Abstract
Background
Magnetotactic bacteria produce membrane-enveloped magnetite crystals (magnetosomes) whose formation is controlled primarily by a gene island termed the magnetosome island (MAI). Characterization of single gene and operon function in MAI has elucidated in part the genetic basis of magnetosome formation. The mamX gene, located in the mamXY operon, is highly conserved in the MAI of all Magnetospirillum strains studied to date. Little is known regarding the function of mamX in the process of biomineralization.
Results
A mamX deletion mutant (∆mamX) and its complemented strain (CmamX) by conjugation in M. gryphiswaldense strain MSR-1 were constructed. There were no striking differences in cell growth among ∆mamX, CmamX, and wild-type strain (WT). ∆mamX displayed a much weaker magnetic response than WT. Transmission electron microscopy revealed the presence of irregular, superparamagnetic magnetite particles in ∆mamX, in contrast to regular, single-domain particles in WT and CmamX. The phenotype of ∆mamX was similar to that of an ftsZ-like deleted mutant and mamXY operon deleted mutant reported previously. Quantitative real-time RT-PCR (qPCR) results indicated that the deletion of mamX had differential effects on the transcription levels of the other three genes in the operon.
Conclusions
The MamX protein plays an important role in controlling magnetosome size, maturation, and crystal form. The four MamXY proteins appear to have redundant functions involved in magnetosome formation. Our findings provide new insights into the coordinated function of MAI genes and operons in magnetosome formation.
Keywords: Magnetospirillum gryphiswaldense, mamXY operon, mamX, Magnetosome, Crystal maturation
Background
Magnetotactic bacteria (MTB) produce nano-sized membrane-enveloped magnetic organelles termed magnetosomes, consisting of single-domain magnetite (Fe3O4) or greigite (Fe3S4) crystals that are integrated into one to several chains depending on the species [1,2]. MTB are aquatic prokaryotes that utilize the magnetosomes to align themselves relative to magnetic fields and swim toward favorable low-oxygen, nutrient-rich environments. This behavior is called magneto-aerotaxis [1,3].
Many studies over the past several decades have focused on the molecular mechanism of magnetosome formation and revealed several important facts. Magnetosome-related genes are concentrated in a structure called the “magnetosome island” (MAI) in the genomes of MTB [4,5]. In Magnetospirillum strains such as M. gryphiswaldense MSR-1, M. magneticum AMB-1, and M. magnetotacticum MS-1, the MAI conservatively contains four common gene operons: mms6, mamGFDC, mamAB, and mamXY[2,6]. The mamXY operon is also conserved in Magnetococcus sp. MC-1 [7]. Mms6, a tightly bound protein found in the magnetosome membrane, plays an essential role in the control of magnetite crystallization and crystal size [8-10]. The MamGFDC proteins have partially redundant and collective functions in the control of magnetosome size [11]. The mamAB operon is a large cluster containing most of the MTB-specific genes, including those that encode the proteins MamE (involved in the localization of magnetosome membrane protein [MMP]), MamK (actin-like protein involved in the alignment of magnetosome chains), and MamJ (interacts with MamK, an important factor in magnetosome chain formation) [12-15]. Recent studies have shown that the mamAB operon is necessary and sufficient for magnetite biomineralization [16,17].
The mamXY operon received less attention than mms6, mamGFDC, and mamAB. mamXY is the last cluster in the MAI and contains four sequential genes termed mamY, mamX, mamZ, and ftsZ-like, identified as a polycistronic transcription unit [18]. The MamXY proteins were shown to play crucial roles in magnetite biomineralization through whole operon deletion in MSR-1 [16]. Such effect was less obvious in AMB-1 [14]. MamY was reported to constrict the magnetosome membrane in AMB-1 [19]. Deletion of FtsZ-like resulted in smaller superparamagnetic particles [18]. MamZ has been predicted (without direct evidence to date) to be an ortholog of MamH and likely a permease belonging to the major facilitator superfamily. MamX has similarities to the serine-like proteases MamE and MamS, but there have been no systematic studies of its function to date. In view of the high conservation of mamXY in MTB, functional studies of this operon are needed to elucidate the entire MAI and its role in the mechanism of magnetosome formation. The present study is focused on the highly conserved but hitherto uncharacterized MamX protein.
Results
Deletion of the mamX gene had no effect on cell growth
To elucidate the function of mamX in the absence of polar effect, MSR-1 was subjected to in-frame gene deletion (to produce strain ∆mamX) and complementation of mamX (to produce strain CmamX) as described in Methods. We validated the construction of the mutant and complemented strains, detected the genes in the MAI, and measured cell growth and magnetic responses. There were no notable differences in the growth curves of WT, ∆mamX, and CmamX (Figure 1A), although the OD565 of ∆mamX was slightly lower than that of WT and CmamX at each sample point. The maximal OD565 values for WT, ∆mamX, and CmamX were 1.33, 1.24, and 1.29, respectively, and were reached by 24 hr in each case.
Figure 1.
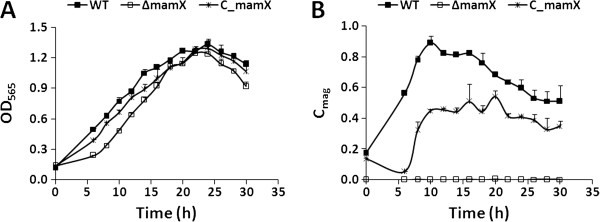
Comparison of cell growth and magnetic response (Cmag) in WT, mutant (∆mamX), and complemented strains (CmamX). All experiments were performed in triplicate. A: There were no striking differences among the growth curves of the three strains. B: The Cmag value of ∆mamX was consistently zero. The Cmag value of WT increased from 0.17 at 0 hr to a maximum of 0.89 at 10 hr and then gradually decreased. The Cmag value of CmamX increased from 0.14 at 0 hr to 0.45 at 10 hr.
