Abstract
Chromosome conformation capture approaches have shown that interphase chromatin is partitioned into spatially segregated Mb-sized compartments and sub-Mb-sized topological domains. This compartmentalization is thought to facilitate the matching of genes and regulatory elements, but its precise function and mechanistic basis remain unknown. Cohesin controls chromosome topology to enable DNA repair and chromosome segregation in cycling cells. In addition, cohesin associates with active enhancers and promoters and with CTCF to form long-range interactions important for gene regulation. Although these findings suggest an important role for cohesin in genome organization, this role has not been assessed on a global scale. Unexpectedly, we find that architectural compartments are maintained in noncycling mouse thymocytes after genetic depletion of cohesin in vivo. Cohesin was, however, required for specific long-range interactions within compartments where cohesin-regulated genes reside. Cohesin depletion diminished interactions between cohesin-bound sites, whereas alternative interactions between chromatin features associated with transcriptional activation and repression became more prominent, with corresponding changes in gene expression. Our findings indicate that cohesin-mediated long-range interactions facilitate discrete gene expression states within preexisting chromosomal compartments.
The regulated transcription of mammalian genomes packaged into nuclei six orders of magnitude smaller than the length of chromosomal DNA requires a complex organization. The underlying mechanisms are beginning to be explored in terms of the biophysical properties of the DNA polymer and associated chromatin (Fudenberg and Mirny 2012) and nuclear landmarks such as nuclear lamina that provide genomic scaffolds (Guelen et al. 2008; McCord et al. 2013; Meuleman et al. 2013). Of particular interest are functional interactions that operate within these constraints to facilitate long-range interactions between gene regulatory elements (Merkenschlager and Odom 2013).
Genome-scale chromosome conformation capture has shown that interphase chromatin is organized into Mb-scale compartments that are “open,” gene-rich, highly transcribed, and interactive (A) or “closed” (B), gene-poor, and less transcriptionally active (Lieberman-Aiden et al. 2009). Compartments are composed of topologically associated domains (TADs), which comprise hundreds of kilobases and are relatively invariant between different cell types (Lieberman-Aiden et al. 2009; Dixon et al. 2012; Nora et al. 2012; Gibcus and Dekker 2013). This architecture is thought to facilitate the regulation of gene expression by constraining the number of regulatory elements a given gene is likely to encounter to those that are colocated within the same compartment or domain (Gibcus and Dekker 2013). It is unknown how compartments and domains are built and how they contribute to the precise regulation of gene expression.
The cohesin complex is essential for genome integrity in cycling cells, where it facilitates post-replicative DNA repair and sister chromatid cohesion by controlling chromosome topology (Nasmyth and Haering 2009; Aragon et al. 2013). In addition, cohesin contributes to the regulation of gene expression by mechanisms thought to involve long-range interactions between its binding sites at regulatory elements associated with CTCF (Parelho et al. 2008; Rubio et al. 2008; Stedman et al. 2008; Wendt et al. 2008) or with active promoters and enhancers (Misulovin et al. 2008; Kagey et al. 2010; Schmidt et al. 2010; Faure et al. 2012; Dorsett and Merkenschlager 2013). Taken together these properties suggest a role for cohesin in genome organization.
Elucidating the global contribution of cohesin to the organization of the genome remains a challenge, not least because the depletion of cohesin from proliferating cells interferes with DNA replication, DNA repair, and chromosome segregation (Nasmyth and Haering 2009). Here we use a genetic approach (Seitan et al. 2011) to define the contribution of cohesin to the organization of the genome and the regulation of gene expression in noncycling cells in vivo.
Contrary to expectation, we find no major role for cohesin in the maintenance of architectural features of genome organization in our experimental system. However, cohesin depletion reduced long-range interactions between cohesin-bound sites, and the resulting chromosomal interaction landscape was characterized by alternative interactions between chromatin features associated with transcriptional activation and repression. Interestingly, this reorganization of long-range interactions was accompanied by changes in gene expression in which genes at the low end of the expression spectrum were preferentially up-regulated, whereas genes at the high end of the expression spectrum were preferentially down-regulated. Our data indicate that the organization of the genome into architectural compartments and the random assortment of genes and regulatory elements within them are insufficient for the precise regulation of gene expression. Rather, cohesin enables discrete gene expression states by promoting cohesin-based interactions within a preexisting architectural framework.
