmic half of the pore, below the selectivity filter. Some of these suppressors also affected channel gating (channel open time and opening frequency determined in single-channel analyses), revealing intriguing interplay between ion permeation and channel gating.
An essential feature of all K+ channels is their ability to discriminate between K+ and Na+, the most abundant alkali metal ions in nature. Experimental studies and theoretical analysis thus far focus on the role of the selectivity filter formed by the K+-channel signature sequence (TXGYG) (1–6). The K+-channel signature sequence, however, is not sufficient for K+ selectivity, because K+ selectivity may be abolished by mutations outside the K+-channel signature sequence (7), and the hyperpolarization-activated cation channels responsible for the pacemaker current Ih have a K+-channel signature sequence (CIGYG) but show little discrimination between Na+ and K+ ions (8). It thus remains unclear why some channels bearing a K+-channel signature sequence are selective for K+ whereas others are not.
Inwardly rectifying potassium (Kir) channels exhibit the same transmembrane helix packing as KirBac1.1 (PDB ID code 1P7B), which contains four membrane-associated helices: the pre-M1 (first transmembrane segment) helix (slide helix), the M1 helix (outer helix), followed by the pore helix and the second transmembrane segment (M2) helix (inner helix) (9–11). Below the selectivity filter, several M2 residues line the pore and may interact with magnesium and polyamines that enter the pore from the cytoplasmic side and cause inward rectification by blocking outward K+ flow (12–17).
In this study, we carried out second-site suppressor experiments in yeast to examine which channel features other than the K+-channel signature sequence are important for K+ selectivity by relying on the growth advantage conferred by K+ selectivity caused by the ubiquitous requirement for high K+ levels inside the cell.
Methods
Random Library Construction. G protein-coupled Kir 3.2(GIRK2) (18) was cloned into the HindIII–XhoI sites of a derivative of pYES2 vector harboring the Met-25 promoter (9). GIRK2 mutant library was generated by means of DNA shuffling (19) with some modifications (7). Approximately 120,000 independent clones were screened and sequenced, revealing an error rate of 0.57%. In vitro backcrossing was performed by means of DNA shuffling with PCR performed at high stringency (Pfu Turbo DNA polymerase, Stratagene) to achieve an error rate of 0.2%.
Yeast Selection. The yeast strain SGY1528 was transformed with randomly mutagenized S177W GIRK2 by using lithium acetate. After ≈72 h of growth on plates with 100 mM potassium at 30°C, colonies replica-plated to 2 mM potassium were replica-plated ≈48 h later onto plates with 0.1 mM potassium. After yeast selection, plasmids were isolated, sequenced, and transformed again into fresh yeast to confirm growth rescue.
Site-Directed Mutagenesis. Site-directed mutagenesis by means of PCR of the PYES2 plasmid was done by using the high-fidelity Pfu Turbo DNA polymerase (Stratagene) and verified by sequencing the entire cDNA. GIRK2 segments containing the mutations then were subcloned into HindIII–XhoI sites of pGEM vector (20).
Western Blotting. Channel proteins expressed in yeast were extracted by cracking the membrane with glass beads in 10 mM Tris·HCl, pH 7.4/1 mM EDTA/1% SDS/0.1% 2-mercaptoethanol/1 mM PMSF/1 mM aprotinin/1 mM pepstatin A. After centrifugation at 20,800 × g for 10 min at 4°C, the supernatant was combined with a loading buffer, incubated at 50°C for 30 min, loaded on 10% polyacrylamide gel, blotted, and probed with GIRK2 antibody (Upstate Biotechnology, Lake Placid, NY) and horseradish peroxidase-coupled goat anti-rabbit antibody (Jackson ImmunoResearch).
Oocyte Expression. Capped cRNAs were generated from pGEM constructs linearized at the AflII restriction site by using Amplicap T7 High-Yield Message Maker kit (Epicentre Technologies, Madison, WI). Stage V–VI oocytes were injected with 2–5 ng of cRNA and maintained for 3–6 days at 16°C in ND96 solution (96 mM NaCl/2 mM KCl/1 mM MgCl2/5 mM Hepes, pH 7.4) or analogous solutions in which Na+ was replaced with K+ or N-methyl-d-glucamine (NMDG).
Two-Electrode Voltage Clamp. Macroscopic currents were recorded from oocytes (GeneClamp 500B, Axon Instruments, Foster City, CA; electrode resistance, 0.1–0.6 MΩ) in 90 mM XCl (X = K, Na, NMDG)/2 mM MgCl2/10 mM Hepes (pH 7.4 with XOH, or HCl for NMDG) in a small chamber (Warner Instruments, Hamden, CT) with fast perfusion system (ValveLink 16, AutoMate Scientific, San Francisco) at 22–25°C.