∆mamX showed decreased intracellular iron content and magnetic response
Cmag can be used as an efficient value for measuring the magnetosome content of MTB [20]. For WT, Cmag increased from 0.17 at 0 hr to a maximum of 0.89 at 10 hr and gradually decreased thereafter (Figure 1B), while the Cmag value of ∆mamX remained zero throughout the culture period. This observation indicates a complete loss of magnetism in ∆mamX. CmamX partially recovered its Cmag value, which increased from 0.14 at 0 hr to 0.45 at 10 hr (Figure 1B). The complemented plasmid may exist as a free plasmid in cytoplasm rather than being integrated into the MSR-1 genome, resulting in an unstable phenotype. To further characterize the mamX mutant, we measured the iron content in cells. The intracellular iron content of ∆mamX (0.20%) was much lower than that of WT and CmamX (both 0.47%) (Figure 2); this difference was statistically significant (P < 0.01, by T-test).
Figure 2.
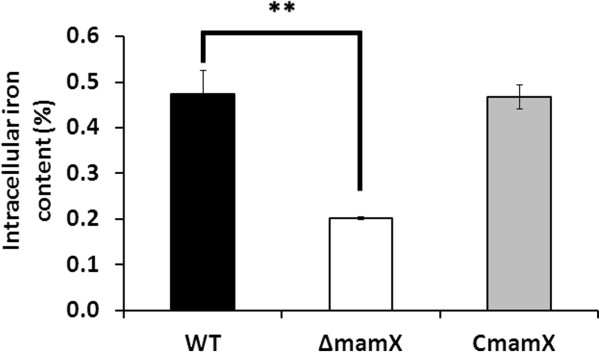
Intracellular iron contents during culture of WT, ∆mamX, and CmamX. The intracellular iron content was much lower for ∆mamX (0.20%) than for WT and CmamX (both 0.47%). **, The difference between WT and ∆mamX was statistically significant (P < 0.01, by t test).
The deletion of mamX resulted in irregular and smaller crystals
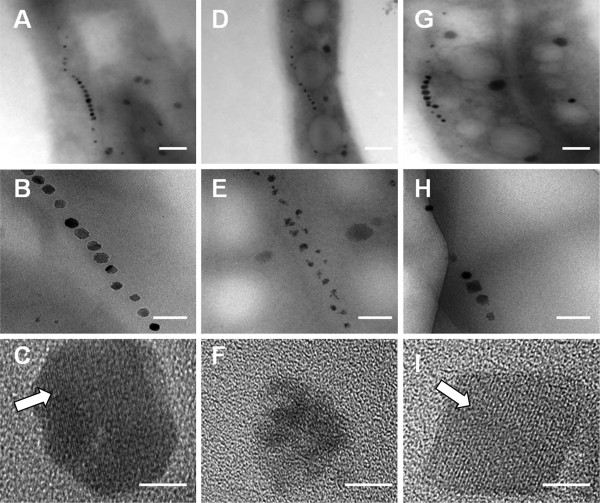
Phenotypic changes in the mutant cells and magnetosomes were observed by HR-TEM. WT had regular cubo-octahedral magnetosomes (mean crystal diameter 41.25±10.46 nm) (Table 1), mature chains (Figure 3A-C), and a standard magnetite crystal lattice (Figure 3C, arrow). In ∆mamX, the magnetosomes were much smaller (mean crystal diameter 26.11±9.92 nm) (Table 1) and irregularly shaped, and the crystal lattice was very poorly developed, although the chains were organized normally (Figure 3D-F). CmamX showed a normal crystal size and phenotype (mean crystal diameter 48.42±11.82 nm) (Table 1) and a typical magnetite crystal lattice (Figure 3I, arrow). The mean numbers of crystals per cell were 15.35±3.06 for WT, 20.85±3.91 for ∆mamX, and 6.55±1.88 for CmamX (Table 1). The number of intracellular magnetosomes was slightly higher in ∆mamX than in the other two strains. An energy-dispersive spectroscopic analysis showed that iron and oxygen were the primary elemental components of magnetosomes in ∆mamX, the same as in WT and CmamX (data not shown).
Table 1.
Magnetosome diameters and numbers in three MSR-1 strains
|
Strains |
Maximum |
Minimum |
Mean |
Mean |
|---|---|---|---|---|
| |
crystal diameter |
crystal diameter |
crystal diameter |
crystal number |
| (nm) | (nm) | (nm) | ||
| WT |
70.08 |
21.99 |
41.25 ± 10.46 a |
15.35 ± 3.06 b |
| ∆mamX |
58.93 |
8.49 |
26.11 ± 9.92 |
20.85 ± 3.91 |
| CmamX | 74.91 | 18.14 | 48.42 ± 11.82 | 6.55 ± 1.88 |
For each strain, 20–30 cells and 250–300 crystals were visualized and measured. a: there is significant difference between the mean crystal diameter of WT and ∆mamX (P < 0.01, by Student t-test); b: there is significant difference between the mean crystal number of WT and ∆mamX (P < 0.01, by Student t-test).
Figure 3.
HR-TEM observation of different cells. HR-TEM of WT (A, B, C), ∆mamX(D, E, F), and CmamX(G, H, I). A, D, G: cell phenotype and magnetosome location. B, E, H: magnetosome chain organization. C, F, I: crystal lattice structure. Arrows: standard Fe3O4 crystal lattice. Scale bars: A, D, G: 200 nm; B, E, H: 100 nm; C, F, I: 10 nm.
To further characterize the magnetosome crystals, we performed rock magnetic measurements on whole-cell samples of WT, ∆mamX and CmamX strains (Figure 4). The WT sample had a pot-bellied hysteresis loop with the hysteresis parameters coercivity Bc, remanence coercivity Bcr, and remanence ratio Mrs/Ms being 5.91 mT, 10.76 mT, and 0.38, respectively. This indicated that the WT cell formed dominant single domain particles and small portion of superparamagnetic particles. The domain states of WT strain formed magnetosomes were further demonstrated by its corresponding first-order reversal curves (FORCs) diagram: a set of concentrated contours distributes around a peak of coercivity at Hc ≈ 9 mT and slightly intersects with the Hb axis. In contrast, the ∆mamX sample had a wasp-waist hysteresis loop; and its FORCs diagram slightly expanded in the horizontal distribution, but strongly intersected with the Hb axis with the peak coercivity reducing to ~2 mT. These features indicated an increased heterogeneity in microcoercivity (i.e., crystal size, morphology, and/or crystallinity) and a larger portion of superparamagnetic particles than in the WT sample [21,22]. The CmamX sample had Stoner-Wohlfarth-type hysteresis loop with the Mrs/Ms value being 0.45; its FORC diagram was characterized by a set of closed contours concentrated around the peak coercivity of ~16 mT narrowly along the horizontal axis. These features, similar to whole-cell samples of other MTB [22-24], were typical behaviors of a randomly oriented array of non-interacting uniaxial single-domain particles [25,26]. The stronger magnetic properties (e.g., higher values of Bc, Bcr and Mrs/Ms) exhibited by CmamX than WT, associated with better magnetosome formation like larger crystal size (Table 1) and/or higher crystallinity within the former than the later, was probably due to the over expression of MamX. This result, consistent with our previous study on C_ftsZ-like strain of MSR-1 [18], further demonstrated that the mamX play a role in controlling the crystal size and/or crystallinity of magnetosomes within MSR-1.
