Abstract
The structures of RNA molecules are often important for their function and regulation1-6, yet there are no experimental techniques for genome-scale measurement of RNA structure. Here, we describe a novel strategy termed Parallel Analysis of RNA Structure (PARS), which is based on deep sequencing fragments of RNAs that were treated with structure-specific enzymes, thus providing simultaneous in-vitro profiling of the secondary structure of thousands of RNA species at single nucleotide resolution. We apply PARS to profile the secondary structure of the mRNAs of the budding yeast S. cerevisiae and obtain structural profiles for over 3000 distinct transcripts. Analysis of these profiles reveals several RNA structural properties of yeast transcripts, including the existence of more secondary structure over coding regions compared to untranslated regions, a three-nucleotide periodicity of secondary structure across coding regions, and a relationship between the efficiency with which an mRNA is translated and the lack of structure over its translation start site. PARS is readily applicable to other organisms and to profiling RNA structure in diverse conditions, thus enabling studies of the dynamics of secondary structure at a genomic scale.
Existing experimental methods for measuring RNA structure can only probe a single RNA structure per experiment and are typically limited in the length of the probed RNA (Supplemental note 1). To simultaneously measure structural properties of many different RNAs, we extracted poly-adenylated transcripts from log-phase growing yeast, renatured the transcripts in vitro, and treated the resulting pool with RNase V1 and separately, with RNase S1. RNase V1 preferentially cleaves phosphodiester bonds 3′ of double-stranded RNA, while RNase S1 preferentially cleaves 3′ of single-stranded RNA7. Thus, data from these two complementary enzymes should allow us to measure the degree to which each nucleotide was in a single- or double-stranded conformation (Fig. 1). We chose renaturation and enzymatic cleavage conditions under which the cleavage reactions occur with single-hit kinetics (Supplementary Fig. 1a,b), and where intramolecular, but not intermolecular, RNA-RNA interactions are dominant (Supplementary Fig. 1c,d). As a control, we also added two short RNA domains from HOTAIR, a human non-coding RNA8, and from the structurally known Tetrahymena group I intron ribozyme9.
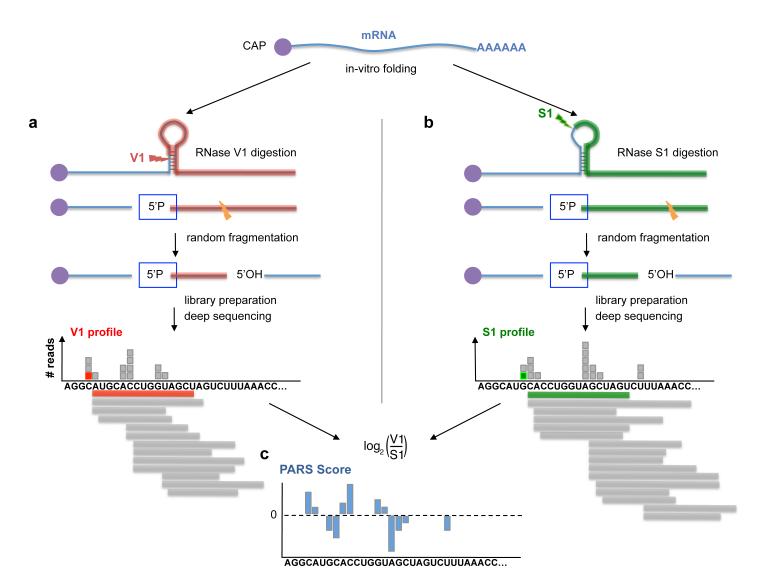
Figure 1. Measuring structural properties of RNA by deep sequencing.
(a) RNA molecules are cleaved by RNase V1, which cuts 3′ of double-stranded RNA, leaving a 5′ phosphate (5′P). One such cut is illustrated by a red arrow. Following random fragmentation, V1-generated fragments are specifically captured and subjected to deep sequencing. Each aligned sequence provides structural evidence about a single base. The marked red square illustrates the evidence obtained from one mapped sequence (red). Additional evidence (gray boxes) is collected by mapping more sequences (gray horizontal bars). A large number of reads aligned to the same base indicates that the base is cleaved multiple times by RNase V1 and is thus more likely to be in double stranded conformation. (b) Same as (a), but when the RNA sample is treated with RNase S1, which cuts 3′ of single-stranded RNA. Collected reads in this case suggest that the base was unpaired in the original RNA structure. (c) By combining the data extracted from the two complementary experiments (a) and (b), we obtain a nucleotide-resolution score representing the likelihood that the inspected base was in a double- or single-stranded conformation.
