Abstract
In vertebrates, trunk neural crest (NC) generates glia, neurons, and melanocytes. In addition, it yields mesectodermal derivatives (connective tissues, chondrocytes, and myofibroblasts lining the blood vessels) in the head. Previous in vitro clonal analyses of avian NC cells unraveled a hierarchical succession of highly pluripotent, followed by various intermediate, progenitors, suggesting a model of progressive restrictions in the multiple potentialities of a totipotent stem cell, as prevails in the hematopoietic system. However, which progenitors are able to self-renew within the hierarchy of the NC lineages is still undetermined. Here, we explored further the stem cell properties of quail NC cells by means of in vitro serial subcloning. We identified types of multipotent and oligopotent NC progenitors that differ in their developmental repertoire, ability to self-maintain, and response to exogenous endothelin 3 according to their truncal or cephalic origin. The most striking result is that bipotent progenitors are endowed with self-renewal properties. Thus glia–melanocyte and glia–myofibroblast progenitors behave like stem cells in that they are able both to self-renew and generate a restricted progeny. In our culture conditions, glia–myofibroblast precursors display a modest capacity to self-renew, whereas glia–melanocyte precursors respond to endothelin 3 by extensive self-renewal. These findings may explain the etiology of certain multiphenotypic NC-derived tumors in humans. Moreover, the presence of multiple stem cell phenotypes along the NC-derived lineages may account for the rarity of the “totipotent NC stem cell” and may be related to the large variety and widespread dispersion of NC derivatives throughout the body.
Keywords: multipotency, quail embryo, clonal culture, neuron, glia
The neural crest (NC) is a transient structure of the vertebrate embryo formed by the lateral borders of the neural primordium. Its constitutive cells, after losing their epithelial arrangement, migrate away through embryonic tissues to stop at elected sites where they differentiate into many various cell types. These cell types include neurons of the peripheral and enteric ganglia, their associated glial cells, and Schwann cells lining peripheral nerves. The NC is also at the origin of melanocytes and endocrine cells, such as adrenomedullary cells and calcitonin-producing cells. The NC cephalic domain yields mesenchymal cells (forming the “mesectoderm”) that differentiate into the cartilages and bones that form most of the skull and the entire facial and visceral skeleton. The mesectoderm also provides head connective-tissue cells and the vascular smooth-muscle cells associated with the vessels derived from the aortic arches and with the vessels irrigating the forebrain and face (1–3).
Several attempts aimed at elucidating how and when the different NC-lineages become segregated during ontogeny were made by using in vitro cultures of single NC cells (NCC) (4–14) or by labeling single NCC with vital dyes either in the embryo (15, 16) or in culture (17). It was established that, before and during their migration, the NCC population contains pluripotent progenitors as well as early restricted precursors. When isolated in vitro, NCC exhibit a striking heterogeneity in their developmental potentials. Some NCC yield colonies containing nearly all of the phenotypes represented in the normal NC progeny, whereas others give rise to only some of them or even to a single cell type (10).
The growth and survival factors present at the sites where NCC migrate are critical in choosing among these potentialities those that fit with the type of NC derivatives that develop in each part of the body. Such factors include BMP2/4, which drives NCC to autonomic sympathetic-like neuronal fate (18–20); neuregulin-1 and Notch ligands, which favor gliogenesis (21–23); and endothelin 3 (ET3), which promotes survival and proliferation of glial–melanocytic (GM) bipotent precursors as well as committed melanocytic and glial cells (24–26).
The NCC, therefore, may differentiate according to a developmental scheme, which is reminiscent of the hierarchical model for generation of blood cells (10, 27–29). Self-renewing hematopoietic stem cells give rise to multiple lineage-restricted precursors whose survival, proliferation, and further maturation depend on defined sets of cytokines (30–32). In the quail NC, studies have evidenced rare, highly multipotent cells in the head NC, but their self-renewing capacity has not yet been demonstrated (10). In mammals, trunk NCC comprise pluripotent progenitors for neurons, glial cells, and myofibroblasts, which self-renew in vitro and in vivo and, hence, are stem cells (13, 14). It is, however, uncertain whether these cells represent totipotent trunk NC stem cells because they were not proven to be endowed with the melanogenic potential.
In the present work, we have systematically investigated the differentiation potential of cephalic and trunk quail NCC derived from single-cell primary cultures subjected to successive subcloning. Five cell phenotypes representing the main NC-derived lineages were examined, leading us to characterize multipotent mesectodermal-neural progenitors in the cephalic NC and a totipotent trunk NCC lying upstream of previously identified precursors. Moreover, we show that bipotent GM precursors and progenitors for glia–myofibroblasts (GF) self-renew in vitro.
Materials and Methods
Cultures of NCC. NCC were isolated from explant cultures of trunk and mesencephalic–anterior rhombencephalic neural tubes that were taken from quail embryos at the 20–25 and 6–8 somite stage, respectively. After 24 h of culture, migratory NCC were harvested for subsequent cell cloning, as described (9, 26). Individual cells were seeded under microscopic control in 96-well culture plates (Nunc) either on rat-tail collagen (Biomedical Technologies, Stoughton, MA) or on a feeder layer of growth-inhibited mouse 3T3 fibroblasts (9). Colonies were maintained in cloning medium, alone or supplemented with 100 nM ET3 (Sigma) (26), and incubated at 37°C in a humidified 5% CO2/95% air atmosphere.