Results
To define the contribution of cohesin to the organization of the genome and the regulation of gene expression in noncycling cells, we deleted the locus encoding the cohesin subunit RAD21 by the activity of a CD4Cre transgene when developing thymocytes exit the cell cycle as part of their developmental program in vivo. Developing thymocytes arrest at the G1 phase of the cell cycle when DNA is unreplicated and chromosomes are present as single copies, not as sister chromatids. In contrast to some model organisms, homologous chromosomes are not paired in mammalian interphase, and cohesin is not tasked with holding sister chromatids or homologs together. Rad21lox/lox CD4Cre CD4+ CD8+ small double positive thymocytes (referred to as “cohesin-deficient thymocytes” below) showed a significant reduction in total RAD21 protein expression (79%–93%, depending on the reference used for normalization) (Supplemental Fig. S1). Most of the cohesin present in thymocytes was associated with chromatin, and the decrease in chromatin-associated RAD21 in Rad21-deleted thymocytes reflected that of total RAD21 (Supplemental Fig. S1). ChIP and qPCR showed that RAD21 binding to known cohesin sites was reduced by 80%–90% (Supplemental Fig. S1). This system allowed us to investigate the role of cohesin in gene expression and the structural and functional organization of the genome in noncycling cells without resorting to cell lines or in vitro culture systems.
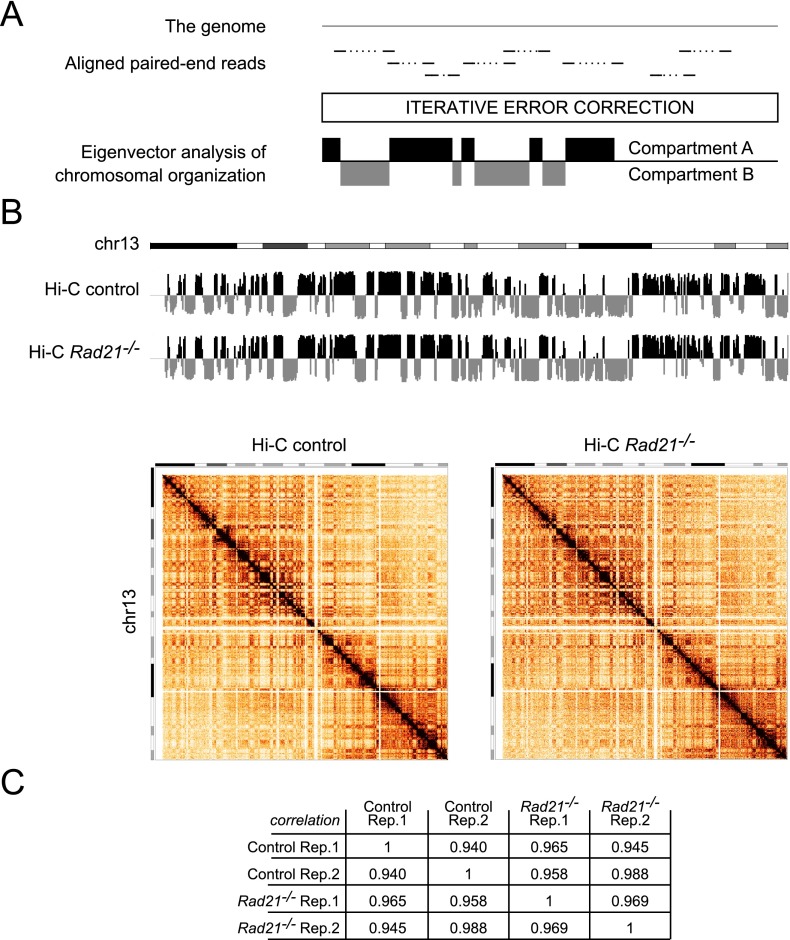
We prepared Hi-C libraries from two biological replicates of control and cohesin-deficient thymocytes and obtained a total of ∼392M unique valid pairs, ∼203M for control and ∼188M for cohesin-deficient thymocytes. To define chromosomal compartments, we applied eigenvector analysis of chromosomal organization to iteratively corrected genome-wide maps (Fig. 1A; Imakaev et al. 2012) at 140-kb resolution. Strikingly, the assignment of chromosomal compartments was highly correlated for control and cohesin-deficient thymocytes (Fig. 1B,C). We conclude that, unexpectedly, Mb-scale architectural compartments assigned by eigenvector analysis of chromosomal organization are resilient to reduced RAD21 protein expression in interphase.
Figure 1.
Chromosomal compartments are resilient to the depletion of the cohesin subunit RAD21 from noncycling thymocytes in vivo. (A) Eigenvector analysis of chromosomal organization. (B) Compartment tracks (top) and interaction matrices (bottom) for chr13 at 140-kb resolution in control and cohesin-deficient thymocytes. Of 17,548 regions evaluated, only two showed a consistent compartment change from A to B and two from B to A (change in eigenvector value >0.03): chr18: 3640001 and chr18: 33600001. (C) Hi-C compartment data are highly correlated for control and cohesin-deficient thymocytes.