We devised a ligation method to specifically ligate V1- and S1-cleaved RNA to adaptors, and converted them into cDNA libraries suitable for deep sequencing (Supplementary Fig. 2). As both enzymes leave a 5′ phosphate at the cleavage point and since only 5′ phosphoryl-terminated RNA are capable of ligating to our adaptors, we enrich for V1- and S1-cleaved fragments and select against random fragmentation and degradation products that typically have 5′ hydroxyl (Supplementary Fig. 3). Thus, each observed cleavage site provides evidence that the cut nucleotide was in a double-stranded (for V1-treated samples) or single-stranded (for S1-treated samples) conformation. As a quantitative measure at nucleotide resolution representing the degree to which a nucleotide was in a double- or single-stranded conformation, we take the log-ratio between the number of sequence reads obtained for each nucleotide in the V1 and S1 experiments. A higher (lower) log-ratio, or PARS score, thus denotes a higher (lower) probability for a nucleotide to be in a double-stranded conformation.
We performed four independent V1 experiments and three independent S1 experiments, which were highly reproducible across replicates (correlation=0.60-0.93, Supplementary Table 1), resulting in a total of ~85 million sequence reads that map to the yeast genome, of which ~97% mapped to annotated transcripts (Supplementary Table 2). At an average nucleotide coverage above 1.0, we obtained structural information for over 3000 yeast transcripts (Supplementary Table 3, Supplementary Fig. 4a), covering in total over 4.2 million transcribed bases, which is ~100-fold more than all published RNA footprints to date.
We used several tests to check for biases in our method, and found that RNase cleavage, adaptor ligation, and cDNA conversion do not introduce significant sequence biases (Supplementary Fig. 5), that our protocol has a very small bias towards particular regions along the transcript (Supplementary Fig. 6), and that we capture RNA fragments in proportion to their abundance in the initial pool (Supplementary Fig. 4b,c). Finally, we confirmed that signals generated by RNase V1 are highly distinct from those generated by RNase S1. Global inspection across all transcripts revealed that ~7% of the V1 and S1 peaks are shared (Methods and Supplementary Table 4 and Supplementary Fig. 7). Those joint peaks could be the result of experimental noise introduced by nonspecific enzymatic activity, but could also correspond to dynamic RNA regions or transcripts that fold into more than one stable conformation.
To test whether PARS accurately measures RNA structures, we first confirmed that its signals are similar to those obtained with traditional footprinting. To this end, we carried out ten separate footprinting experiments with either RNase V1 or S1, on two domains from the Tetrahymena ribozyme, two domains from the human HOTAIR non-coding RNA, which we doped into our samples and two domains of endogenous yeast mRNAs. In all cases, we found high agreement between our PARS signals and footprinting (correlations=0.63-0.97, Fig. 2 and Supplementary Fig. 8-10). Notably, due to length limitations of footprinting, we had to select short domains from each of the above transcripts, in vitro transcribe them, and then apply footprinting. Thus, footprinting may be inaccurate, since due to long-range interactions, the excised fragment could fold differently when taken out of context. In contrast, PARS can probe RNAs in their full-length context.
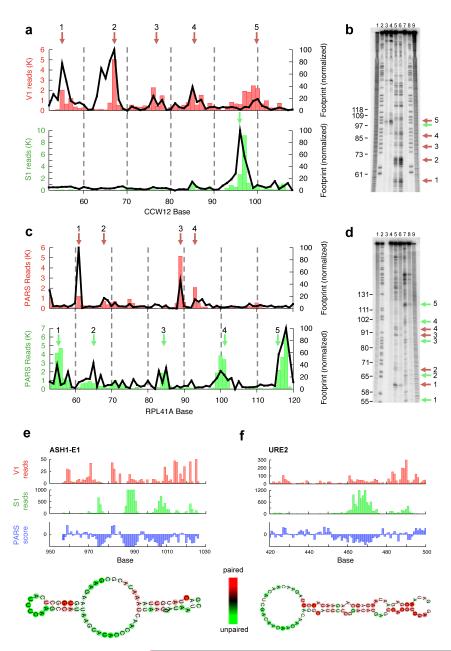
Figure 2. PARS correctly recapitulates results of RNA footprinting and known structures.