Subcloning Procedure. Subcloning was exclusively performed on colonies cultured on collagen to avoid contamination by cells of the feeder layer. NCC were harvested from day 9 cultures (containing 50–1,000 cells per clone) after treatment with trypsin/EDTA (Sigma). For each subcloned colony, a minimum of 40 subcultures was performed. This subcloning procedure was repeated until it became precluded because of the small number of cells per colony. The identity of the colonies used for subcloning was determined by analyzing the phenotypes of the cells remaining in the suspension.
Immunocytochemistry and Culture Analysis. NCC grown on 3T3 fibroblasts were detected by bisbenzimide nuclear staining with Hoeschst 33342 (Sigma) (9). The phenotypes of NCC were identified by using lineage-specific markers. Detailed procedures are described in refs. 26, 36, and 37. Pigment cells were recognized by the presence of melanin and unpigmented melanocytes by staining with the melanoblast–melanocyte early marker (MelEM) mAb (33). Cartilage aggregates were identified by phase–contrast microscopy, and glial cells were identified by immunostaining with the Schwann cell myelin protein (SMP) marker (11, 34). Nonadrenergic and adrenergic neurons were labeled by using Abs against 200-kDa neurofilament protein (Sigma) and quail tyrosine hydroxylase (35), respectively. Myofibroblasts were identified by immunoreactivity to α-smooth-muscle actin (α-SMA; clone 1A4, Sigma). Fluorescence was observed under an X70 microscope (Olympus, Melville, NY). Differences in cell and colony numbers between ET3-treated and control cultures were analyzed by χ2 test (GraphPad, San Diego) and considered to be statistically significant at P < 0.05.
Results
Characterization of Cephalic and Trunk NCC Repertoire. To characterize the extent of NCC pluripotency, we analyzed the progeny of single cephalic and trunk quail NCC grown on a feeder layer of 3T3 fibroblasts, which favors all main phenotypes of NCC to be expressed (9–11). The influence of ET3 on the diverse precursors was investigated also because this factor proved to promote melanocytic and glial outcome in trunk NCC (24, 26).
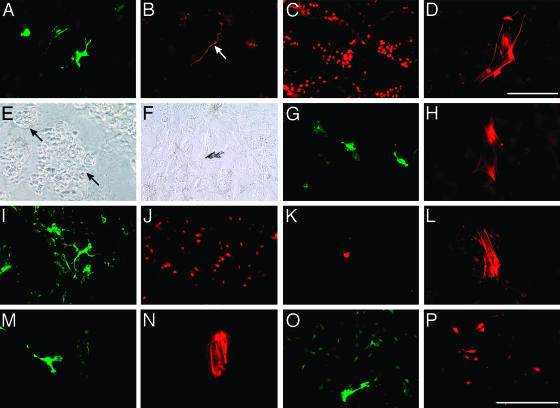
Cephalic NCC in control medium provided 17% of clonogenic cells that generated progeny containing 50–8,000 cells in day 9 cultures. Eight types of clonogenic precursors were recovered (Table 1). Unipotent glial and bipotent glial–neuronal (GN) cells were the most frequent. In the latter case, GN precursors gave rise to mostly nonadrenergic neurons. Other precursors produced two (GM and GF) or three (GMC and GNM) distinct phenotypes. Several highly pluripotent progenitors unknown from experiments (7, 10) were found also, such as GNMF (Fig. 1 A–D) and GMFC (forming multiphenotypic clones with cartilage but not neurons) (Fig. 1 E–H).
Table 1. Phenotypic analysis of cephalic and trunk NC precursors.
| % of clones from total clone number
|
||||
|---|---|---|---|---|
| Cephalic NCC
|
Trunk NCC
|
|||
| Precursor | Control | ET3 | Control | ET3 |
| G | 36 | 50 | 45 | 44 |
| M | 0 | 0 | 0 | 1 |
| F | 0 | 0 | 0 | 0 |
| GN | 36 | 25 | 43 | 7.5* |
| GM | 11 | 3.5 | 2 | 34.5* |
| GF | 2.8 | 18 | 4 | 6 |
| GNM | 5.5 | 0 | 0 | 3 |
| GNF | 0 | 0 | 4 | 1 |
| GMF | 0 | 0 | 2 | 2 |
| GMC | 2.8 | 0 | 0 | 0 |
| GNMF | 2.8 | 0 | 0 | 1 |
| GMFC | 2.8 | 0 | 0 | 0 |
| U | 0 | 3.5 | 0 | 0 |
Control and ET3-treated clonal cultures of NCC grown on 3T3 feeder-layers were analyzed for cell phenotypes. Values were obtained from five and four experiments for cephalic (36 control and 28 treated colonies) and trunk (51 control and 104 treated colonies) NCC, respectively. Statistically significant differences between ET3-treated and control cultures are indicated. *, P < 0.0005. U, unidentified.
Fig. 1.