Cohesin binding predicts perturbed long-range interactions in cohesin-deficient thymocytes
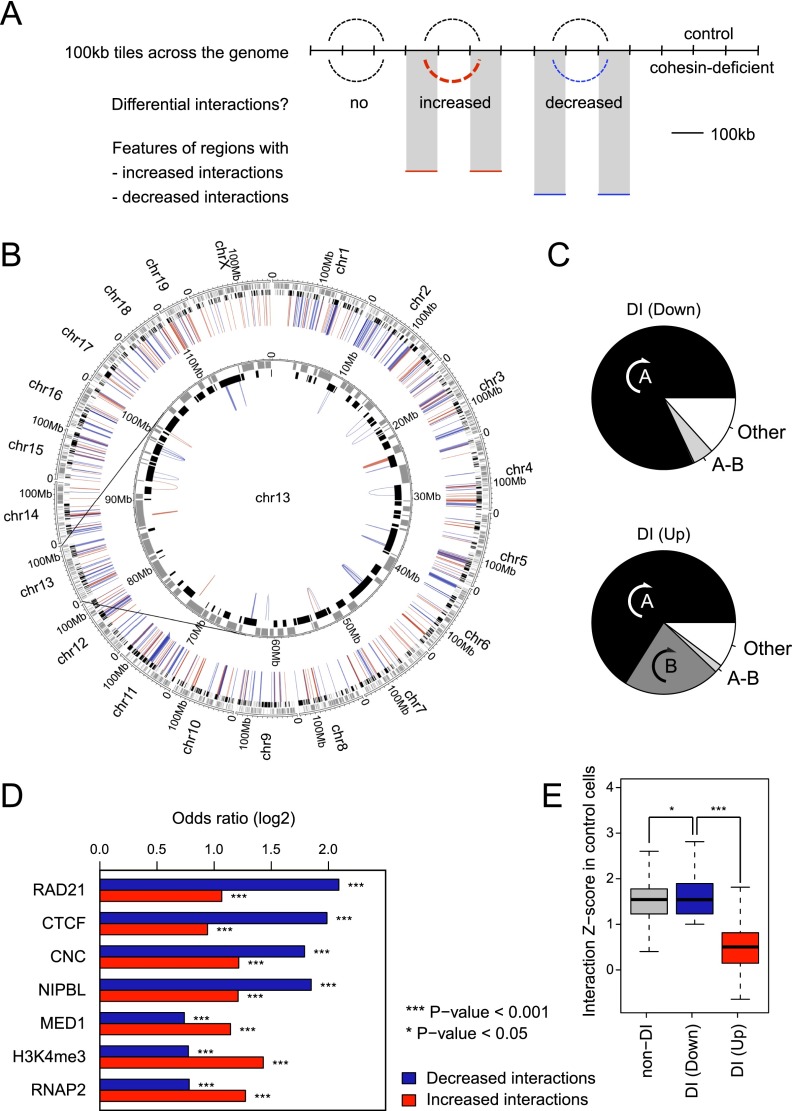
We applied the HOMER pipeline (Heinz et al. 2010) to our Hi-C data (see Methods, section on Hi-C data analysis, for details). We first identified 100-kb genomic regions that interacted significantly with each other in either control or cohesin-deficient cells (FDR = 0.1), taking into account linear genomic distance and sequencing depth. Interactions between these 100-kb regions were then compared between control and cohesin-deficient cells (Fig. 2A). Although the majority of significant interactions were unchanged in cohesin-deficient cells, 10,917 interactions were significantly altered in the pooled data, of which 1476 were found in replicate 1 and 5004 in replicate 2 (P < 0.05). There was a highly significant overlap of 502 differential interactions between replicates (P < 10−15, odds ratio of 5.25 for 224 increased interactions and 9.96 for 278 decreased interactions) (Fig. 2B). All differential interactions were intra-chromosomal, and the great majority were contained within individual A compartments (Fig. 2C; see Supplemental Table 1 for a list of differential interactions determined by HOMER).
Figure 2.