Figure 4.
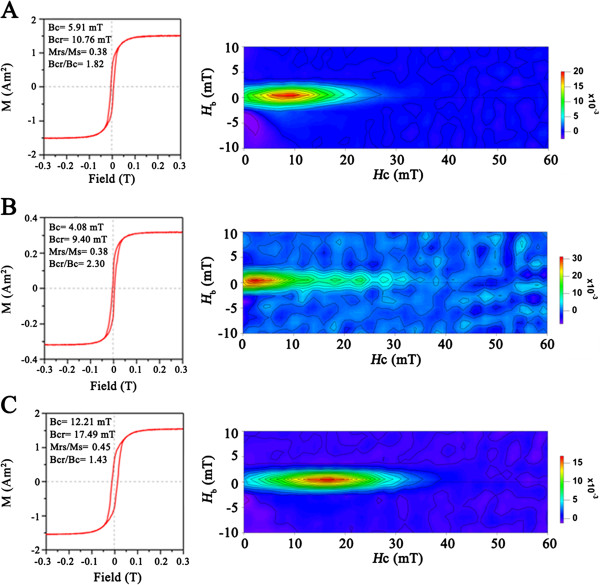
Measurements of magnetism in deferent cells. (A):WT, (B): ΔmamX and (C): CmamX. Left: room-temperature hysteresis loops. Right: FORCs diagrams.
mamXY gene transcription levels were affected by mamX deletion
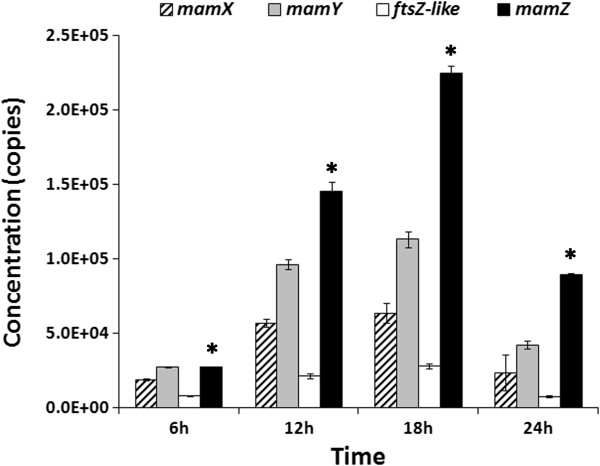
mamXY gene transcription levels were evaluated in the three strains. In WT, each of the four genes (mamY, mamX, mamZ, and ftsZ-like) in the mamXY operon showed high transcription levels from 12 to 18 hr in absolute qPCR assay (Figure 5). This period corresponds to the log phase of growth, which is the period of rapid cell growth and magnetosome synthesis. The transcription level of mamZ was much higher than those of the other three genes at each of the four time points (Figure 5); i.e., the level of mamZ was 3–6 times that of mamY, 4–11 times that of mamX, and 10–36 times that of ftsZ-like (Table 2). These findings suggest that the MamZ protein plays a crucial role during cell growth.
Figure 5.
Absolute qPCR results for transcription levels of the four genes (mamY , mamX, mamZ, ftsZ-like) in the mamXY operon in WT. Each of the genes had a high transcription level from 12 to 18 hr, corresponding to the log phase of growth. The transcription level of mamZ was much higher than those of the other three genes at all four sampling times. *, 1/3 of original transcription level of mamZ in the figure was showed for better display of the other gene transcriptions.
Table 2.
Ratio of transcription levels of MamZ to other MamXY proteins in WT and ΔmamX strains, based on qPCR results
| WT | ΔmamX | |||||
|---|---|---|---|---|---|---|
| Ratios | |
MamZ / |
MamZ / |
MamZ / |
MamZ / |
MamZ / |
| MamX |
MamY |
FtsZ-like |
MamY |
FtsZ-like |
||
| 6 hr |
4.0 |
3.0 |
10.0 |
0.4 a |
0.2 e |
|
| 12 hr |
8.0 |
5.0 |
21.0 |
1.6 b |
2.2 f |
|
| 18 hr |
11.0 |
6.0 |
24.0 |
0.8 c |
0.2 g |
|
| 24 hr | 11.0 | 6.0 | 36.0 | 2.9 d | 1.0 h | |
a, b, c and d: ratios of taranscription level MamZ/MamY have significant differences between in WT and in ΔmamX strain at all the four time points (all P < 0.01, by t test); e, f, g and h: ratios of taranscription level MamZ/FtsZ-like have significant differences between in WT and in ΔmamX strain at all the four time points (all P < 0.01, by t test).
We used qPCR to measure the transcription levels of mamY, mamZ, and ftsZ-like in ∆mamX. The relative transcription level of mamY was similar in ∆mamX and WT at 6 and 12 hr but was twice as high in ∆mamX as in WT at 18 hr (Figure 6A). The transcription level of mamZ was much higher than those of the other three genes at all four sampling points in WT (Figure 5) but was only slightly different in ∆mamX (Table 2). As a result of the loss of mamX in the mutant, the transcription of mamY and ftsZ-like increased. The transcriptional disparity between mamZ and the other three genes was large in WT but much smaller in ∆mamX (Figure 6B; Table 2). Regardless of whether mamX was knocked out, the transcription level of mamZ was highest during the period of cell growth and high magnetosome synthesis. ftsZ-like showed dramatic changes of transcription level during cell growth in ∆mamX. Its level was twice as high as in WT at 6 hr, decreased 6-fold by 12 hr, increased >4-fold by 18 hr, and then gradually declined until 24 hr (Figure 6C). The phase of old cell division and new cell formation presumably places a high demand on the protein FtsZ-like. In summary, the deletion of mamX evidently resulted in higher expression of mamY and ftsZ-like, particularly at later cell growth phases, but had no major effect on the expression of mamZ. It should be noted that gene expression in the complemented strain CmamX was not identical to that in WT.