The relative permeability ratios were determined (21) by using the Goldman–Hodgkin–Katz equation: ΔErev = Erev,B – Erev,A = (RT/zF)ln(PB[B]o/PA[A]o). Reversal-potential (Erev) measurements were corrected for junction potential (4 mV). Only oocytes that expressed currents showing inward rectification were used for analysis.
Patch-Clamp Recordings. Single-channel currents were recorded at room temperature from cell-attached patch, clamped at –100 mV, of oocytes 3–6 days after cRNA injection (7) with an Axopatch 200A patch-clamp amplifier (Axon Instruments) and low-pass-filtered (3 dB, 1 kHz) with an 8-pole Bessel filter (Frequency Devices, Haverhill, MA). Single-channel data, digitized at 20 kHz on-line by using clampex 8 software (Axon Instruments) with a 16-bit analog/digital converter (Digidata acquisition board 1200A; Axon Instruments), were detected by using fetchan 6.06 (events list) of pclamp (Axon Instruments) with 50% threshold crossing criterion and analyzed with interval5 (Barry S. Pallotta, University of North Carolina, Chapel Hill). Only patches with infrequent multiple-channel activity were used for analysis, performed at the main conductance level (≈27–32 pS), for duration histograms (22) and fits with exponential functions (23, 24). Mean durations were corrected for missed events by taking the sum of the relative area of each exponential component in the duration frequency histogram multiplied by the time constant of the corresponding component.
Results
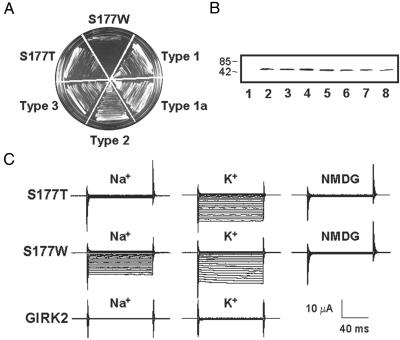
K+ Selectivity Is Essential for Rescue of K+ Transport-Deficient Yeast. Whereas the constitutively active S177T mutant GIRK2 channels support yeast growth (7), S177W channels failed to rescue yeast (Fig. 1A) despite robust protein expression in yeast (Fig. 1B) and constitutive channel activity, presumably because of their substantial permeability to Na+ (Fig. 1C and Fig. 6, which is published as supporting information on the PNAS web site). In support of this notion, the K+-selective S177T channels supported robust yeast growth in medium containing 1 M NaCl, whereas the nonselective S177W channels did not (data not shown).
Fig. 1.
S177W mutation in M2 abolishes K+ selectivity of GIRK2. (A) Wild-type GIRK2 did not rescue yeast growth due to low probability of opening in the absence of mammalian Gβγ. Constitutively active but nonselective S177W GIRK2 failed to rescue yeast growth on a plate supplemented with a low concentration of K+ (0.5 mM KCl), whereas K+-selective S177T GIRK2 supported yeast growth, as did four mutant types each containing multiple mutations in addition to S177W. (B) Comparable channel protein-expression levels of wild type (lane 2), S177T (lane 3), S177W (lane 4), and the four mutant types (lanes 5–8) in yeast. The predicted mass of the GIRK2 monomer is 47.5 kDa. Anti-GIRK2 antibody does not recognize Kir2.1 (IRK1) channel expressed in the same yeast strain (lane 1). (C) Representative current traces obtained from Xenopus oocytes expressing wild-type GIRK2, S177T, or S177W mutant channels bathed in 90 mM K+, 90 mM Na+, or 90 mM NMDG. Currents were recorded at membrane potentials ranging from +40 to –150 mV in 10-mV increments.
To determine the channels' permeability for Na+ relative to that for K+ (PNa/PK), we measured the membrane potential at which there is no net current flow (the reversal potential, Erev) in biionic conditions with primarily Na+ ions in the extracellular solution and K+ ions in the intracellular solution (Table 3, which is published as supporting information on the PNAS web site), yielding an estimate of PNa/PK = 0.047 ± 0.002 for S177T (n = 16) and PNa/PK = 0.5 ± 0.01 for S177W (n = 31). To control for the contribution from endogenous channels, we applied the reversible channel blocker tertiapin (25) to block GIRK2 channels in high-Na+ solution and then in high-K+ solution (Fig. 7, which is published as supporting information on the PNAS web site). The corrected permeability ratio using tertiapin-sensitive GIRK2 currents from the same oocyte expressing S177T is even smaller [PNa/PK = 0.006 ± 0.0009 (n = 4)], because (unlike in high-K+ solutions) in high-Na+ solutions the endogenous currents are comparable with the negligible S177T Na+ current (Fig. 7). The tertiapin subtraction, however, did not alter the PNa/PK measurement for the nonselective S177W channels, because the mutant GIRK2 currents carried by either Na+ or K+ were at least 15-fold greater than the endogenous currents (Fig. 7). We therefore used current measurements without tertiapin subtraction to obtain estimates for PNa/PK for the large number of mutants derived from yeast screens and always expressed S177T and S177W channels in the same batch of oocytes as control. As noted above, these measurements are more likely to underestimate the K+ selectivity for channels that are highly selective for K+.