Cohesin binding predicts perturbed long-range interactions in cohesin-deficient thymocytes. (A) Schematic of approach for the mapping of differentially interacting regions. (B) Circos plot (Krzywinski et al. 2009) illustrating the chromosomal position of differential interactions in the context of chromosomal compartments. The HOMER software suite was used to determine significant interactions between 100-kb genomic regions in either control or cohesin-deficient thymocytes (FDR = 0.1; replicates pooled). Of 10,917 interactions that were significantly altered in the pooled samples, 1476 interactions changed in replicate 1 and 5004 in replicate 2 (P < 0.05; the remaining interactions were only seen in the pooled data). Of 502 differential interactions that were shared between replicates, 278 were decreased (blue) and 224 were increased (red). All differential interactions were intra-chromosomal. Compartment assignment is indicated at the base of the interactions: black for open compartments and gray for closed compartments. (C) Differential interactions in cohesin-deficient cells are largely contained within preexisting chromosomal compartments. (A) Differential interactions entirely contained within the same A compartment. (B) Differential interactions entirely contained within the same B compartment (the frequency of intra-B interactions was <2% among down-regulated interactions). (A-B) Differential interactions bridging A and B compartments. (Other) interacting regions are either unassigned (i.e., inconsistent between replicates) or bridge two distinct A or B compartments. Down-regulated interactions (top) and up-regulated interactions (bottom) are shown separately. (D) Features enriched in 100-kb regions that show differential interactions in cohesin-deficient thymocytes. Differential interactions between control and cohesin-deficient samples involved 946 unique 100-kb regions (510 involved in decreased interactions, 427 in increased interactions, and 9 involved in both) that participated in 502 unique differential interactions (278 that were decreased and 224 that were increased) and that were shared between replicates (P < 0.05). We tested whether differentially interacting 100-kb regions were enriched for the presence of Rad21, CTCF, NIPBL, MED1, H3K4me3, and RNAP2 binding events. See Supplemental Figure S1C,D for validation of the MED1 and NIPBL antibodies and ChIP, and Supplemental Figure S1E for an analysis of the relationship between RAD21, CTCF, and NIPBL ChIP-seq peaks. Differentially interacting regions were significantly enriched for the binding of the cohesin subunit RAD21, both with and without CTCF (cohesin-non-CTCF; CNC), and of the cohesin-associated factors CTCF and NIPBL, and features of transcriptional activity including MED1, H3K4me3, and RNA Pol II. Differential interactions were further classified into decreased and increased interactions. (E) Strength distribution of cohesin-dependent interactions. Using the number of Hi-C reads as an indicator of the strength of interactions, differential interactions that are decreased in cohesin-depleted thymocytes are similar in strength to unchanged interactions in control cells, whereas increased interactions tend to be weak before cohesin depletion (Mann-Whitney U-test P < 10−15). Outliers are not depicted.
To explore the relationship between cohesin binding and cohesin-dependent differential interactions, we focused on the 946 distinct 100-kb genomic regions that participated in the 502 differential interactions. These differentially interacting regions (Supplemental Table 1) were significantly enriched for the binding of the cohesin subunit RAD21, CTCF, the cohesin loading factor NIPBL, and the mediator subunit MED1 (Fig. 2D; Supplemental Fig. S1C,D for validation of the MED1 and NIPBL antibodies; ChIP and Supplemental Fig. S1E for an analysis of the relationship between RAD21, CTCF, and NIPBL ChIP-seq peaks).
At the 100-kb level, decreased interactions in particular were enriched for cohesin, both with and without CTCF (cohesin-non-CTCF or CNC in Fig. 2D), CTCF, and NIPBL (Fig. 2D). Increased interactions were enriched for marks of transcriptional activity, including MED1, H3K4me3, and RNA Pol II (Fig. 2D). The observed decrease in interactions between regions rich in cohesin and CTCF binding is intuitive based on reduced interactions between cohesin and CTCF binding sites in cohesin-depleted cells (Hadjur et al. 2009; Kagey et al. 2010; Seitan et al. 2011). Increased interactions between regions rich in features associated with active genes (MED1, H3K4me3, and RNA Pol II) could suggest a role for transcriptional activity in such interactions (Fraser and Bickmore 2007), although the 100-kb resolution achieved in the analysis of such regions is insufficient to pinpoint the role of individual features in driving differential interactions (see below).
Interactions that decreased upon cohesin depletion were strong in control cells, whereas interactions that increased following cohesin depletion were significantly weaker than average in control cells where cohesin was present (Fig. 2E).