Phenotypic analysis of NCC in primary clones on 3T3 cells (A–L) and in subclones on collagen (M–P). (A–H) Cephalic NCC in control medium. (A–D) GNMF clone with cells expressing SMP (A), neurofilament (B, arrow), MelEM (C), and α-SMA (D) antigens. (E–H) GMFC clone showing cartilage islets (arrows in E, phase–contrast), a pigment cell (F, bright field), and cells positive for SMP (G) and α-SMA (H). (I–L) Trunk NCC in presence of ET3. GNMF clone with cells expressing SMP (I), MelEM (J), tyrosine hydroxylase (K), and α-SMA (L) is shown. (M and N) Cephalic NCC secondary cloning in the presence of ET3. GF subclone with cells immunoreactive for SMP (M) and α-SMA (N) is shown. (O and P) Trunk NCC tertiary cloning in the presence of ET3. GM subclone with cells positive for SMP (O) and MelEM (P) is shown. (Scale bar in P represents 50 μm; scale bar in D is 15 μm for D only.)
In the presence of ET3, the outcome of cephalic NCC did not change significantly in terms of cloning efficiency and cell phenotypes (Table 1). However, ET3 increased the proportion of “large” colonies (>500 cells) to 69% (as compared with 32% in controls; P < 0.001). In addition to precursors of identified phenotypes, NCC included a few cells that generated unidentified progeny (negative for all of the markers tested); such progeny may be formed by not-yet-differentiated cells or may belong to NC sublineages other than those identified here.
Trunk NCC in control medium generated six different types of colonies deprived of cartilage (Table 1). The most frequent derived from glial and GN precursors. The latter produced mainly neurons of adrenergic phenotype (Fig. 1K). Myofibroblasts derived from GF, GMF, and GNF precursors. ET3 significantly enhanced clonal efficiency (65% versus 32% in control cultures; P < 0.0001), cell proliferation (59% of large clones versus 29% in controls; P < 0.0001), and development of GM precursors while reducing the frequency of GN cells, as described (26). Exposure to ET3 also unraveled the presence of pluripotent GNM and GNMF progenitors (Fig. 1 I–L), the latter being, thus, able to generate all of the main trunk NC derivatives.
Propagation of NC Precursors by Serial Subcloning. To evaluate the ability of the various precursors to be propagated, NCC were serially subcloned in collagen-coated wells in the absence or presence of ET3, and the resulting subclones were analyzed after 9 days for cell phenotypes.
In control medium, cephalic NCC exhibited 27% of clonal efficiency (versus 17% on 3T3 cells) and generated primary clones of smaller size than on 3T3 cells (3.5% classified as large). They did not produce neurons or cartilage, and only unipotent or bipotent progenitors were recorded (Table 2). Altogether, myofibroblastic and GF precursors provided 80% of the clones, whereas glial, unidentified, and GM precursors developed in low proportions. Subsequent subcloning of primary progeny yielded clones II and III without significant change in clonal efficiency, but the total number of cells per clone decreased rapidly to a few cells. GM and unidentified progenitors propagated until cloning II but were absent in cloning III. Glial, GF, and myofibroblasts were the only types of progenitors maintained until cloning III, at which myofibroblast precursors became prominent (yielding 71% of total colonies) (Table 2).
Table 2. Propagation of cephalic NC precursors.
| % of total clones (no. of cells per clone)
|
|||||||
|---|---|---|---|---|---|---|---|
| Control
|
ET3
|
||||||
| Colony | I | II | III | I | II | III | IV |
| G | 7.5 (50) | 9 (5) | 4 (4) | 10 (100) | 41.5† (50) | 18.5‡ (10) | 0 |
| M | 0 | 15.5 (1) | 0 | 4 (80) | 17† (80) | 11 (15) | 67‡ (3) |
| F | 49 (15) | 40 (15) | 71* (1) | 40 (15) | 9.5‡ (15) | 17 (10) | 0 |
| GM | 5 (200) | 4.5 (4) | 0 | 1.5 (200) | 4.5 (200) | 34‡ (30) | 33 (8) |
| GF | 31 (80) | 22 (60) | 25 (3) | 36.5 (50) | 15‡ (30) | 13 (15) | 0 |
| U | 7.5 (1) | 9 (1) | 0 | 8 (1) | 12.5 (1) | 6.5 (1) | 0 |
The types of colony derived from cephalic NCC through serial subcloning were analyzed and quantified (% of total clone number) at successive clonings I-IV. The mean cell number per colony is indicated in parentheses. The number of colonies used for subcloning was 3 (cloning I and II) in control medium and 16 (cloning I), 10 (cloning II), and 1 (cloning III) in the presence of ET3. Values were obtained from six independent experiments. Total numbers of clones are as follows: 78, 45, and 24 for control I-III, respectively, and 78, 202, 76, and 24 for ET3 I-IV, respectively. Statistically significant differences between two subsequent clonings are indicated. *, P < 0.05; †, P < 0.001; ‡, P < 0.0001.
In the presence of ET3, cephalic NCC primary progeny did not differ significantly from the controls in cloning efficiency, cell proliferation, and precursor types, except that some melanocytic precursors were recovered (Table 2). Four cloning rounds were successful. The proliferation rate decreased less than in control medium in the successive steps. The clonal efficiency rose from 24% to 54.5% between cloning I and IV (P < 0.001). Myofibroblastic and GF precursors (Fig. 1 M and N) decreased in number between cloning I and II, but they were still present in cloning III, as were glial and unidentified precursors. By contrast, GM and melanocytic progenitors that were present in very low proportions in primary progeny were subsequently expanded until cloning IV (Table 2).