Figure 6.
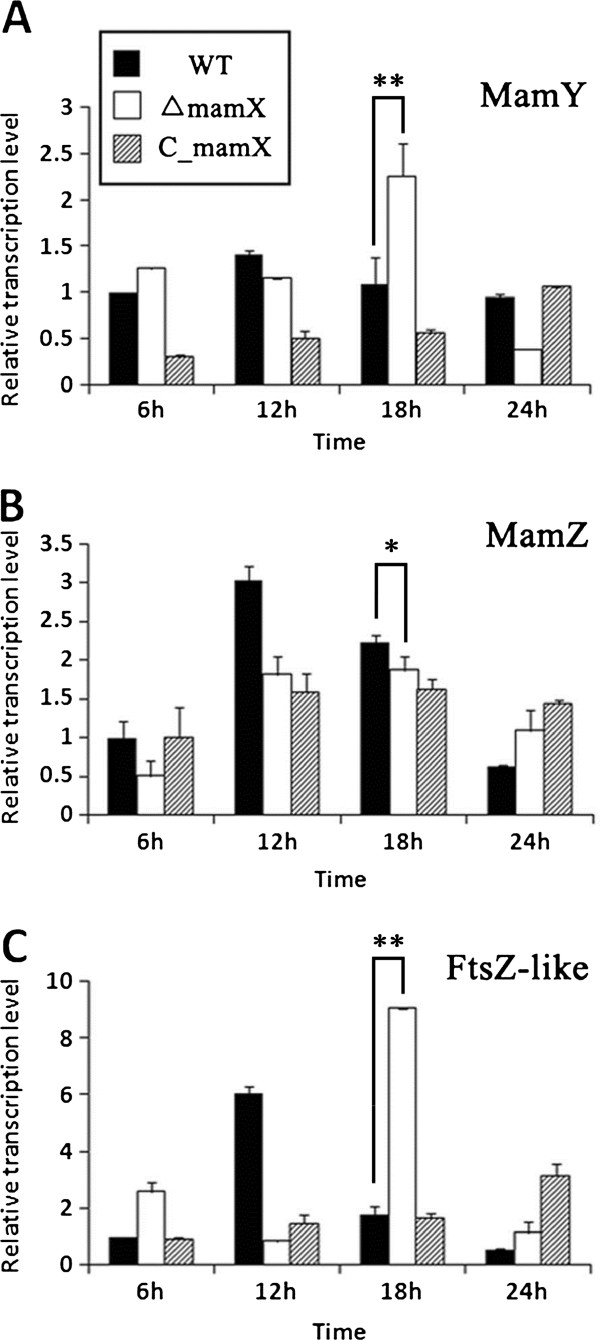
Transcription levels of four genes in WT, ΔmamX, and CmamX strains. All experiments were performed in triplicate. A: The content of MamY was similar in ∆mamX and WT at 6 and 12 hr but was twice as high in ∆mamX as in WT at 20 hr. B: Deletion of mamX had no striking effect on mamZ transcription. The transcriptional disparity between mamZ and the other three genes was large in WT but much smaller in ∆mamX. C: The level of ftsZ-like showed dramatic changes during cell growth in ∆mamX. The level was twice as high as in WT at 6 hr, decreased 6-fold by 12 hr, increased >4-fold by 18 hr, and then gradually declined until 24 hr. For the highest transcription of all four genes appeared at 18h in WT (see Figure 5), the Student t-test was used to analyze the differences between transcription levels of WT and ∆mamX at this time point. *, the difference was statistically significant (P < 0.05, by t test). **, the difference was statistically extremely significant (P < 0.01, by t test).
Discussion
MamX is involved in magnetite crystal maturation in MSR-1 cells
To elucidate the function of the highly conserved MamX protein in MTB, we constructed mamX deletion mutant (∆mamX) and complemented (CmamX) strains of M. gryphiswaldense MSR-1. For ∆mamX, the Cmag value was zero and intracellular iron content was significantly reduced, although cell growth was similar to that of WT (Figure 1). HR-TEM observations revealed that the magnetite particles in ∆mamX were irregularly shaped, small (26.11±9.92 nm), and predominantly superparamagnetic, whereas those in WT were symmetrically cuboid, large (41.25±10.46 nm), and predominantly single-domain. These findings indicate that MamX plays an essential role in the control of magnetosome morphology and that mamX is involved in magnetite crystal maturation in MSR-1.
There was a notable reduction of intracellular iron content in ∆mamX, corresponding to a crystal diameter much smaller than that in WT. The observed alteration of the crystal lattice may account for the reduction of Cmag in ∆mamX and result in a phenotype similar to that of a mamXY operon knock-out in MSR-1 [16]. Surprisingly, the mean crystal number per cell for ∆mamX (20.85±3.91) was 36% higher than that for WT (15.35±3.06). This finding may be due to the fact that crystals in the mutant strain were smaller; i.e., equivalent amounts of materials (iron, MMP, electrons, ATP, etc.) in the cells may have been capable of producing more crystals, as supported by HR-TEM observations (Figure 3E).