Cohesin depletion perturbs gene expression in open compartments
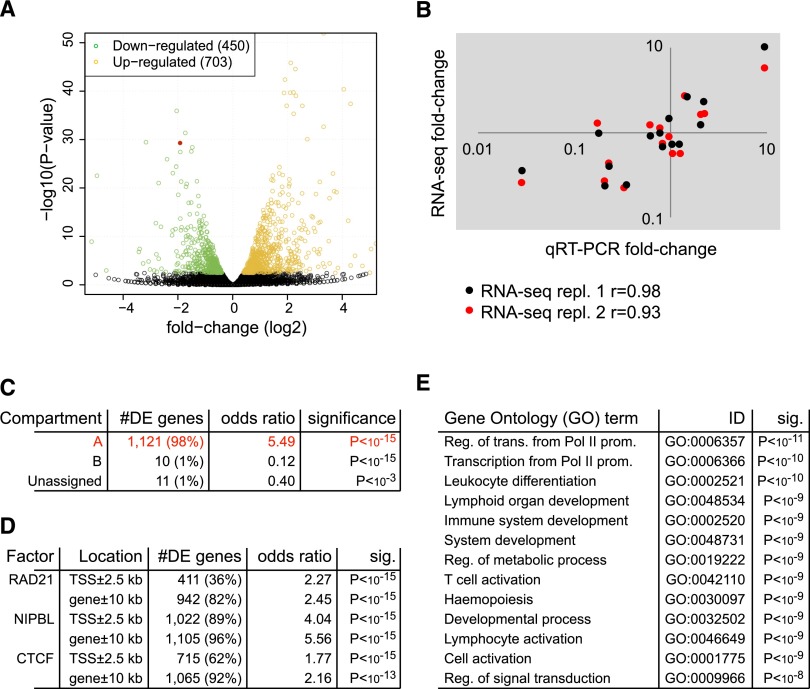
RNA-seq analysis of two independent biological replicates at a depth of 144M total reads defined 1153 genes that were differentially expressed between control and cohesin-deficient thymocytes (FDR = 0.05; 703 up-regulated, 450 down-regulated) (Fig. 3A). We validated 15 of these by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) and found that both methods were in close agreement (Fig. 3B). Comparison of RNA-seq data with Hi-C compartment analysis showed that 98% of deregulated genes reside in open (A) compartments (P < 10−15, odds ratio = 5.49) (Fig. 3C).
Figure 3.
Gene expression in cohesin-deficient thymocytes. (A) Volcano plot of RNA-seq data. Of the 17,849 genes assayed, 1153 are significantly differentially expressed in cohesin-deficient thymocytes (FDR = 0.05), 450 of which are up-regulated (orange) and 703 down-regulated (green). The red dot represents Rad21. Eight genes above the −log10(P-value) = 50 threshold (all up-regulated) were omitted from the plot. (B) Validation of RNA-seq data for 15 transcripts over a wide range of expression levels. Shown is fold-change measured by qRT-PCR (x-axis, two independent biological replicates) against the fold-change measured by RNA-seq (y-axis) and the Pearson's correlation coefficient (r) for two independent RNA-seq experiments. (C) Genes affected by cohesin depletion are associated with open compartments. Approximately 98% of differentially expressed genes that could be assigned to compartments (1121 of 1142 autosomal and X-linked genes) reside in open (A) compartments, compared with 91.2% (16,255 of 17,819) of genes included in our analysis. Genes spanning more than one compartment were assigned to A when they overlapped at least partially with open compartments; genes overlapping compartments that could not be clearly defined as A or B because they were inconsistent between replicates were called unassigned. Only 21 deregulated genes were found outside open compartments. Of these, 11 were located in unassigned compartments and just 10 of 1142 deregulated genes were located in closed (B) compartments. This corresponds to a highly significant depletion of deregulated genes in B compartments (P < 10−15, odds ratio = 0.12). (D) Cohesin-regulated genes are bound by cohesin and associated factors. Associations are shown for ChIP-seq peaks for RAD21, NIPBL, and CTCF (Shih et al. 2012) within 2.5 kb of transcription start sites (TSS) and within 10 kb of canonical gene bodies. (E) Gene Ontology (GO) analysis of genes that are differentially expressed in cohesin-deficient thymocytes. Representative GO biological process terms with adjusted P < 10−8 are shown (the complete list can be found in Supplemental Table 2).
Genes that are sensitive to cohesin dosage are bound by cohesin, CTCF, and NIPBL
The great majority of genes that were deregulated in cohesin-deficient thymocytes were bound by RAD21, NIPBL, or CTCF (Fig. 3D), indicating that most are direct targets of cohesin-mediated regulation. In agreement with this, the most highly enriched gene ontology (GO) terms (adjusted P < 10−8) included functions related to development, transcription, signal transduction, lymphocyte activation, and differentiation as well as hematopoiesis and the immune system (Fig. 3E). There was no enrichment for cell cycle related terms including cell cycle, DNA replication, chromosome segregation, and checkpoint activation. Terms relating to DNA damage, DNA repair, and cell division that were highly enriched in previous studies using RNAi-mediated depletion of cohesin from dividing ES cells (Kagey et al. 2010) were not enriched in cohesin-deficient thymocytes, validating our rationale for an experimental system based on nondividing cells.