(a) The PARS signal obtained for bases 50-110 of the yeast gene CCW12 using the double-stranded cutter RNase V1 (red bars) or single-stranded cutter RNase S1 (green bars) accurately matches the signals obtained by traditional footprinting of that same transcript domain (black lines). PARS signal is shown as the number of sequence reads which mapped to each nucleotide; footprinting results are obtained by semi-automated quantification of the RNase lanes shown in (b). The red arrows indicate RNase V1 cleavages and the green arrows indicate RNase S1 cleavages as shown in the gel (b). (b) Gel analysis of RNase V1 (lanes 5,6) and S1 (lanes 3,4) probing of CCW12. Additionally, RNase T1 ladder (lanes 2,8), alkaline hydrolysis (lanes 1,9), and no RNase treatment (lane 7) are shown. (c) The PARS signal obtained from bases 50-120 of the yeast gene RPL41A matches the signals obtained by traditional footprinting. (d) RNase V1 (lanes 5,6) and S1 (lanes 7,8) probing of RPL41A, RNase T1 ladder (lane 2), alkaline hydrolysis (lanes 1,9), and no RNase treatment (lane 4). (e-f) Raw number of reads obtained using RNase V1 (red bars) or RNase S1 (green bars) and the resulting PARS score (blue bars) along one inspected domain of ASH1 (e) and URE2 (f). Also shown are the known structures of the inspected domains with nucleotides color-coded according to their computed PARS score.
Next, we compared PARS to reported structures of yeast coding and non-coding RNAs, and found that it correctly reproduces the known secondary structure of three structured RNA domains of ASH110, of a structural element in URE2 mRNA11 and of the glu-tRNA (Fig. 2e-f and Supplementary Fig. 11-12). This suggests that PARS can provide structural information of transcripts in their full-length context and endogenous abundance from within a complex RNA pool. Taken together, our analyses demonstrate that PARS recapitulates results obtained by low-throughput methods with high accuracy, and also has advantages over existing methods, stemming from its ability to probe structures of long RNAs.
As another independent validation of PARS, we compared it to computational predictions of RNA structure, by applying the Vienna package12 to the 3000 transcripts that we analyzed. We found a significant correspondence between these predictions and our PARS scores, whereby nucleotides with high (low) double-stranded PARS score had a significantly higher (lower) average predicted pairing probability (p<10−200, Fig. 3a and Supplemental Fig. 13). Despite this significant global correspondence, there are large differences between PARS and predictions, in part due to noise in our approach but also due to known inaccuracies of folding algorithms. We thus suggest that genome-wide PARS data can be used to constrain folding algorithms and improve their accuracy, as previously shown for specific RNAs13,14 (Supplementary Fig. 15).
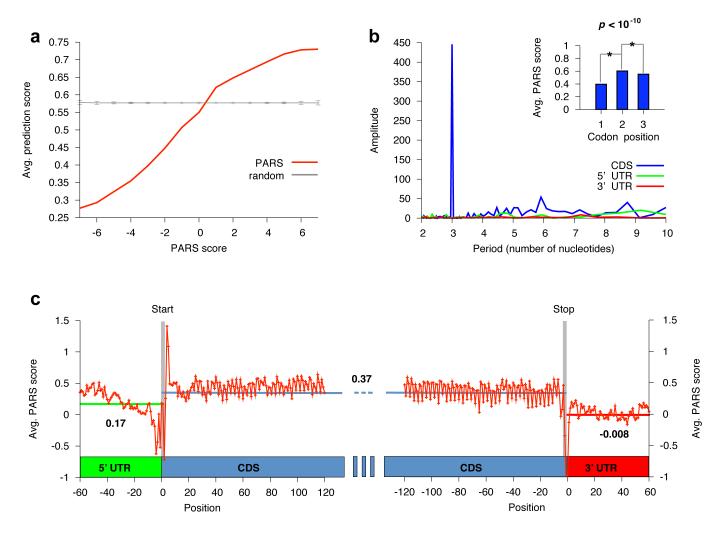
Figure 3. Functional units of the transcript are demarcated by distinct properties of RNA structure.