In control medium, trunk NCC showed 13.5% (versus 32% on 3T3 cells) of clonal efficiency and yielded small primary colonies (≤50 cells). Most of them derived from glial, melanocytic, GF, and GM precursors. Some myofibroblastic, unidentified, and GMF precursors developed also (Table 3). Thus, melanocytic and GM progenitors were more frequent than myofibroblastic and GF progenitors. Trunk NCC showed very little ability to propagate in control medium, yielding a few tiny melanocytic and unidentified secondary subclones only (data not shown).
Table 3. Propagation of trunk NC precursors.
| % of total clones (no. of cells per clone)
|
||||||
|---|---|---|---|---|---|---|
| Control
|
ET3
|
|||||
| Colony type | I | I | II | III | IV | V |
| G | 29.5 (15) | 13 (30) | 20 (30) | 20.5 (30) | 23 (5) | 0 |
| M | 23.5 (8) | 4.5 (40) | 18* (30) | 22 (30) | 7‡ (10) | 80 (10) |
| F | 6 (8) | 16 (15) | 5† (5) | 2 (2) | 0 | 0 |
| GM | 17.5 (50) | 45.5 (1500) | 50 (150) | 52 (80) | 66 (30) | 20 (10) |
| GF | 11.5 (30) | 15 (40) | 4.5* (10) | 0 | 0 | 0 |
| GMF | 6 (50) | 3 (2000) | 0 | 0 | 0 | 0 |
| U | 6 (6) | 3 (30) | 2.5 (20) | 3.5 (1) | 4 (2) | 0 |
The types of colony produced by trunk NCC through serial subcloning were quantified as the percentage of total clone number at successive cloning I to V. The mean cell number per clone is indicated in parentheses. In control medium, primary colonies could not be propagated by subcloning (data not shown). In presence of ET3, the number of colonies used for successive subclonings was 13 (cloning I), 10 (cloning II), 6 (cloning III), and 1 (cloning IV). Total numbers of clones are as follows: 17 (for control I) and 68, 195, 174, 112 and 15 (for ET3 I-V, respectively). Values were obtained from six independent experiments. Differences between two subsequent rounds of cloning are indicated. *, P < 0.05; †, P < 0.001; ‡, P < 0.0001.
In the presence of ET3, the number of clonogenic trunk NCC rose to 42.5%. Similar types of primary progeny, but a higher frequency of GM precursors, were found, as compared with control cultures. ET3 also increased the proportion of large clones (29.5%), mostly originating from GM and GMF cells (Table 3). Trunk NCC exposed to ET3 could be cloned until five successive times while maintaining a high clonal efficiency (Table 3). Certain founder cells disappeared at different times along the subsequent clonings: first, GMF (at cloning II); second, GF (at cloning III); and third, myofibroblasts (at cloning IV). Only melanocytic (80% of the clones) and GM (20% of the clones) (Fig. 1 O and P) founder cells were still present at cloning V (Table 3).
Evidence for Self-Renewal of Multipotent NCC. The results described above, showing that GM and GF progenitors can be serially propagated, are consistent with the self-renewal of these progenitors. In a subsequent step, we considered the successive clones generated by identified multipotent founders and examined whether they contained cells with the same multiple potentials. Quantitative analysis of the self-renewal of identified cephalic and trunk progenitors is provided by Tables 4–8 and Supporting Text, which are published as supporting information on the PNAS web site.
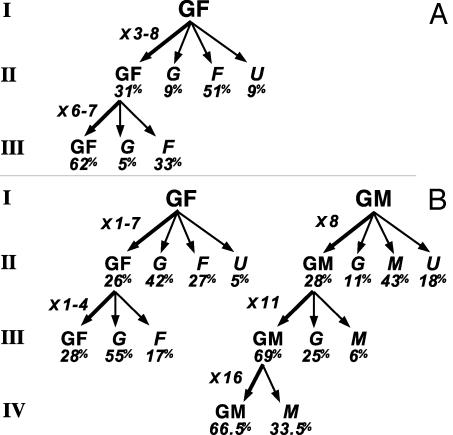
Analysis of the propagation of cephalic GF and GM precursors is summarized in Fig. 2 (see also Tables 4 and 5 and Supporting Text).
Fig. 2.
Self-renewal of cephalic NCC. Primary clones (I) from GF and GM founders were subcloned serially in control (A) and ET3-supplemented (B) media. At each generation, the different types of clones (and mean percentage of total clones), as well as the fold increase in the number of GF or GM produced by self-renewing founder cells, are indicated. The subcloned colonies were two GF I and GF II (control); and five GF I, two GF II, and one GM at each subcloning I–III (ET3) (see Tables 4 and 5 and Supporting Text).
In control medium (Fig. 2 A), two GF primary clones (I) generated three and eight GF subclones (II); hence, the corresponding GF founders have expanded between cloning I and II. Further subcloning of GF secondary clones (II) gave rise to several GF subclones (forming 62% of clones III). This result indicates that initial GF founder cells have been amplified by several symmetric cell divisions. Because self-renewing GF progenitors gave rise to restricted (glial, myofibroblastic, and unidentified) progeny, they must also have undergone at least one asymmetric stem cell division.