Random Mutagenesis and Yeast Screen for Suppressors. We isolated second-site suppressors by screening a library of mutagenized GIRK2 S177W for those that support yeast growth in low-K+ medium by using DNA shuffling for random mutagenesis and the in vitro backcross to reduce spurious mutations (19). Of ≈40,000 yeast colonies transformed with the GIRK2 mutant sequences after in vitro backcross, ≈200 (0.5%) grew after replica-plating onto plates containing low K+ (0.1 mM KCl), yielding four mutant types carrying (in addition to S177W) a number of mutations: type 1 = T80P N94Y F98Y M123I F147Y S177W N263S D346N; type 1a = type 1 + T136I; type 2 = F98I Y102N M123L S177W; and type 3 = M19T P26L N94I S177W N184D K219R Y336H. The number of independent clones sequenced was 19 for type 1, 1 for type 1a, 3 for type 2, and 1 for type 3.
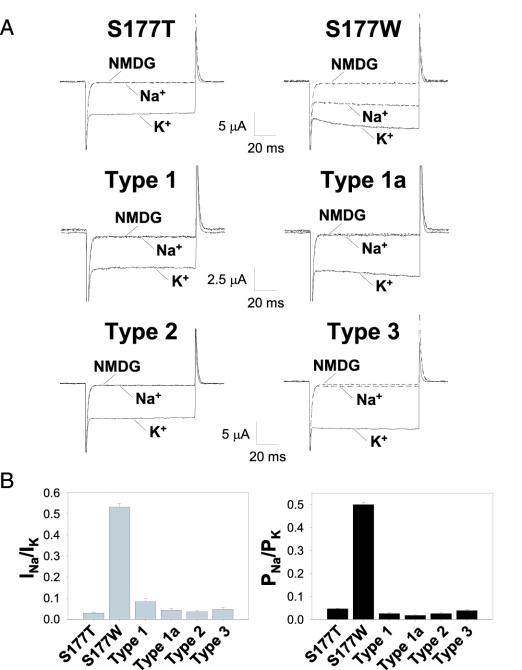
Second-Site Suppressor for K+ Selectivity. For the four mutant types that rescued yeast growth (Fig. 1 A), we examined the properties of these channels expressed in Xenopus oocytes by using two-electrode voltage clamp (TEVC). Similar to S177W, channels with suppressor mutations generated ≈6–10 times larger basal currents than wild-type GIRK2 (Fig. 2A), even though Western blots revealed similar levels of channel protein expression (Fig. 1B). In contrast to GIRK2 S177W, channels isolated from the yeast screen did not produce significant inward current in 90 mM Na bath solution (Fig. 2 A and Fig. 8, which is published as supporting information on the PNAS web site), yielding much smaller INa/IK (Fig. 2B). The estimates of PNa/PK also were much smaller than that of the S177W single mutant (Fig. 2B and Table 3). Thus, these four mutant types capable of rescuing yeast not only are constitutively active but have regained K+ selectivity.
Fig. 2.
Four mutant types recovered from yeast selection restore K+ selectivity. (A) S177W mutant channels but not S177T or the four S177W-containing mutant types sustained Na+ currents (at –100 mV). Current traces were elicited with voltage pulses from +40 to –150 mV (in 10-mV increments) from a holding potential of 0 mV in 90 mM K (solid line), 90 mM Na (long-dash line), and 90 mM NMDG (short-dash line). (B) S177W but not S177T or the S177W-containing mutant types displayed significant Na+ permeability, as indicated by current (INa/IK) and permeability (PNa/PK) ratios deduced from the difference in reversal potential (Erev) measured in 90 mM Na and 90 mM K solutions.