MamX has conserved double heme-binding motifs
MamX is conserved in not only spirillum strains such as M. gryphiswaldense MSR-1 (MGR_4149), M. magneticum AMB-1 (amb1017), and M. magnetotacticum MS-1 (MMMS1v1_36310026) but also in vibrio and cocci strains such as Magnetovibrio MV-1 (mv1g00028) and Magnetococcus sp. MC-1 (Mmc1_2238). A comparative genomic analysis showed that mamX is one of a set of 28 genes that are specifically associated with the magnetotactic phenotype [7]. The ubiquity and specific presence of MamX within MTB suggest that this protein plays a role in magnetotaxis. The results of the present study indicate that MamX is involved in magnetite crystal maturation but do not clarify its exact function. A protein sequence blast search using PROSITE (http://prosite.expasy.org/) showed that MamX contains two CXXCH heme-binding motifs that are typical of c-type cytochromes (Additional file 1: Figure S1). Similar double heme-binding motifs were found recently in the magnetosome proteins MamE, MamP, and MamT [27,28]. Site-directed mutagenesis of the two motifs in MamE resulted in the production of smaller magnetite crystals [27]. These motifs were suggested to be involved in electron transport or as a redox buffer during magnetite formation [28]. Such a function could explain the specific requirement of redox potential for magnetite formation in several MTB strains [29,30] and may be related to the function of the double heme-binding motif in MamX.
The four proteins encoded by the mamXY operon may have a close relationship
The qPCR results showed that the four genes in the mamXY operon were all highly expressed during the log phase of growth, supporting previous findings that the log phase is an essential period for MMP function and magnetosome synthesis [31]. The expression of mamZ was much higher than that of the other three genes at each of the sampling times (Figure 5; Table 2), indicating that mamZ plays a crucial role during growth. MamZ is a highly hydrophobic protein with a predicted weight of 71.7 kDa and contains a major facilitator superfamily domain (predicted by PROSITE), a ferric reductase-like transmembrane component (Pfam; http://pfam.janelia.org/search), and up to 17 transmembrane helices (HMMTOP; http://www.enzim.hu/hmmtop). It is therefore possible that MamZ is involved in ferric iron reduction, although there is no direct experimental evidence to date for such a function. The results of the relative qPCR assay indicated that deletion of mamX resulted in a notable increase in mamY and ftsZ-like transcription but had no effect on mamZ transcription. These findings suggest some redundancy among the functions of mamX, mamY, and ftsz-like.
Application of the online tool STRING (http://string-db.org) predicted interactions among the four proteins encoded by the mamXY operon (Additional file 2: Figure S2). According to this predicted network view, the four MamXY proteins undergo intrinsic interactions with each other and are also associated with certain proteins related to cell division (MGR-2076, MGR-3226, MGR-1090, MGR-2217) and to cell wall formation (MGR-0063, MGR-1112, MGR-1092, MGR-2078, MGRGRv1-0136, MGRGRv1-0133) through FtsZ-like. These associated proteins in strain AMB-1 have predicted functions similar to those in MSR-1(Additional file 3: Table S1). Further experiments are needed to test this model.
Interestingly, the phenotypes of a mamX mutant, ftsZ-like mutant, and mamXY operon deleted mutant in MSR-1 are similar in that they produce magnetosomes that are small and irregularly shaped in comparison with those of WT [16,18]. In view of the previous finding that MamGFDC proteins have partially redundant and collective functions in controlling magnetosome size [11], and the results of the present study, we propose that the four genes in the mamXY operon have redundant functions involved in the complex process of magnetosome formation. A recent study showed that a single deletion of the mamAB operon in MSR-1 resulted in the complete loss of magnetosome synthesis, whereas deletion of the conserved mms6, mamGFDC, and mamXY operons led to severe defects in the morphology, size, and organization of magnetite crystals [16]. The MamP, MamS, MamR, and MamT proteins were shown to function in the regulation of crystal number, size, and shape [14]. Magnetite biocrystallization in MTB is clearly a complex process in which many proteins are involved. It is appropriate now to consider completing the model of MMP functions and magnetosome formation that was proposed previously [14,32].
Conclusions
The results of the present study show that the MamX protein plays an important role in controlling magnetosome size, maturation, and crystal form. Previous studies have shown that a single gene deletion in mamXY and knock-out of the entire operon result in very similar phenotypic characteristics. The MamXY proteins may therefore have redundant functions involved in magnetosome synthesis. These findings are important for further elucidation of the biomineralization process in MTB.
Methods
Bacterial strains and growth conditions
The bacterial strains and plasmids used are listed in Table 3. Escherichia coli strains were cultured in Luria broth (LB) at 37°C. M. gryphiswaldense and its mutant strains were cultured in liquid optimized flask medium (OFM) at 30°C [33]. Sterile ferric citrate was added to OFM as an iron source after autoclaving. For conjugation, M. gryphiswaldense was cultured on a selection medium plate [34]. The antibiotics used were as follows: for E. coli, 50 μg/ml chloromycetin (Cm), 20 μg/ml gentamicin (Gm), 12.5 μg/ml tetracycline (Tc); for M. gryphiswaldense, the same antibiotics at concentrations of 5 μg/ml. The biomass of MSR-1 cells during culture was measured in terms of OD565. The magnetism of cells was measured as Cmag value as described previously [20].
Table 3.