In presence of ET3 (Fig. 2B), identified GF primary colonies generated 26% (versus 31% in control medium) of GF subclones (II) and, therefore, derived from self-renewing founder cells. One GF precursor self-renewed until cloning III. Among restricted progeny derived from GF founders, glial cells prevailed over myofibroblastic cells (42% and 55% versus 27% and 17% of subclones II and III, respectively; Fig. 2B), whereas the reverse was found in controls (Fig. 2 A).
Self-renewal of identified GM precursors was evidenced only in the presence of ET3 (Fig. 2B). GM founder cells generated GM clones along three successive subclonings with high frequency (69% and 66.5% of clones III and IV, respectively), indicating that the clonogenic cells were strongly biased to the GM type.
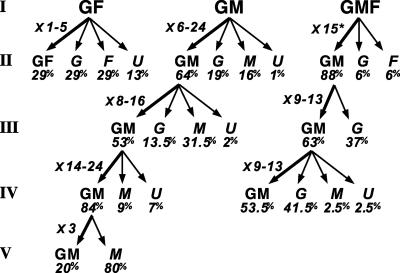
Similar analysis was performed on trunk GF, GM, and GMF precursors (see Tables 6–8 and Supporting Text) subcloned in presence of ET3. In control medium, none of these precursors were propagated (Table 3).
The progeny of identified GF founder cells yielded one to five small GF secondary colonies (forming 29% of clones II) (Fig. 3).
Fig. 3.
Self-renewal of trunk NC precursors in presence of ET3. The progeny of identified GF, GM, and GMF precursors was analyzed during serial subcloning. The different types of subclones (and mean percentage of total clones), as well as the fold increase in the number of parental-like progeny produced by individual founders, are indicated. The increase in GM progeny is given for GMF founders (*). The subcloned colonies were as follows: three GF I; six GM I, five GM II, two GM III, and one GM IV; and one GMF (I) and two GM at cloning II and III (see Tables 6–8 and Supporting Text).
By contrast, subcloning of GM primary clones produced GM colonies at high frequency; GM colonies, thus, represented 64%, 53%, and 84% of total colonies recorded after three subsequent subclonings (Fig. 3). GM stem cells could self-maintain with a significant rate of cell divisions up to cloning V and were highly expanded by successive cloning.
Also, we analyzed the progeny of trunk GMF precursors in presence of ET3. Subcloning one GMF primary clone produced no parental-like progeny but a high proportion (88%) of GM secondary founder cells that self-maintained at high frequency and proliferation rate over two subsequent rounds of cloning (Fig. 3).
Discussion
In this work, we have further investigated the developmental repertoire of avian cephalic and trunk NCC to determine which subsets of precursors display stem cell properties (i.e., multipotency and self-renewal). By analyzing the cell types generated by serial propagation of single quail NCC, we found highly multipotent as well as oligopotent progenitors. Moreover, we have shown that intermediate bipotent precursors are able to self-renew in vitro. In addition, intrinsic differences in repertoire expression, self-renewal, proliferation potential, and response to the ET3 environmental factor were demonstrated between NCC, depending on the level of the neuraxis (cephalic or truncal) from which they originate at the onset of migration.
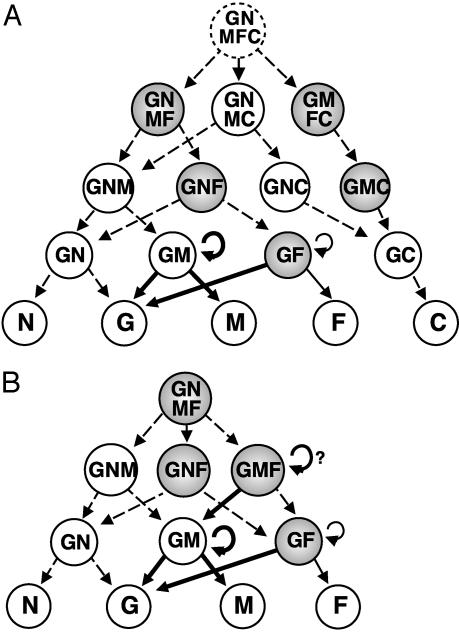
In the cephalic NC, the existence of multipotent progenitors that are able to yield glial cells, neurons, melanocytes, and cartilage (GNMC precursors) (10), as well as others giving rise to neurons, cartilage, and myofibroblasts (7), were demonstrated. Here, we provide evidence for other types of highly pluripotent cells that had not yet been evidenced: GNMF and GMFC precursors. These findings reinforce the idea that mesectodermal lineages (such as chondrocytes and myofibroblasts) are not yet segregated from the neural and melanocytic NC lineages at the early migratory stage. This diversified pluripotency makes likely the existence of a rare GNMFC totipotent stem cell lying upstream of the various progenitors that have already been identified in the cephalic NC (Fig. 4A).
Fig. 4.
Model for cell-lineage segregation in cephalic (A) and trunk (B) NC. Progenitors are classified according to the number of developmental potentials, and those identified for the first time in the present study are shown in shaded circles. Data from in vitro clonal analysis (refs. 9–12 and 26 and this study) indicate that differentiation of neurons, glia, melanocytes, myofibroblasts, and cartilage from totipotent quail NCC involves progressive developmental restrictions yielding several intermediate oligopotent precursors. Here, we identified a highly pluripotent GNMF precursor that could be a totipotent trunk NCC. The existence of a totipotent cephalic progenitor (GNMFC) and the filiations (dashed arrows) are hypothetical. The GM and GF precursors behave as stem cells. GF precursors self-renew independently of ET3 (curved arrows), whereas GM (and possibly GMF) progenitors display ET3-induced high self-renewal activity (bold curved arrows). ET3 also promotes GM differentiation into glial and melanocytic cells and biased GF stem cell progeny toward a glial fate (bold arrows).