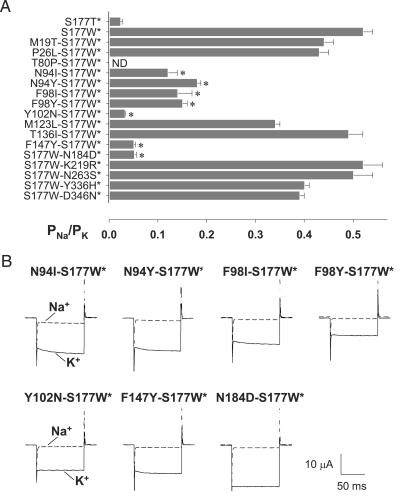
Mutations of Five Residues in the Transmembrane Domain Suppressed S177W and Partially Restored K+ Selectivity. To search systematically for second-site suppressors, we examined each of the mutations included in the four mutant types for its ability to reinstate K+ selectivity of GIRK2 S177W by combining each mutation individually with S177W along with the D228N mutation to eliminate the possibility of channel activation by cytoplasmic sodium (18, 26, 27). The inclusion of D228N did not alter their PNa/PK [0.023 ± 0.004 for S177T D228N (n = 8); 0.52 ± 0.02 for S177W D228N (n = 18); and 0.18 ± 0.008 for N94Y S177W D228N (n = 8)] (Fig. 9, which is published as supporting information on the PNAS web site). We therefore have consistently maintained the D228N mutation in the background as a safeguard and call these channels GIRK2*. Of the 16 mutations tested, 7 (N94I/Y, F98I/Y, Y102N, F147Y, and N184D) reduced PNa/PK by >2-fold, relative to GIRK2* S177W (Fig. 3A and Table 3). Of these, Y102N exerted the strongest suppression, reducing PNa/PK of GIRK2* double mutants to levels comparable with that of the K+-selective GIRK2* S177T [PNa/PK = 0.032 ± 0.002 for GIRK2* Y102N S177W (n = 11)]. All seven GIRK2* double mutants, when hyperpolarized to –100 mV, gave rise to large inward currents in high-K+ but not high-Na+ solution (Fig. 3B). These experiments thus identify seven mutations of five residues outside the P loop (Fig. 4), each capable of partially suppressing the loss of K+-selectivity phenotype of S177W.
Fig. 3.
Seven mutations of five residues in the transmembrane domain partially restore K+ selectivity of the nonselective S177W channel. (A) The PNa/PK permeability ratio of S177W-containing double mutants for each of the mutations included in the four mutant types. Seven mutations, marked with asterisks (*) in the bar graph, reduced PNa/PK by >50%. ND indicates that PNa/PK could not be determined because the current was indistinguishable from leak current. (B) Suppression of the Na+ current caused by the S177W mutation by second-site suppressors. Current traces were elicited with voltage pulses from +40 to –150 mV (in 10-mV increments) from a holding potential of 0 mV in 90 mM K (solid line) and 90 mM Na (dashed line). Only traces at –150 mV are shown. Whereas currents recorded in 90 mM K solution from “double mutants” (in the background of the D228N mutation) were comparable in amplitude, some exhibited a slower time course of current activation.
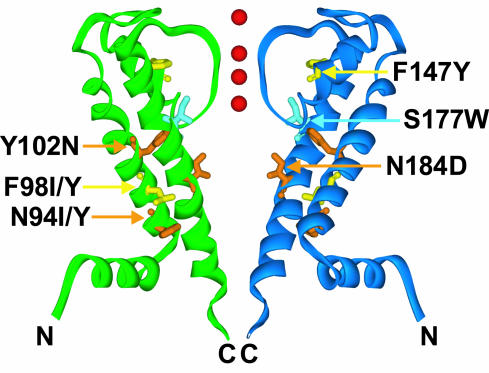
Fig. 4.
Suppressors are located on helices in the transmembrane domain of GIRK2. Suppressors are mapped onto the KirBac1.1 structure in two opposing KirBac monomers. S177W (pale blue), affecting a pore-lining residue of the inner helix M2, abolishes K+ selectivity. Suppressors N94Y/I, F98Y/I, and Y102N affect M1 residues, F147Y affects a pore–helix residue, and N184D affects a pore-lining residue in M2. F98Y/I and F147Y mutations (yellow) restored K+ selectivity to S177W-containing channels, as did N94Y/I, Y102N, and N184D mutations (orange), which also impacted channel gating. Red spheres represent K+ ions in the pore.