(a) Significant correspondence between PARS and computational predictions of RNA structure. We used the Vienna package12 to fold the 3000 yeast mRNAs used in our analysis, and extracted the predicted double-stranded probability of each nucleotide. Shown is the average predicted double-stranded probability of each nucleotide (y-axis), where nucleotides were sorted by their PARS score (x-axis). Average and standard deviation from 1000 shuffle experiments in which a random prediction score was assigned to each probed base are shown in gray. (b) Discrete Fourier transform of average PARS score across the coding region, 3′ UTR and 5′ UTR. Inset shows PARS score obtained for each of the three positions of every codon, averaged across all codons. (c) PARS score across the 5′ UTR, the coding region, and the 3′ UTR, averaged across all transcripts used in our analysis. Transcripts were aligned by their translational start and stop sites for the left and right panel, respectively; start and stop codons are indicated by gray bars; horizontal bars denote the average PARS score per region.
We used the obtained structural profiles to investigate five global properties of yeast transcripts. First, examining the average PARS score across the coding regions and untranslated regions (UTRs), we found that coding regions exhibit significantly more pairing than 5′ and 3′ UTRs (p<10−30 and p<10−50 respectively, Fig. 3c). Notably, the start and stop codons each exhibit local minima of PARS scores, indicating reduced tendency for double-stranded conformation and increased accessibility. These findings agree with previous computational predictions for mouse and human genes15. The evolutionary conservation of this global organization of mRNA secondary structure suggests that it may have functional importance. An overall decreased pairability in UTRs may allow functional elements to stand out and conversely, highly paired domains along coding regions may protect against ectopic translation initiation, or regulate ribosome translocation and protein folding, as recently postulated13.
Second, aligning our measured transcripts about their start codon and applying a discrete Fourier transform analysis to the average PARS signal, we detected a periodic structure signal across coding regions with a cycle of three nucleotides, such that on average, the first nucleotide of each codon is least structured and the second nucleotide is most structured. Notably, this periodic signal is only found in coding regions, and not in UTRs (Fig. 3b), and the degree of three-nucleotide periodicity in transcripts is significantly associated with ribosome density in vivo16 (Supplementary Fig. 14), suggesting that the three-nucleotide periodicity may directly or indirectly facilitate translation.
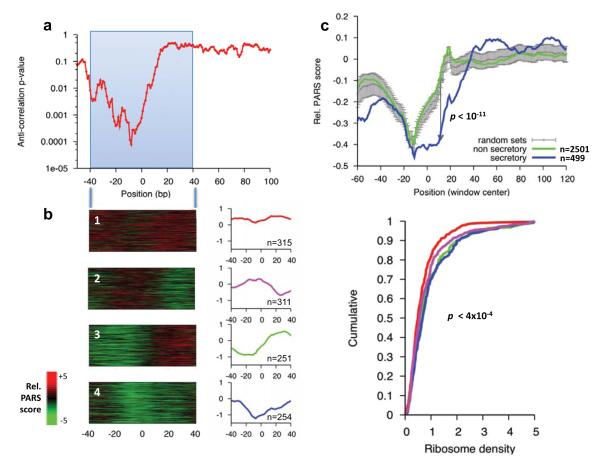
Third, we tested whether there is a correlation between mRNA structure around the translation start site and translation efficiency. Such a relationship has long been hypothesized17 and recently shown for one reporter protein in E. coli18. We found a small but significant anti-correlation between PARS scores at the region located ~10bp upstream of the translation start site and ribosome density throughout the transcript16, a proxy for translational efficiency (correlation=-0.1, p<10−4, Fig. 4a). Intriguingly, the −10bp region corresponds to the 5′ position of the first ribosome on yeast mRNAs16. To examine this relationship in more detail, we applied k-means clustering (k=4) to the PARS structural profile of the ±40bp surrounding the translation start site. Notably, genes found in clusters 3 and 4, exhibit significantly less structure in their 5′ UTR than in the beginning of their coding region, as well as a higher ribosome density (Fig. 4b). Overall, these results provide the first genome-wide experimental validation for the suggestion that mRNA secondary structure around the start codon may reduce translational efficiency17, although the low correlation we found implies that in vivo, translational efficiency is determined by additional factors.