Unlike their cephalic counterparts grown under the same conditions, trunk NCC did not generate cartilage, thus providing additional evidence for their lack of skeletogenic potential (see ref. 38 for a review), although some recent experiments of long-term in vitro cultures challenged this idea (39, 40). The trunk NC progenitors identified here are, thus, very similar to the nonchondrogenic ones isolated from cephalic NCC (Fig. 4B). The neurons generated by trunk NC precursors, however, belong mostly to the adrenergic type, whereas nonadrenergic neurons prevailed in cephalic NCC progeny. Although myofibroblasts do not differentiate from trunk NCC developing in vivo (see refs. 1 and 2 for reviews), they were shown to do so in culture (13, 18, 41) or after unilateral transplantation into the embryonic head region (42). The present study has revealed four types of oligopotent myofibroblast progenitors: GF, GMF, and GNMF, as well as the GNF precursors that had been characterized in mammals (18, 21, 41). The GNMF progenitors are, thus, the most pluripotent trunk NCC that have been characterized.
Identified cephalic and trunk NC progenitors can be ordered according to their differentiation options, suggesting filiations from highly pluripotent to more and more restricted progenitors (Fig. 4). It is remarkable that, of all possible combinations of these options, those combinations devoid of the potential to yield glia were never found. All of the 12 different pluripotent (i.e., with more than one option) progenitors identified so far in cephalic and trunk NCC, thus, share the potential to give rise to glial cells, suggesting that the determination of the “NC-state” primarily and necessarily involves the potential to become glia (58). This hypothesis is in agreement with the finding that glial cells occupy a central position in the CNS lineage because they can switch to either the stem cell or the neuronal phenotype (43).
Serial subcloning led us to gain insight on how some of the precursors are amplified during transition from pluripotent to differentiated NCC. The present experiments did not allow us to examine whether the most highly pluripotent NCC are able to self-maintain because these precursors could be identified only from clonal cultures carried out on 3T3 feeder layers. The drawback of this system is that it does not permit us to isolate cells for subculture. Subcloning was achieved only from cultures on collagen-coated dishes devoid of feeder layers. We could demonstrate that oligopotent GF, GM, and possibly GMF precursors self-renew in vitro. Moreover, GF and GM stem cell properties varied in growth-factor requirement and also with the axial level of precursor origin.
GF progenitors were present at low frequency and showed a limited ability to propagate in trunk-derived NCC. They were significantly enriched and could self-renew over two generations independently of the presence of ET3 in cephalic NCC. ET3, however, biased glial and myofibroblastic outcome in GF progeny toward a glial fate. Factors different from ET3, therefore, are likely to control self-renewal and differentiation of myofibroblastic NC precursors. Several candidate signaling molecules are platelet-derived growth factors (44, 45), ET1 (46–49), or members of the TGFβ/BMPs family (18, 50–52), which were shown to influence the development of NCC into vascular smooth-muscle cells.
In contrast to GF, GM progenitors strictly depended on ET3 to self-renew and could undergo many cell divisions over several rounds of cloning. Such dependency is consistent with our findings (26) that GM precursors of trunk origin are a privileged target for the survival- and proliferation-promoting effects of ET3 in vitro. The strong potential of ET3 to trigger expansion of GM stem cells may also underlie the capacity of differentiated glial cells and melanocytes to reverse their phenotypic program and transdifferentiate reciprocally in vitro (36, 37, 53). GM cells isolated from the cephalic NC showed a delayed response to ET3 because they were unaffected by treatment during the first cloning. Nevertheless, these GM precursors, which formed a small subset of the initial clonogenic cell population, were highly expanded by means of stem cell divisions over subsequent cloning, similar to the GM stem cells of trunk NC origin.
That oligopotent NCC are able to self-renew has several implications for normal and altered NCC development. An extended proliferative potential, thought to enhance susceptibility to accumulate mutations leading to transformation, may account for the possibility that GM stem cells are the target of transformation in several human tumors involving overproduction of glia and melanocytes, such as neurofibromas and melanotic Schwannomas (54, 55). Similarly, GF stem cells could be at the origin of tumors that affect both neural and mesectodermal cells, such as Ewing's sarcomas (56, 57). During ontogeny, an initially limited pool of NCC must be expanded to contribute large populations of distinct differentiated cells in the multiple NC-derived tissues. Therefore, aside from the putative self-maintenance of rare multipotent stem cells, an alternative strategy for expanding particular NC sublineages locally may involve the amplification of self-renewing intermediate precursors under the control of environmental signals. How environmental signals interact with intrinsic determinants (58, 59) to regulate self-renewal of the various NC stem cells remains to be understood.