Strains and plasmids used in this study
| Strains and plasmids | Description | Source or reference |
|---|---|---|
|
Strains |
|
|
|
M. gryphiswaldense MSR-1 |
wild-type, Nxr |
DSM6361 |
|
M. gryphiswaldense MSR-1 ΔmamX |
mamX deficient mutant, Nxr Gmr |
present study |
|
M. gryphiswaldense MSR-1 CmamX |
complementation of ΔmamX, NxrGmrTcr |
present study |
|
E. coli DH5α |
endA1 hsdR17 (r- m+) supE44 thi-1 recA1 gyrA (NalR) recA1 Δ (lacZYA-argF)U169 deoR [Ø80ΔdlacZ ΔM15] |
[35] |
|
E. coli S17-1 |
thi endA recA hsdR with RP4-2-Tc::Mu-Km::Tn7 integrated in chromosome, Smr |
[36] |
|
Plasmids |
|
|
| pUCGm |
pUC1918 carrying the aacC1 gene, Gmr |
[37] |
| pSUP202 |
suicide vector for M. gryphiswaldense MSR-1, CmrTcr Ampr |
[38] |
| pSUPpX2 |
pSUP202 derivative for mamX deletion, GmrCmrAmpr |
present study |
| pRK415 |
Cloning vector, pRK290 derivative, Tcr |
[39] |
| pRK415X | pRK415 derivative for mamX expression, Tcr | present study |
Construction of the mamX deletion mutant and complemented strains
The mamX deletion mutant was constructed by conjugation and subsequent homologous recombination in MSR-1. (i) The 5′ flank (1003 bp; primers: mamX-5F, CGCGGATCCAT GTTGATGAACTTTGTCAA; mamX-5R,CGAGCTCGGGAGTTCGACTGTGGTCAA3) and 3′ flank (1043 bp; primers: mamX-3F, CGAGCTCGTGCCCTGCGTGACGACCAT; mamX-3R, ACGCGTCGACAACATTCCGAGCCAGATATA) of the mamX gene in the MSR-1 genome were amplified by PCR (restriction sites are underlined). The aacC1 gene that confers Gm resistance (Gmr) was digested from plasmid pUCGm by SacI sites. (ii) The digested and purified 5′ flank, Gmr, and 3′ flank were cloned into plasmid pSUP202 by BamHI, SacI, and Sal I sites to obtain the suicide plasmid pSUPpX2. (iii) E. coli strain S17-1 transformed with pSUPpX2 was conjugated with MSR-1 as described previously [18]. The final Gmr CmS colonies, confirmed by PCR, comprised a double-crossover recombination mamX deletion mutant (∆mamX). To complement the mutant, the mamX gene (primers: X-F, 5′AACTGCAGTTGACCACAGTCGAACTCCC3′; X-R, 5′CGCGGATCCTATTCCATTG GGTGGGAGCG3′) was cloned into pRK415 by PstI and BamHI sites, and the resulting plasmid pRK415X was transferred into E. coli S17-1 (restriction sites are underlined). The subsequent conjugation was performed as described above. The Gmr Tcr colonies, confirmed by PCR, were complemented strains (termed CmamX).
Transmission electron microscopy
Cells were placed on a copper grid, washed twice with distilled water, dried, and observed by TEM (Philips Tecnai F30, Eindhoven, Netherlands). For HR-TEM (JEOL 2010, Tachikawa, Tokyo), a carbon grid was used.
Measurement of iron content
Each strain was cultured microaerobically at 30°C in OFM. After the cultures reached stationary phase, 10-ml samples were centrifuged at 10,000 x g for 2 min. The pellets were washed three times with distilled water, dried to a constant weight and nitrified in 1 ml nitric acid for 3 hr as described previously [40]. Intracellular iron content was assayed using an Inductively Coupled Plasma Optical Emission Spectrometer (ICP-OES; Optima 5300DV; Perkin Elmer, Waltham, MA, USA). The iron percentage of cells was calculated as iron content divided by dry weight.
Rock magnetic measurements
Cell cultures were centrifuged (10,000 x g) at 4°C for 5 min, and the pellets were subjected to magnetic measurements. Room-temperature hysteresis loops and first-order reversal curves (FORCs) were measured by an Alternating Gradient Force Magnetometer Model MicroMag 2900 (Princeton Measurements Corp., Princeton, NJ, USA; sensitivity 1.0×10−11 Am2) as described previously [22].
Quantitative real-time RT-PCR (qPCR)
Total RNA was purified using TRIzol Reagent (Invitrogen Corp., Carlsbad, CA, USA) according to the manufacturer’s instructions. The remaining genomic DNA in RNA preparations was degraded by DNase I (Takara, Shiga, Japan). cDNA synthesis was performed using M-MLV reverse transcriptase, dNTPs, and random primers (Promega Corp., San Luis Obispo, CA, USA) according to the manufacturer’s instructions.
A LightCycler 480 Instrument II (Roche, South San Francisco, CA, USA) was used for qPCR. The LightCycler 480 SYBR Green I Master kit (Roche) was used as the manual. In a 20-μl PCR system, the template cDNA content was set below 500 ng and that of each oligo as 0.5 μM. The reaction program consisted of initial denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 sec, annealing at 62°C for 5 sec, extension at 72°C for 15 sec, and fluorescence measurement at 76°C for 3 sec. The reactions were repeated three more times with template cDNA serially 10-fold diluted (1/10, 1/100, and 1/1000 concentrations) to ensure that the final cDNA concentrations were between 103 and 106 copies. The crossing point values (Cp) were converted to absolute copies of cDNA using standard curves. The relative expressions of the target genes were calculated by dividing the absolute number of copies of cDNA by that of the reference gene rpoc (which encodes RNA polymerase subunit ß') in the same batch reactions. The primer sequences for qPCR are listed in Additional file 4: Table S2.
Abbreviations
HR-TEM: High-resolution transmission electron microscopy; MAI: Magnetosome island; MMP: Magnetosome membrane protein; MTB: Magnetotactic bacteria; qPCR: Quantitative real-time RT-PCR; WT: Wild-type; OFM: Optimized flask medium; FORCs: First-order reversal curves.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JY and YL were involved in the study design. JY, SL, and XH performed the mutant construction. JL and YP performed the magnetic measurements. JY, SL, and LL performed all the other measurements. JY, SL, and YL performed the data analysis. JY and YL wrote the draft manuscript. All of the authors read and approved the final manuscript.
Supplementary Material
Alignments of MamX in five MTB strains. M. magneticum AMB-1 (amb1017), M. magnetotacticum MS-1 (MMMS1v1_36310026), M. gryphiswaldense MSR-1 (MGR_4149), Magnetococcus sp. MC-1 (Mmc1_2238), and Magnetovibrio MV-1 (mv1g00028). Identical residues are highlighted in dark gray and less conserved residues in light gray. The two boxes indicate two conserved CXXCH heme-binding motifs that are typical of c-type cytochromes in MamX.
Predicted interactions among MamX, MamY, MamZ, FtsZ-like, and related proteins. See Discussion/ “The four proteins encoded by the mamXY operon …” for details. Top: mamXY organized as a whole operon with the same promoter. Middle: molecular weights of MamXY proteins in MSR-1. Bottom: bioinformatic prediction of interactions within and outside of MamXY of MSR-1. The network nodes are proteins (green, MamY; brown, MamX; pink, MamZ; red, FtsZ-like; white, MamXY-associated proteins). The lines between two nodes represent predicted associations between two proteins. Stronger associations are represented by thicker lines.
Predicted proteins associated with FtsZ-like in MSR-1, and the corresponding homolog proteins in M. magneticum AMB-1.
Primer sequences used for quantitative real-time RT-PCR (qPCR).