Allele-Specific Suppression of the Loss of K+-Selectivity Phenotype. Both the S177 residue, yielding the loss of K+-selectivity mutations, and the N94 residue, yielding suppressors, are critical for holding the channel in the closed conformation, as is the E152 residue that also yields nonselective mutants (7). Whereas the “outer pair” of S177 and E152 controls channel gating in a manner that is independent and distinct from the action of the “inner pair” of N94 and V188 (7, 28), there is remarkably long-range interaction between N94 and S177 with regard to K+-selective permeation. The specificity of such functional interactions is underscored by the allele-specific suppression by N94 mutations of the yeast growth (Table 1) and K+-selectivity phenotypes (Fig. 10, which is published as supporting information on the PNAS web site, and Table 3) induced by S177 and E152 mutations. Moreover, none of the five N94 mutations rescued growth of yeast-expressing channels carrying the G156S weaver mutation (Table 1) or enhanced K+ selectivity of G156S-containing double-mutant channels (Fig. 10 and Table 3; refs. 21 and 29–32). These double-mutant studies raise the possibility of covariance between N94 in M1 and S177 in M2 or E152 in the pore helix (but probably not G156 in the P loop) in achieving/maintaining K+ selectivity.
Table 1. Allele-specific suppression of S177 and E152 mutations but not the weaver mutation by N94 mutations.
| N94Y | N94F | N94H | N94M | N94D | |
|---|---|---|---|---|---|
| S177D | + | + | - | - | - |
| S177G | - | + | + | - | - |
| S177F | + | + | + | - | - |
| S177P | - | - | - | - | - |
| S177Y | + | + | + | - | - |
| S177N | + | + | + | - | - |
| S177H | - | - | - | - | - |
| E152T | - | + | - | - | - |
| E152V | - | - | - | - | - |
| E152A | + | + | + | - | - |
| E152H | - | - | - | - | - |
| E152G | + | + | + | - | - |
| weaver | - | - | - | - | - |
Eight substitutions at S177 in M2 (D, G, F, W, P, Y, N, and H), five substitutions at E152 in the pore helix (T, V, A, H, and G), and the G156S identified in the pore loop of the gene encoding for the GIRK2 channel of the weaver mouse were found to confer Na+ permeability to GIRK2 channels (Fig. 10 and Table 3). These mutations (rows 2-14) were combined with five substitutions of N94 in M1 (Y, F, H, M, and D, columns 2-6), and the double-mutant GIRK2 channels were tested for their ability to rescue yeast growth on 0.5 mM K+ medium.
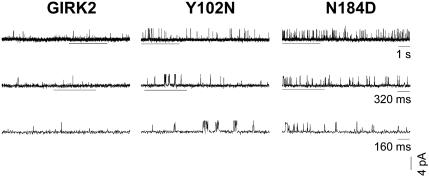
Effects of the Suppressor Mutations on Channel Gating. To probe at the coupling between permeation and gating further, we asked whether suppressors other than mutations of N94 also affected gating. Of the seven double mutants tested, only N94Y S177W, Y102N S177W, and N184D S177W rescued yeast (Table 3), indicating that Y102N and N184D also may have increased constitutive activity of the double mutant. To examine the gating effects of Y102N and N184D, we carried out single-channel analyses of cell-attached patch recordings from Xenopus oocytes. Indeed, the mean open time more than doubled in Y102N and N184D single mutants (Table 2). The N184D mutation also significantly reduced the mean closed time, causing a significant increase of both the opening frequency and the opening probability (Fig. 5 and Table 2). As control, we also examined the single-channel properties of the F147Y single mutant and found no significant effects of this mutation on single-channel properties (Table 2). Thus, three of the five residues yielding suppressor mutations on K+ selectivity seem to be involved in channel gating.
Table 2. Single-channel properties of GIRK2 wild-type and mutant channels.
| GIRK2 | Y102N | F147Y | N184D | |
|---|---|---|---|---|
| Channel opening probability, % | 0.34 ± 0.13 | 1.08 ± 0.39 | 0.33 ± 0.09 | 1.56 ± 0.47* |
| Opening frequency, s-1 | 3.68 ± 1.00 | 5.50 ± 2.04 | 4.36 ± 0.85 | 9.02 ± 2.07* |
| Mean open duration, ms | 0.67 ± 0.14 | 1.82 ± 0.19*** | 0.50 ± 0.10 | 1.45 ± 0.17** |
| Mean closed duration, ms | 453.3 ± 119.8 | 459.3 ± 183.1 | 309.5 ± 73.93 | 162.0 ± 33.64* |
| No. of patches | 8 | 7 | 6 | 8 |
| No. of openings | 4,346 | 5,675 | 4,863 | 9,144 |
Single-channel recordings were made from cell-attached patches from Xenopus oocytes injected with cRNAs encoding for GIRK2 wild-type and mutant channels. Patch membrane potential was clamped at -100 mV. Comparisons were made by performing one-way analysis of variance on data obtained from GIRK2, Y102N, F147Y, and N184D GIRK2 channels followed by Bonferroni's multiple-comparison tests. Data are presented as mean ± SEM. Significance levels are: ***, P < 0.0005; **, P < 0.005; *, P < 0.05; other values are not significantly different from wild type.