Supplementary Material
Acknowledgments
We thank S. Gournet, M. Fromaget, and F. Beaujean for help in preparing the illustrations. This work was supported by the Centre National de la Recherche Scientifique, the Collège de France, and Institut National de la Santé et de la Recherche Médicale Grant N°4CS13H. A.T. was the recipient of a postdoctoral fellowship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Abbreviations: NC, neural crest; NCC, NC cells; ET3, endothelin 3; SMP, Schwann cell myelin protein; MelEM, melanoblast–melanocyte early marker; G, glia(l); F, myofibroblast(ic); M, melanocyte/melanocytic; N, neuron(al); C, cartilage; α-SMA, α-smooth-muscle actin.
References
- 1.Le Douarin, N. M. (1982) The Neural Crest (Cambridge Univ. Press, Cambridge).
- 2.Le Douarin, N. M. & Kalcheim, C. (1999) The Neural Crest (Cambridge Univ. Press, New York), 2nd Ed..
- 3.Etchevers, H. C., Vincent, C., Le Douarin, N. M. & Couly, G. F. (2001) Development (Cambridge, U.K.) 128, 1059–1068. [DOI] [PubMed] [Google Scholar]
- 4.Sieber-Blum, M. & Cohen, A. M. (1980) Dev. Biol. 80, 96–106. [DOI] [PubMed] [Google Scholar]
- 5.Sieber-Blum, M. (1989) Science 243, 1608–1611. [DOI] [PubMed] [Google Scholar]
- 6.Sieber-Blum, M. (1991) Neuron 6, 949–955. [DOI] [PubMed] [Google Scholar]
- 7.Ito, K. & Sieber-Blum, M. (1991) Dev. Biol. 148, 95–106. [DOI] [PubMed] [Google Scholar]
- 8.Ito, K., Morita, T. & Sieber-Blum, M. (1993) Dev. Biol. 157, 517–525. [DOI] [PubMed] [Google Scholar]
- 9.Baroffio, A., Dupin, E. & Le Douarin, N. M. (1988) Proc. Natl. Acad. Sci. USA 85, 5325–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baroffio, A., Dupin, E. & Le Douarin, N. M. (1991) Development (Cambridge, U.K.) 112, 301–305. [DOI] [PubMed] [Google Scholar]
- 11.Dupin, E., Baroffio, A., Dulac, C., Cameron-Curry, P. & Le Douarin, N. M. (1990) Proc. Natl. Acad. Sci. USA 87, 1119–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupin, E. & Le Douarin, N. M. (1995) Dev. Biol. 168, 529–548. [DOI] [PubMed] [Google Scholar]
- 13.Stemple, D. L. & Anderson, D. J. (1992) Cell 71, 973–985. [DOI] [PubMed] [Google Scholar]
- 14.Morrison, S. J., White, P. M., Zock, C. & Anderson, D. J. (1999) Cell 96, 737–749. [DOI] [PubMed] [Google Scholar]
- 15.Bronner-Fraser, M. & Fraser, S. E. (1988) Nature 335, 161–164. [DOI] [PubMed] [Google Scholar]
- 16.Bronner-Fraser, M. & Fraser, S. E. (1989) Neuron 3, 755–766. [DOI] [PubMed] [Google Scholar]
- 17.Henion, P. D. & Weston, J. A. (1997) Development (Cambridge, U.K.) 124, 4351–4359. [DOI] [PubMed] [Google Scholar]
- 18.Shah, N. M., Groves, A. K. & Anderson, D. J. (1996) Cell 85, 331–343. [DOI] [PubMed] [Google Scholar]
- 19.Shah, N. M. & Anderson, D. J. (1997) Proc. Natl. Acad. Sci. USA 94, 11369–11374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider, C., Wicht, H., Enderich, J., Wegner, M. & Rohrer, H. (1999) Neuron 24, 861–870. [DOI] [PubMed] [Google Scholar]
- 21.Shah, N. M., Marchionni, M. A., Isaacs, I., Stroobant, P. & Anderson, D. J. (1994) Cell 77, 349–360. [DOI] [PubMed] [Google Scholar]
- 22.Morrison, S. J., Perez, S. E., Qiao, Z., Verdi, J. M., Hicks, C., Weinmaster, G. & Anderson, D. J. (2000) Cell 101, 499–510. [DOI] [PubMed] [Google Scholar]
- 23.Wakamatsu, Y., Maynard, T. M. & Weston, J. A. (2000) Development (Cambridge, U.K.) 127, 2811–2821. [DOI] [PubMed] [Google Scholar]
- 24.Lahav, R., Ziller, C., Dupin, E. & Le Douarin, N. M. (1996) Proc. Natl. Acad. Sci. USA 93, 3892–3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stone, J. G., Spirling, L. I. & Richardson, M. K. (1997) J. Cell Sci. 110, 1673–1682. [DOI] [PubMed] [Google Scholar]
- 26.Lahav, R., Dupin, E., Lecoin, L., Glavieux, C., Champeval, D., Ziller, C. & Le Douarin, N. M. (1998) Proc. Natl. Acad. Sci. USA 95, 14214–14219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson, D. J. (1989) Neuron 3, 1–12. [DOI] [PubMed] [Google Scholar]