Contributor Information
Jing Yang, Email: yj-yj2005@163.com.
Shuqi Li, Email: lsq_1116@sina.com.
Xiuliang Huang, Email: hxlool@live.cn.
Jinhua Li, Email: lijinhua@mail.iggcas.ac.cn.
Li Li, Email: llcs0229@163.com.
Yongxin Pan, Email: yxpan@mail.iggcas.ac.cn.
Ying Li, Email: yingli528@vip.sina.com.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant No. 30970041 and 31270093) and the Undergraduate Student Innovation Program of China Agricultural University (Grant No. 2010-BKS-16). The authors thank Dr. Xin Gao (Testing Center, University of Science and Technology of China) for the HR-TEM observations, and Dr. S. Anderson for English editing of the manuscript.
References
- Komeili A. Molecular mechanisms of compartmentalization and biomineralization in magnetotactic bacteria. FEMS Microbiol Rev. 2012;36:232–255. doi: 10.1111/j.1574-6976.2011.00315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jogler C, Schüler D. Genomics, genetics, and cell biology of magnetosome formation. Annu Rev Microbiol. 2009;63:501–521. doi: 10.1146/annurev.micro.62.081307.162908. [DOI] [PubMed] [Google Scholar]
- Bazylinski DA, Frankel RB. Magnetosome formation in prokaryotes. Nat Rev Microbiol. 2004;2:217–230. doi: 10.1038/nrmicro842. [DOI] [PubMed] [Google Scholar]
- Grunberg K, Wawer C, Tebo BM, Schüler D. A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl Environ Microbiol. 2001;67:4573–4582. doi: 10.1128/AEM.67.10.4573-4582.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich S, Kube M, Schubbe S, Reinhardt R, Schüler D. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J Bacteriol. 2005;187:7176–7184. doi: 10.1128/JB.187.21.7176-7184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jogler C, Kube M, Schubbe S, Ullrich S, Teeling H, Bazylinski DA, Reinhardt R, Schüler D. Comparative analysis of magnetosome gene clusters in magnetotactic bacteria provides further evidence for horizontal gene transfer. Environ Microbiol. 2009;11:1267–1277. doi: 10.1111/j.1462-2920.2009.01854.x. [DOI] [PubMed] [Google Scholar]
- Richter M, Kube M, Bazylinski DA, Lombardot T, Glockner FO, Reinhardt R, Schüler D. Comparative genome analysis of four magnetotactic bacteria reveals a complex set of group-specific genes implicated in magnetosome biomineralization and function. J Bacteriol. 2007;189:4899–4910. doi: 10.1128/JB.00119-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakaki A, Webb J, Matsunaga T. A novel protein tightly bound to bacterial magnetic particles in Magnetospirillum magneticum strain AMB-1. J Biol Chem. 2003;278:8745–8750. doi: 10.1074/jbc.M211729200. [DOI] [PubMed] [Google Scholar]
- Wang L, Prozorov T, Palo PE, Liu X, Vaknin D, Prozorov R, Mallapragada S, Nilsen-Hamilton M. Self-assembly and biphasic iron-binding characteristics of Mms6, a bacterial protein that promotes the formation of superparamagnetic magnetite nanoparticles of uniform size and shape. Biomacromolecules. 2012;13:98–105. doi: 10.1021/bm201278u. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Mazuyama E, Arakaki A, Matsunaga T. MMS6 protein regulates crystal morphology during nano-sized magnetite biomineralization in vivo. J Biol Chem. 2011;286:6386–6392. doi: 10.1074/jbc.M110.183434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffel A, Gardes A, Grunberg K, Wanner G, Schüler D. The major magnetosome proteins MamGFDC are not essential for magnetite biomineralization in Magnetospirillum gryphiswaldense but regulate the size of magnetosome crystals. J Bacteriol. 2008;190:377–386. doi: 10.1128/JB.01371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- Scheffel A, Gruska M, Faivre D, Linaroudis A, Plitzko JM, Schuler D. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature. 2006;440:110–114. doi: 10.1038/nature04382. [DOI] [PubMed] [Google Scholar]
- Murat D, Quinlan A, Vali H, Komeili A. Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proc Natl Acad Sci USA. 2010;107:5593–5598. doi: 10.1073/pnas.0914439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitraki A, Sonkaria S, Fuentes G, Verma C, Narang R, Khare V, Fischer A, Faivre D. Insight into the assembly properties and functional organisation of the magnetotactic bacterial actin-like homolog MamK. PLoS ONE. 2012;7:e34189. doi: 10.1371/journal.pone.0034189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohsse A, Ullrich S, Katzmann E, Borg S, Wanner G, Richter M, Voigt B, Schweder T, Schuler D. Functional analysis of the magnetosome island in Magnetospirillum gryphiswaldense: the mamAB operon is sufficient for magnetite biomineralization. PLoS ONE. 2011;6:e25561. doi: 10.1371/journal.pone.0025561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat D, Falahati V, Bertinetti L, Csencsits R, Kornig A, Downing K, Faivre D, Komeili A. The magnetosome membrane protein, MmsF, is a major regulator of magnetite biomineralization in Magnetospirillum magneticum AMB-1. Mol Microbiol. 2012;85:684–699. doi: 10.1111/j.1365-2958.2012.08132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Li J, Liu J, Yang J, Jiang W, Tian J, Li Y, Pan Y. Deletion of the ftsZ-like gene results in the production of superparamagnetic magnetite magnetosomes in Magnetospirillum gryphiswaldense. J Bacteriol. 2010;192:1097–1105. doi: 10.1128/JB.01292-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Arakaki A, Matsunaga T. Identification and functional characterization of liposome tubulation protein from magnetotactic bacteria. Mol Microbiol. 2010;76:480–488. doi: 10.1111/j.1365-2958.2010.07117.x. [DOI] [PubMed] [Google Scholar]
- Schüler D, Uhl R, Bäuerlein E. A simple light scattering method to assay magnetism in Magnetospirillum gryphiswaldense. FEMS Microbiol Lett. 1995;132:139–145. doi: 10.1111/j.1574-6968.1995.tb07823.x. [DOI] [Google Scholar]
- Roberts AP, Pike CR, Verosub KL. First-order reversal curve diagrams: A new tool for characterizing the magnetic properties of natural samples. J Geophys Res. 2000;105(B12):28461–28475. doi: 10.1029/2000JB900326. [DOI] [Google Scholar]
- Li J, Pan Y, Chen G, Liu Q, Tian L, Lin W. Magnetite magnetosome and fragmental chain formation of Magnetospirillum magneticum AMB-1: transmission electron microscopy and magnetic observations. Geophys J Int. 2009;177(1):33–42. doi: 10.1111/j.1365-246X.2009.04043.x. [DOI] [Google Scholar]
- Fischer H, Mastrogiacomo G, Löffler JF, Warthmann RJ, Weidler PG, Gehring AU. Ferromagnetic resonance and magnetic characteristics of intact magnetosome chains in Magnetospirillum gryphiswaldense. Earth Planet Sci Lett. 2008;270(3–4):200–208. [Google Scholar]
- Li J, Pan Y, Liu Q, Zhang Y, Menguy N, Che R, Qin H, Lin W, Wu W, Petersen N, Yang X. Biomineralization, crystallography and magnetic properties of bullet-shaped magnetite magnetosomes in giant rod magnetotactic bacteria. Earth Planet Sci Lett. 2010;293:368–376. doi: 10.1016/j.epsl.2010.03.007. [DOI] [Google Scholar]
- Li JH, Wu WF, Liu QS, Pan YX. Magnetic anisotropy, magnetostatic interactions and identification of magnetofossils. Geochem Geophys Geosyst. 2012;13(12):1–16. [Google Scholar]
- Li JH, Ge KP, Pan YX, Williams W, Liu QS, Qin HF. A strong angular dependence of magnetic properties of magnetosome chains: implications for rock magnetism and paleomagnetism. Geochem Geophys Geosyst. 2013. doi:10.1002/ggge. 20228.