Taken together with the observation that architectural compartments remain largely intact in cohesin-deficient thymocytes, these results suggest that deregulated gene expression occurs at the level of individual loci, and is not secondary to a global collapse in genome organization.
Examples of deregulated genes located in differentially interacting regions as determined by HOMER analysis of Hi-C data included the region just 3′ of the T cell receptor locus Tcra on chr14 and around the Cd3 gene cluster on chr9 (see Supplemental Table 1 for a list of differentially expressed genes within differentially interacting regions in cohesin-deficient thymocytes).
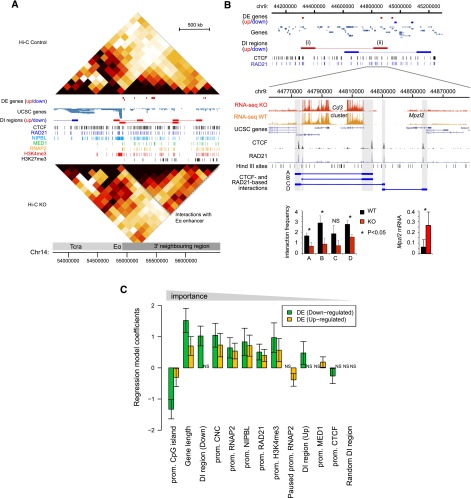
The Tcra locus is highly expressed in developing thymocytes, undergoes V(D)J recombination, and is separated from the 3′ flanking region by CTCF and cohesin sites with CTCF-dependent insulator activity (Zhong and Krangel 1999; Magdinier et al. 2004). Depletion of cohesin results in increased interactions between Tcra and the 3′ flanking region, which are visible in the Hi-C data and consistent with a scenario in which deregulated interactions with the Tcra enhancer (Eα) affect the expression of genes in the region (Fig. 4A).
Figure 4.
Features and predictors of cohesin-dependent gene expression. (A) The T cell receptor locus Tcra and 3′ flanking region of chr14 are shown alongside Hi-C heat maps at 140-kb resolution for control (top) and cohesin-deficient thymocytes (bottom), epigenomic features, and genes that are up- (red) or down-regulated (blue, no examples in this region). Differentially interacting (DI) regions as determined by HOMER analysis of Hi-C data are shown in red (up-regulated) or blue (down-regulated). Dashed gray lines demarcate increased interactions with the region 3′ of Tcra. (B) HOMER-identified increased and decreased interactions around the Cd3 gene cluster on chr9. (Top) increased interactions marked as (i) chr9:44300000–44400000 and (ii) chr9:44800000–44900000 contain the Bcl9 and Mpzl2 genes that are up-regulated in cohesin-deficient thymocytes. (Bottom) Gene expression and 3C analysis of region (ii). Hind III fragments containing CTCF and RAD21 binding sites are shaded gray. (Left inset) Primary 3C data (mean ± SD, n = 3). (Right inset) RT-PCR validation of Mpzl2 expression (mean ± SD, n = 3). (C) Multinomial logistic regression model integrating gene expression, Hi-C, and ChIP-seq data to predict up-regulated, down-regulated, and unchanged genes. We tested each gene for the following features: (1) the presence or absence of ChIP-seq peaks near the promoter (TSS ± 2.5 kb); (2) location within 100-kb differentially interacting regions (divided into interactions that were stronger, DI region [Up], or weaker, DI region [Down], in cohesin-deficient thymocytes); (3) the presence of the H3K4me3 histone modification; (4) the binding of RAD21, CTCF, NIPBL and Mediator (MED1), RNA Pol II (RNAP2), paused RNA Pol II at the promoter (Hendrix et al. 2008); (5) the presence of a promoter CpG island (CGI); and (6) gene length. Error bars represent 95% confidence intervals. Variables are ranked by coefficient significance from left to right. Of 17,849 genes assayed by RNA-seq, 1461 resided in DI regions. Of 450 down-regulated genes, 83 resided in DI regions, which represents a strong enrichment (P-value < 10−11, odds ratio 2.63). Up-regulated genes were only slightly enriched in DI regions (P < 0.05, odds ratio 1.27).