Figure 4. Structure around start codons correlates with low translational efficiency.
(a) Sliding window analysis of local PARS score and ribosome density16. Shown is the significance (p-value) of the anti-correlation between average PARS score along a 40bp-wide window and the reported ribosome density. (b) Left: k-means clustering of PARS scores across the ±40bp window surrounding the translation start site of all transcripts for which enough coverage was obtained. The average structural profile and number of member genes is shown to the right of each cluster. Right: Cumulative distribution plot of ribosome occupancy for each cluster and the associated Kolmogorov-Smirnoff test p-value between the distribution of cluster 1 and 4. (c) Tendency for less RNA structure in the first 30 bases of ORFs encoding predicted secretory proteins. While structure typically builds up immediately upon entry to the coding sequence (CDS), genes predicted to code for secretory proteins retain low structure in the first ~30 bases of the CDS, consistent with the dual function SSCR having structural features of UTR rather than CDS20. Shown are the average relative PARS scores (Methods) across a 30bp sliding window for the 499 genes coding for secretory proteins (blue), the remaining 2501 genes (green) and the mean and standard deviation obtained from 1000 shuffle experiments in which sets of 499 genes were randomly selected (gray).
Fourth, we asked whether genes with shared biological functions or cytotopic localizations19 tend to have similar PARS scores, indicative of similar degrees of secondary structures. We found a rich picture of biological coordination (Supplementary Fig. 16 and Supplementary Table 5), including increased RNA structure, especially in coding regions, in transcripts whose encoded proteins localize to distinct cellular domains or participate in distinct metabolic pathways, and found that mRNAs with the least secondary structure in their 5′ UTR and CDS encode subunits of the ribosome.
Finally, we examined the PARS score of transcripts predicted to encode a signal peptide, since a recent study showed that RNA sequences encoding the signal sequence (termed the SSCR) of secretory proteins function as RNA elements that promote RNA nuclear export20. We found that the 5′ UTR region and first ~30 coding nucleotides of signal peptide transcripts have lower PARS signal, indicating increased single-stranded propensity, as compared to other transcripts (p<10−11, Fig. 4c). Since SSCRs typically reside in the beginning of the coding region, these results suggest that specific secondary RNA structure around gene starts may assist in the cytotopic localization of mRNAs and their resulting proteins. More generally, we suggest that PARS can be used to both generate and test hypotheses regarding signals of secondary structure that may characterize and have functional importance for classes of mRNAs.
In summary, we introduced PARS, the first high-throughput approach for genome-wide experimental measurement of RNA structural properties, and showed that it recovers structural profiles with high accuracy and at nucleotide resolution. Like most existing methods, one limitation of PARS is that it maps RNA structures in vitro, and its reported structures may thus differ significantly from the in vivo conformations. This may be addressed in the future using reagents that can probe RNA structure in living cells7, but will require new methods to adapt to deep sequencing. Overall, PARS transforms the field of RNA structure probing into the realm of high-throughput, genome-wide analysis and should prove useful both in determining the structure of entire transcriptomes of other organisms as well as in systematically measuring the effects of diverse conditions on RNA structure. Probing RNA structure in the presence of different ligands, proteins, or in different physical or chemical conditions may provide further insights into how RNA structures control gene activity.
METHODS SUMMARY
Sample preparation
Total RNA was extracted from yeast grown at 30C to exponential phase in YPD medium by using hot acid phenol. Poly(A)+ RNA was obtained by purifying twice using the Poly(A) purist Kit. A diagram showing the PARS protocol is provided in Supplementary Fig. 2.
Sequencing library construction
RNA was folded and probed for structure using 0.01U of RNase V1 (Ambion), or 1000U of S1 nuclease (Fermentas), in a 100ul reaction volume. A modified version (see supplementary methods) of the SOLiD Small RNA Expression Kit was used to convert fragments into a sequencing library.
SOLiD™ Sequencing and mapping
cDNA libraries were amplified onto beads and subjected to emulsion PCR, according to the standard protocol described in the SOLiD Library Preparation Guide. Obtained sequences were truncated to 35bp, and required to map uniquely to either the yeast genome or transcriptome, allowing up to one mismatch and no insertions or deletions.