- Quinlan A, Murat D, Vali H, Komeili A. The HtrA/DegP family protease MamE is a bifunctional protein with roles in magnetosome protein localization and magnetite biomineralization. Mol Microbiol. 2011;80(4):1075–1087. doi: 10.1111/j.1365-2958.2011.07631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siponen MI, Adryanczyk G, Ginet N, Arnoux P, Pignol D. Magnetochrome: a c-type cytochrome domain specific to magnetotatic bacteria. Biochem Soc Trans. 2012;40(6):1319–1323. doi: 10.1042/BST20120104. [DOI] [PubMed] [Google Scholar]
- Frankel RB, Blakemore RP. Precipitation of Fe3O4 in magnetotactic bacteria. Trans R Soc London Ser B. 1984;304:567–573. doi: 10.1098/rstb.1984.0047. [DOI] [Google Scholar]
- Zhang WJ, Chen CF, Li Y, Song T, Wu LF. Configuration of redox gradient determines magnetotactic polarity of the marine bacteria MO-1. Environ Microbiol Rep. 2010;2(5):646–650. doi: 10.1111/j.1758-2229.2010.00150.x. [DOI] [PubMed] [Google Scholar]
- Li J, Pan Y. Environmental factors affect magnetite magnetosome synthesis in Magnetospirillum magneticum AMB-1: implications for biologically controlled mineralization. Geomicrobio J. 2012;29:362–373. doi: 10.1080/01490451.2011.565401. [DOI] [Google Scholar]
- Greene SE, Komeili A. Biogenesis and subcellular organization of the magnetosome organelles of magnetotactic bacteria. Curr Opin Cell Biol. 2012;24:490–495. doi: 10.1016/j.ceb.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JB, Zhao F, Tang T, Jiang W, Tian JS, Li Y, Li JL. High-yield growth and magnetosome formation by Magnetospirillum gryphiswaldense MSR-1 in an oxygen-controlled fermentor supplied solely with air. Appl Microbiol Biotechnol. 2008;79:389–397. doi: 10.1007/s00253-008-1453-y. [DOI] [PubMed] [Google Scholar]
- Rong C, Huang Y, Zhang W, Jiang W, Li Y, Li J. Ferrous iron transport protein B gene (feoB1) plays an accessory role in magnetosome formation in Magnetospirillum gryphiswaldense strain MSR-1. Res Microbiol. 2008;159:530–536. doi: 10.1016/j.resmic.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotech. 1983;1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- Schweizer HD. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques. 1993;15:831–834. [PubMed] [Google Scholar]
- Rock JL, Nelson DR. Identification and characterization of a hemolysin gene cluster in Vibrio anguillarum. Infect Immun. 2006;74:2777–2786. doi: 10.1128/IAI.74.5.2777-2786.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen NT, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- Rong C, Zhang C, Zhang Y, Qi L, Yang J, Guan G, Li Y, Li J. FeoB2 functions in magnetosome formation and oxidative stress protection in Magnetospirillum gryphiswaldense strain MSR-1. J Bacteriol. 2012;194:3972–3976. doi: 10.1128/JB.00382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignments of MamX in five MTB strains. M. magneticum AMB-1 (amb1017), M. magnetotacticum MS-1 (MMMS1v1_36310026), M. gryphiswaldense MSR-1 (MGR_4149), Magnetococcus sp. MC-1 (Mmc1_2238), and Magnetovibrio MV-1 (mv1g00028). Identical residues are highlighted in dark gray and less conserved residues in light gray. The two boxes indicate two conserved CXXCH heme-binding motifs that are typical of c-type cytochromes in MamX.
Predicted interactions among MamX, MamY, MamZ, FtsZ-like, and related proteins. See Discussion/ “The four proteins encoded by the mamXY operon …” for details. Top: mamXY organized as a whole operon with the same promoter. Middle: molecular weights of MamXY proteins in MSR-1. Bottom: bioinformatic prediction of interactions within and outside of MamXY of MSR-1. The network nodes are proteins (green, MamY; brown, MamX; pink, MamZ; red, FtsZ-like; white, MamXY-associated proteins). The lines between two nodes represent predicted associations between two proteins. Stronger associations are represented by thicker lines.
Predicted proteins associated with FtsZ-like in MSR-1, and the corresponding homolog proteins in M. magneticum AMB-1.
Primer sequences used for quantitative real-time RT-PCR (qPCR).