HOMER identified differential interactions around the highly expressed Cd3 gene cluster (Fig. 4B, top panel). The Bcl9 and Mpzl2 genes are located in regions of increased interactions and are up-regulated in cohesin-deficient thymocytes (Fig. 4B, top panel). Chromosome conformation capture (3C) analysis within the differentially interacting 100-kb region containing the Cd3d, Cd3e, Cd3g, and Mpzl2 (Fig. 4B, top panel [ii]) showed that the depletion of cohesin significantly reduced local interactions between CTCF and RAD21 sites (Fig. 4B, bottom panel) for three of four interactions tested (Fig. 4B, inset, bottom left). Reduced local interactions may facilitate the increased long-range interactions detected at the 100-kb level and could also be relevant to altered gene expression in this region: CTCF sites demarcate the Cd3 gene cluster from the neighboring Mpzl2 gene that is barely expressed in control thymocytes, but up-regulated in cohesin-deficient thymocytes (RNA-seq data in Figure 4B, bottom panel; and RT-PCR validation, inset, bottom right).
Our data causally link cohesin to long-range chromatin interactions. Although the associated changes in gene expression may be rationalized by reduced boundary or insulator function, the link between long-range interactions and deregulated gene expression remains correlative at this time.
Predictive features of genes that show cohesin-dependent expression
To delineate factors associated with gene expression changes in cohesin-deficient thymocytes, we used a regression model that integrated gene expression, Hi-C, and ChIP-seq data. We assigned genes to one of three classes (up-regulated, down-regulated, or unchanged) and tested for the presence or absence of ChIP-seq peaks near each gene promoter (TSS ± 2.5 kb) as well as gene location within 100-kb regions that interact differentially in control and cohesin-deficient thymocytes. Differential interactions were further divided into interactions that were stronger (DI region, Up) or weaker (DI region, Down) in cohesin-deficient thymocytes. We also considered the presence of the H3K4me3 histone modification; the binding of RAD21, CTCF, NIPBL, and Mediator, RNA Pol II, paused RNA Pol II at the promoter (Hendrix et al. 2008); the presence of promoter CpG islands (CGI); and gene length. We determined the relative importance of each variable in the multivariate model and ranked the corresponding coefficients by their statistical significance. The presence of a CpG island (CGI) at the promoter, gene length, the presence near promoters of RAD21, particularly without CTCF (CNC), and promoter-associated RNA Pol II emerged as important variables (Fig. 4C; see Supplemental Fig. S2 for the results of a univariate analysis considering each variable separately). Interestingly, location within regions that showed differential interactions in cohesin-deficient thymocytes—but not a control set of interacting regions that showed no differences between control and cohesin-deficient thymocytes (“Random DI region” in Fig. 4C)—was predictive of gene expression changes. Decreased gene expression in particular was associated with differential interactions (P-value < 10−11, odds ratio = 2.63) (Fig. 4C).
Investigating variables that were highlighted by the regression model, we first probed the relationship between deregulated gene expression and differential long-range interactions. This analysis showed a strong association between decreased interactions and down-regulated gene expression (Supplemental Fig. S3A). Further analysis confirmed that genes with increased promoter CpG density were more likely to be up- than down-regulated in cohesin-deficient thymocytes (Supplemental Fig. S3B, left), and also more likely to coincide with differentially interacting regions in cohesin-deficient cells (Supplemental Fig. S3B, right). Moreover, longer genes were more likely to be down-regulated in cohesin-deficient cells (Supplemental Fig. S3C, left) and were found preferentially in regions of reduced interactions (Supplemental Fig. S3C, right). A set of complex loci of significantly greater length than average (Mann-Whitney U-test P < 10−15) overlap ultraconserved noncoding elements, regulatory blocks or archipelagos (Akalin et al. 2009; Montavon and Duboule 2012; Dimitrieva and Bucher 2013). These loci were preferentially down-regulated in cohesin-deficient thymocytes (Supplemental Fig. S3D), which is consistent with a role for cohesin in long-range regulation of genes with complex regulatory inputs.