Fig. 5.
Suppressors with gating mutant phenotypes as revealed by single-channel recordings. Representative single-channel recordings of GIRK2 and the Y102N and N184D single mutants were obtained in the cell-attached configuration from Xenopus oocytes at a holding potential of –100 mV. Channel openings are upward deflections. Bars indicate traces shown in expanded time scales below.
Discussion
Starting with the nonselective S177W GIRK2 channels, we conducted unbiased mutant screens in yeast to isolate seven second-site suppressors, all outside the K+-channel signature sequence. Below we discuss questions emerging from our suppressor analyses: different ways of gaining potassium selectivity, each time involving a mutation in a helix within the transmembrane domain, and functional coupling between permeation and gating.
Evolving Potassium Selectivity. All K+-selective channels contain recognizable K+-channel signature sequences. On the other hand, we have seen both mutant and wild-type channels that contain the K+-channel signature sequence but do not exhibit K+ selectivity. Recognizing the selective pressure for the collection of modern channels in different organisms, including a variety of K+ channels, we seek to learn about the features important for achieving K+ selectivity of a nonselective GIRK2 mutant channel by conducting second-site suppressor experiments in yeast. Of interest to evolutionary considerations is the emergence of two to three suppressor mutations in each of the mutant types that rescued yeast growth, illustrating the cumulative benefits of multiple mutations. One scenario compatible with these observations is that, early on during evolution, channels might have first acquired the K+-channel signature sequence but not the ability to discriminate between Na+ and K+, laying the foundation for subsequent evolution of K+ selectivity by accumulating mutations in the transmembrane domain. With this scenario, the modern-day hyperpolarization-activated cation channels could mirror such an ancestral nonselective channel. It is also conceivable that K+ selectivity may have been gained and then lost multiple times during evolution, so that hyperpolarization-activated cation channels might have emerged from a K+ channel as their immediate ancestor.
Uncovering Several Different Ways of Gaining Potassium Selectivity. To restore K+-selective permeation to channels bearing the S177W mutation of a pore-lining M2 residue (9, 10), the second-site suppressors may correspond to subtle mutations near the K+ selectivity filter, such as the F147Y mutation of a critical residue on the pore helix (Fig. 4; ref. 33). Elsewhere, conservative mutations such as F98I/Y, affecting an M1 residue highly conserved among Kir channels, conceivably could adjust the packing of M1 and M2 to restore K+ selectivity. Indeed, all three M1 residues yielding second-site suppressors are on the face of M1 that packs against M2 (Fig. 4; refs. 9 and 10). It is also possible that these mutations exert nonstructural effects on ion selectivity. For example, point mutations in α-lytic protease can change its substrate specificity in part by affecting characteristic vibrational motions in the binding pocket (34). Given the dynamic nature of ion permeation (the K+ ion interacts with multiple parts of the channel while passing through the pore), ion selectivity should be strongly influenced by motions inherent in the channel.
A more direct effect seems possible for the second-site suppressor N184D, which affects an M2 residue that, similar to S177, lines the pore of both the open and the closed channel (9, 10) and most likely interacts with permeant ions as well as blocking ions that cause inward rectification (13–16, 35–38). Interestingly, biophysical studies of the K+ channel from Streptomyces lividans (KcsA) suggest a 5- to 7-fold preference for K+ over Na+ in the vestibule below the selectivity filter (39). Given that eight different S177 mutations of GIRK2 abolish selectivity between external Na+ and K+, that N184D suppresses S177W and restores K+ selectivity, and that the converse D172N mutation of Kir2.1 alters the ability of the channel to discriminate internal Rb+ and K+ ions (36), it will be interesting to determine, in future studies, whether these and perhaps additional M2 pore-lining residues (16, 17, 40) contribute to K+ selectivity under physiological conditions, thereby preventing cytoplasmic Na+ from entering a K+ channel and blocking K+ flow (39).
Interaction Between Gating and Permeation. Rather than simply having a collection of separate gating mutations to increase channel activity and suppressor mutations to restore K+ selectivity in the S177W-containing mutant types, we found that nearly half of the suppressor mutations affected residues involved in channel gating (N94, Y102, and N184). The prevalence of gating mutations that interact with S177W to restore K+ selectivity highlights the functional link between channel gating and ion permeation.