- 28.Baroffio, A. & Blot, M. (1992) J. Cell Sci. 103, 581–587. [DOI] [PubMed] [Google Scholar]
- 29.Anderson, D. J. (1997) Trends Genet. 13, 276–280. [DOI] [PubMed] [Google Scholar]
- 30.Metcalf, D. (1989) Nature 339, 27–30. [DOI] [PubMed] [Google Scholar]
- 31.Metcalf, D. (1991) Proc. Natl. Acad. Sci. USA 88, 11310–11314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weissman, I. L., Anderson, D. J. & Gage, F. (2001) Annu. Rev. Cell Dev. Biol. 17, 387–403. [DOI] [PubMed] [Google Scholar]
- 33.Nataf, V., Mercier, P., Ziller, C. & Le Douarin, N. M. (1993) Exp. Cell Res. 207, 171–182. [DOI] [PubMed] [Google Scholar]
- 34.Dulac, C., Cameron-Curry, P., Ziller, C. & Le Douarin, N. M. (1988) Neuron 1, 211–220. [DOI] [PubMed] [Google Scholar]
- 35.Fauquet, M. & Ziller, C. (1989) J. Histochem. Cytochem. 37, 1197–1205. [DOI] [PubMed] [Google Scholar]
- 36.Dupin, E., Glavieux, C., Vaigot, P. & Le Douarin, N. M. (2000) Proc. Natl. Acad. Sci. USA 97, 7882–7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dupin, E., Real, C., Glavieux-Pardanaud, C. & Le Douarin, N. M. (2003) Proc. Natl. Acad. Sci. USA 100, 5229–5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santagati, F. & Rijli, F. M. (2003) Nat. Rev. Neurosci. 423, 806–818. [DOI] [PubMed] [Google Scholar]
- 39.McGonnell, I. M. & Graham, A. (2002) Curr. Biol. 12, 767–771. [DOI] [PubMed] [Google Scholar]
- 40.Abzhanov, A., Tzahor, E., Lassar, A. B. & Tabin, C. J. (2003) Development (Cambridge, U.K.) 130, 4567–4579. [DOI] [PubMed] [Google Scholar]
- 41.Jain, M. K., Layne, M. D., Watanabe, M., Chin, M. T., Feinberg, M. W., Sibinga, N. E. S., Hsieh, C.-M., Yet, S.-F., Stemple, D. L. & Lee, M.-E. (1998) J. Biol. Chem. 273, 5993–5996. [DOI] [PubMed] [Google Scholar]
- 42.Nakamura, H. & Ayer-le Lièvre, C. S. (1982) J. Embryol. Exp. Morphol. 70, 1–18. [PubMed] [Google Scholar]
- 43.Doetsch, F. (2003) Nat. Neurosci. 6, 1127–1134. [DOI] [PubMed] [Google Scholar]
- 44.Lindahl, P., Johansson, B. R., Leveen, P. & Betsholtz, C. (1997) Science 277, 242–245. [DOI] [PubMed] [Google Scholar]
- 45.Hoch, R. V. & Soriano, P. (2003) Development (Cambridge, U.K.) 130, 4769–4784. [DOI] [PubMed] [Google Scholar]
- 46.Kurihara, Y., Kurihara, H., Oda, H., Maemura, K., Nagai, R., Ishikawa, T. & Yasaki, Y. (1995) J. Clin. Invest. 96, 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yanagisawa, H., Hammer, R. E., Richardson, J. A., Williams, S. C., Clouthier, D. E. & Yanagisawa, M. (1998) J. Clin. Invest. 102, 22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ballard, V. L. T. & Mikawa, T. (2002) Dev. Biol. 251, 167–177. [DOI] [PubMed] [Google Scholar]
- 49.Clouthier, D., Williams, S. C., Hammer, R. E., Richardson, J. A. & Yanagisawa, M. (2003) Dev. Biol. 261, 506–519. [DOI] [PubMed] [Google Scholar]
- 50.Topouzis, S. & Majesky, M. W. (1996) Dev. Biol. 178, 430–445. [PubMed] [Google Scholar]
- 51.Hirschi, K. K., Rohovsky, S. A. & D'Amore, P. A. (1998) J. Cell Biol. 141, 805–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Darland, D. C. & D'Amore, P. A. (2001) Curr. Top. Dev. Biol. 52, 107–149. [DOI] [PubMed] [Google Scholar]
- 53.Le Douarin, N. M. & Dupin, E. (2003) Curr. Opin. Genet. Dev. 13, 1–8. [DOI] [PubMed] [Google Scholar]
- 54.Riccardi, M. (1991) N. Engl. J. Med. 324, 1283–1285. [DOI] [PubMed] [Google Scholar]
- 55.Ferner, R. E. & O'Doherty, M. J. (2002) Curr. Opin. Neurol. 15, 679–684. [DOI] [PubMed] [Google Scholar]
- 56.Thiele, C. J. (1991) Cancer Metastasis Rev. 10, 311–319. [DOI] [PubMed] [Google Scholar]
- 57.Cavazzana, A. O., Miser, J. S., Jefferson, J. & Triche, T. J. (1987) Am. J. Pathol. 127, 507–518. [PMC free article] [PubMed] [Google Scholar]
- 58.Kim, J., Lo, L., Dormand, E. & Anderson, D. J. (2003) Neuron 38, 17–31. [DOI] [PubMed] [Google Scholar]
- 59.Molofsky, A. V., Pardal, R., Iwashita, T., Park, I-K., Clarke, M. F. & Morrison, S. J. (2003) Nature 425, 962–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.