Computing the PARS Score
The PARS Score is defined as the log2 of the ratio between the number of times the nucleotide immediately downstream to the inspected nucleotide was observed as the first base when treated with RNase V1 and the number of times it was observed in the RNase S1 treated sample. To account for differences in overall sequencing depth between the V1- and S1- treated samples, the number of reads for each nucleotide is normalized prior to the computation of the ratio.
Periodicity
Periodicity analysis was done applying a Discrete Fourier Transform to the average PARS score collected from the following genomic features: last 100 bases of the 5′ UTR, first 200 bases of the coding sequence, 100 first bases of the 3′ UTR.
Supplementary Material
Acknowledgements
We thank D. Herschlag’s group, A. Adler, A. Fire, M. Kay, Life Technologies SOLiD team, M. Rabani, G. Sherlock, and A. Weinberger for assistance and critiques. Supported by NIH grant (RO1HG004361). Y.W. is funded by the Agency of Science, Technology and Research of Singapore. H.Y.C. is an Early Career Scientist of the Howard Hughes Medical Institute. E.S. is the incumbent of the Soretta and Henry Shapiro career development chair.
Footnotes
Online Resources. Nucleotide-resolution raw reads and PARS scores for the 3000 genes included in our analysis can be visualized and downloaded at http://genie.weizmann.ac.il/pubs/PARS10
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions M.K., J.R., H.C. and E.S. conceived the project; Y.W. and H.C. developed the protocol and designed the experiments; Y.W. performed all experiments; M.K., E.M. and E.S. planned and conducted the data analysis; J.R. and R.N. helped with sequencing; M.K., Y.W., E.M., H.C. and E.S. wrote the paper with contribution from all authors.
Author Information Sequencing data has been deposited in the Gene Expression Omnibus (GEO) under accession number GSE22393.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Arava Y, et al. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2003;Vol. 100:3889–94. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y, et al. Precision and functional specificity in mRNA decay. Proc Natl Acad Sci U S A. 2002;99:5860–5. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takizawa PA, DeRisi JL, Wilhelm JE, Vale RD. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000;290:341–4. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- 4.Shepard KA, et al. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc Natl Acad Sci U S A. 2003;100:11429–34. doi: 10.1073/pnas.2033246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Curr Opin Struct Biol. 2005;15:342–8. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;Vol. 39:1278–84. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 7.Ziehler WA, Engelke DR. Probing RNA structure with chemical reagents and enzymes. Curr Protoc Nucleic Acid Chem. 2001:1. doi: 10.1002/0471142700.nc0601s00. Chapter 6, Unit 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;Vol. 129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo F, Gooding AR, Cech TR. Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site. Mol Cell. 2004;16:351–62. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Chartrand P, Meng XH, Huttelmaier S, Donato D, Singer RH. Asymmetric sorting of ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol Cell. 2002;10:1319–30. doi: 10.1016/s1097-2765(02)00694-9. [DOI] [PubMed] [Google Scholar]
- 11.Reineke LC, Komar AA, Caprara MG, Merrick WC. A small stem loop element directs internal initiation of the URE2 internal ribosome entry site in Saccharomyces cerevisiae. J Biol Chem. 2008;283:19011–25. doi: 10.1074/jbc.M803109200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hofacker IL, Fekete M, Stadler PF. Secondary structure prediction for aligned RNA sequences. J Mol Biol. 2002;319:1059–66. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 13.Watts JM, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–6. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathews DH, et al. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci U S A. 2004;101:7287–92. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shabalina SA, Ogurtsov AY, Spiridonov NA. A periodic pattern of mRNA secondary structure created by the genetic code. Nucleic Acids Res. 2006;Vol. 34:2428–37. doi: 10.1093/nar/gkl287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-Wide Analysis In Vivo of Translation with Nucleotide Resolution Using Ribosome Profiling. Science. 2009:1168978v1. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;Vol. 361:13–37. doi: 10.1016/j.gene.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 18.Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;Vol. 324:255–8. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palazzo AF, et al. The signal sequence coding region promotes nuclear export of mRNA. PLoS Biol. 2007;5:e322. doi: 10.1371/journal.pbio.0050322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;Vol. 320:1344–9. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cech TR, Damberger SH, Gutell RR. Representation of the secondary and tertiary structure of group I introns. Nat Struct Biol. 1994;1:273–80. doi: 10.1038/nsb0594-273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.