Of four M2 residues facing the pore in both the open and the closed Kir channels (9, 10), all yielded gating mutants as shown by this study and previous studies (7, 41). Moreover, mutations reducing the electronegativity of the backbone carbonyl oxygen at either glycine of the selectivity filter affect channel gating and subconductance states rather than K+ selectivity (42). Indeed, gating effects are evident in wild-type channels when K+ is replaced with Tl+ or other permeant ions (43, 44), illustrating a clear influence of permeant ions on gating.
Our finding of a set of residues that can strongly influence gating and K+ selectivity suggests that certain positions can control characteristically dynamic properties of the channel, large-scale motions that mediate the opening and closing of the pore, and smaller, more rapid motions involved in the selective passage of ions through the channel.
Supplementary Material
Acknowledgments
We thank F. Chatelain and T. Surti for support and members of the Jan laboratory. In particular, B. Cohen, M. Grabe, O. Wiser, A. Fay, K. Hu, H. Lai, and B. Schroeder gave helpful input at all stages of this project, and S. Barbel and S. Fried provided technical assistance. We are grateful to D. Minor for helpful comments on the manuscript. We also thank F. Lesage and M. Ladzunski for the gift of GIRK2 cDNA, S. Kurtz for providing the yeast strain, and Z. Lu for a generous gift of tertiapin. Y.N.J. and L.Y.J. are Howard Hughes Medical Institute Investigators. This work was supported by National Institute of Mental Health Grant MH65334.
Abbreviations: Kir, inwardly rectifying potassium; M1, first transmembrane segment; M2, second transmembrane segment; GIRK, G protein-coupled Kir; NMDG, N-methyl-d-glucamine.
References
- 1.Heginbotham, L., Lu, Z., Abramson, T. & MacKinnon, R. (1994) Biophys. J. 66, 1061–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doyle, D. A., Morais Cabral, J., Pfuetzner, R. A., Kuo, A., Gulbis, J. M., Cohen, S. L., Chait, B. T. & MacKinnon, R. (1998) Science 280, 69–77. [DOI] [PubMed] [Google Scholar]
- 3.Zhou, Y., Morais-Cabral, J. H., Kaufman, A. & MacKinnon, R. (2001) Nature 414, 43–48. [DOI] [PubMed] [Google Scholar]
- 4.Aqvist, J. & Luzhkov, V. (2000) Nature 404, 881–884. [DOI] [PubMed] [Google Scholar]
- 5.Berneche, S. & Roux, B. (2001) Nature 414, 73–77. [DOI] [PubMed] [Google Scholar]
- 6.Morais-Cabral, J. H., Zhou, Y. & MacKinnon, R. (2001) Nature 414, 37–42. [DOI] [PubMed] [Google Scholar]
- 7.Yi, B. A., Lin, Y. F., Jan, Y. N. & Jan, L. Y. (2001) Neuron 29, 657–667. [DOI] [PubMed] [Google Scholar]
- 8.Roncaglia, P., Mistrik, P. & Torre, V. (2002) Biophys. J. 83, 1953–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minor, D. L., Jr., Masseling, S. J., Jan, Y. N. & Jan, L. Y. (1999) Cell 96, 879–891. [DOI] [PubMed] [Google Scholar]
- 10.Kuo, A., Gulbis, J. M., Antcliff, J. F., Rahman, T., Lowe, E. D., Zimmer, J., Cuthbertson, J., Ashcroft, F. M., Ezaki, T. & Doyle, D. A. (2003) Science 300, 1922–1926. [DOI] [PubMed] [Google Scholar]
- 11.Bichet, D., Haass, F. A. & Jan, L. Y. (2003) Nat. Rev. Neurosci. 4, 957–967. [DOI] [PubMed] [Google Scholar]
- 12.Stanfield, P. R., Nakajima, S. & Nakajima, Y. (2002) Rev. Physiol. Biochem. Pharmacol. 145, 47–179. [DOI] [PubMed] [Google Scholar]
- 13.Lu, Z. & MacKinnon, R. (1994) Nature 371, 243–246. [DOI] [PubMed] [Google Scholar]
- 14.Stanfield, P. R., Davies, N. W., Shelton, P. A., Sutcliffe, M. J., Khan, I. A., Brammar, W. J. & Conley, E. C. (1994) J. Physiol. (London) 478, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wible, B. A., Taglialatela, M., Ficker, E. & Brown, A. M. (1994) Nature 371, 246–249. [DOI] [PubMed] [Google Scholar]
- 16.Yang, J., Jan, Y. N. & Jan, L. Y. (1995) Neuron 14, 1047–1054. [DOI] [PubMed] [Google Scholar]
- 17.Fujiwara, Y. & Kubo, Y. (2002) J. Gen. Physiol. 120, 677–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lesage, F., Duprat, F., Fink, M., Guillemare, E., Coppola, T., Lazdunski, M. & Hugnot, J. P. (1994) FEBS Lett. 353, 37–42. [DOI] [PubMed] [Google Scholar]
- 19.Stemmer, W. P. (1994) Proc. Natl. Acad. Sci. USA 91, 10747–10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liman, E. R., Tytgat, J. & Hess, P. (1992) Neuron 9, 861–871. [DOI] [PubMed] [Google Scholar]
- 21.Slesinger, P. A., Patil, N., Liao, Y. J., Jan, Y. N., Jan, L. Y. & Cox, D. R. (1996) Neuron 16, 321–331. [DOI] [PubMed] [Google Scholar]
- 22.Sigworth, F. J. & Sine, S. M. (1987) Biophys. J. 52, 1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korn, S. J. & Horn, R. (1988) Biophys. J. 54, 871–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McManus, O. B. & Magleby, K. L. (1988) J. Physiol. (London) 402, 79–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin, W. & Lu, Z. (1998) Biochemistry 37, 13291–13299. [DOI] [PubMed] [Google Scholar]
- 26.Ho, I. H. & Murrell-Lagnado, R. D. (1999) J. Physiol. (London) 520, 645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petit-Jacques, J., Sui, J. L. & Logothetis, D. E. (1999) J. Gen. Physiol. 114, 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alagem, N., Yesylevskyy, S. & Reuveny, E. (2003) Biophys. J. 85, 300–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tucker, S. J., Pessia, M., Moorhouse, A. J., Gribble, F., Ashcroft, F. M., Maylie, J. & Adelman, J. P. (1996) FEBS Lett. 390, 253–257. [DOI] [PubMed] [Google Scholar]
- 30.Tong, Y., Wei, J., Zhang, S., Strong, J. A., Dlouhy, S. R., Hodes, M. E., Ghetti, B. & Yu, L. (1996) FEBS Lett. 390, 63–68. [DOI] [PubMed] [Google Scholar]
- 31.Navarro, B., Kennedy, M. E., Velimirovic, B., Bhat, D., Peterson, A. S. & Clapham, D. E. (1996) Science 272, 1950–1953. [DOI] [PubMed] [Google Scholar]
- 32.Kofuji, P., Hofer, M., Millen, K. J., Millonig, J. H., Davidson, N., Lester, H. A. & Hatten, M. E. (1996) Neuron 16, 941–952. [DOI] [PubMed] [Google Scholar]
- 33.Dart, C., Leyland, M. L., Spencer, P. J., Stanfield, P. R. & Sutcliffe, M. J. (1998) J. Physiol. (London) 511, 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, D. W. & Agard, D. A. (1999) J. Mol. Biol. 286, 267–278. [DOI] [PubMed] [Google Scholar]
- 35.Fakler, B., Brandle, U., Bond, C., Glowatzki, E., Konig, C., Adelman, J. P., Zenner, H. P. & Ruppersberg, J. P. (1994) FEBS Lett. 356, 199–203. [DOI] [PubMed] [Google Scholar]
- 36.Reuveny, E., Jan, Y. N. & Jan, L. Y. (1996) Biophys. J. 70, 754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shyng, S. L., Sha, Q., Ferrigni, T., Lopatin, A. N. & Nichols, C. G. (1996) Proc. Natl. Acad. Sci. USA 93, 12014–12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abrams, C. J., Davies, N. W., Shelton, P. A. & Stanfield, P. R. (1996) J. Physiol. (London) 493, 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nimigean, C. M. & Miller, C. (2002) J. Gen. Physiol. 120, 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishida, M. & MacKinnon, R. (2002) Cell 111, 957–965. [DOI] [PubMed] [Google Scholar]
- 41.Sadja, R., Smadja, K., Alagem, N. & Reuveny, E. (2001) Neuron 29, 669–680. [DOI] [PubMed] [Google Scholar]
- 42.Lu, T., Ting, A. Y., Mainland, J., Jan, L. Y., Schultz, P. G. & Yang, J. (2001) Nat. Neurosci. 4, 239–246. [DOI] [PubMed] [Google Scholar]
- 43.Lu, T., Wu, L., Xiao, J. & Yang, J. (2001) J. Gen. Physiol. 118, 509–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choe, H., Sackin, H. & Palmer, L. G. (1998) J. Gen. Physiol. 112, 433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.