Cohesin depletion perturbs long-range interactions within architectural compartments and compresses the dynamic range of gene expression
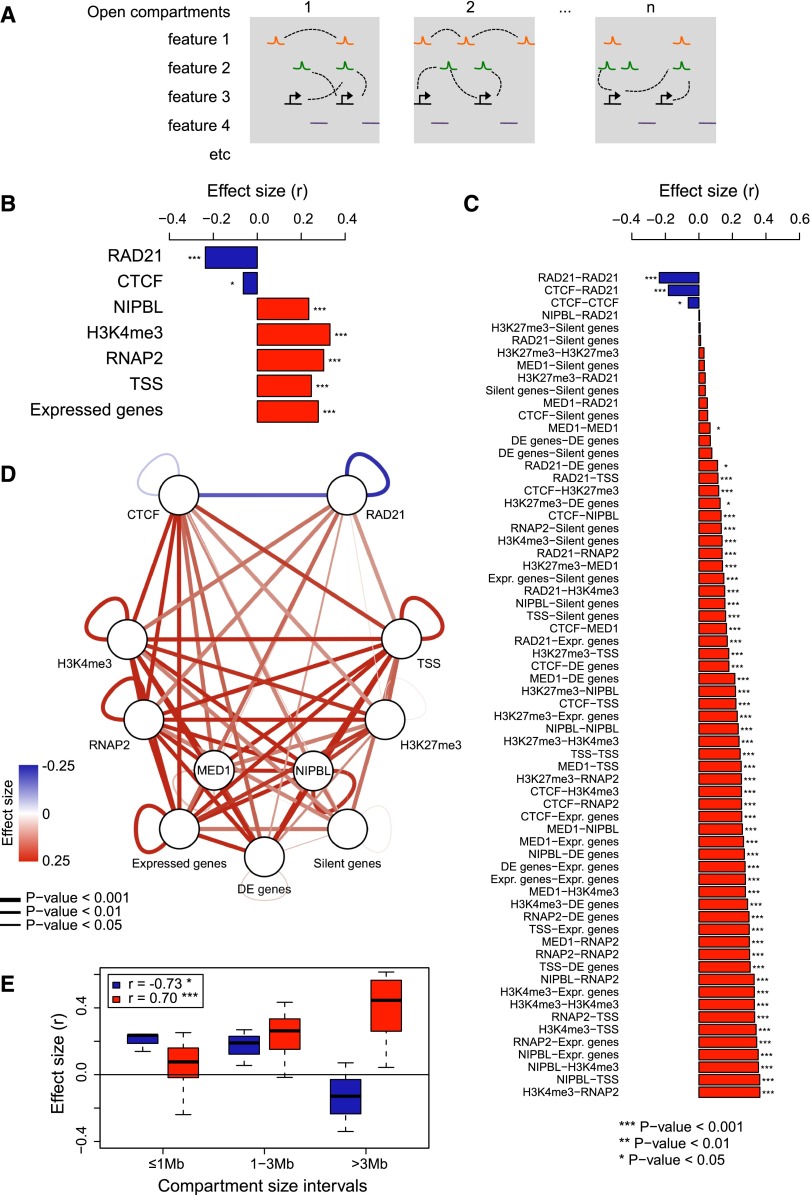
We used Structured Interaction Matrix Analysis (SIMA) (Lin et al. 2012) to obtain a high-resolution view of interactions between specific chromatin features within the chromosomal compartments assigned by eigenvector analysis of chromosomal organization (Fig. 5A). Although the number of Hi-C reads that map to any one specific feature—for example a cohesin-bound site—is too low to assign interactions with confidence, this approach is designed to combine information for multiple occurrences of this feature (Lin et al. 2012). We focused on open (A) compartments, where the great majority of cohesin binding sites and cohesin-regulated genes reside (Fig. 3C), and selected a range of features for analysis. These included ChIP-seq peaks for cohesin and associated factors, histone modifications indicative of active (H3K4me3), and repressed (H3K27me3) chromatin states, transcription start sites of active and silent genes, as well as control sites that did not overlap with these features. For each compartment (1, 2,…, n in Fig. 5A) we counted Hi-C reads connecting features of the same type (homotypic interactions, illustrated for feature 1 in Fig. 5A) and Hi-C reads connecting different features (heterotypic interactions, illustrated for features 2 and 3 in Fig. 5A), associating Hi-C reads that mapped within 10 kb of each feature (Lin et al. 2012). To determine the impact of cohesin on these interactions we compared interactions in control and cohesin-deficient cells for each feature (and pair of features) within open compartments (Methods; Supplemental Table 5).
Figure 5.
The chromatin landscape of cohesin-deficient thymocytes is characterized by the loss of cohesin-based interactions and the detection of alternative interactions. (A) Outline of approach used to score interactions between specific chromatin features located within architectural compartments as assigned by eigenvector analysis of chromosomal organization (SIMA) (Lin et al. 2012). Open compartments (1, 2, …, n) are indicated by gray boxes. Orange, green, and black symbols (features 1, 2, etc.) represent chromatin features, and purple lines indicate “featureless” control sites. Hi-C interactions between features are indicated by dashed lines. For each compartment, we counted Hi-C reads connecting features of the same type (homotypic interactions, illustrated for feature 1), and Hi-C reads connecting different features (heterotypic interactions, illustrated for features 2 and 3), assigning Hi-C reads that mapped within 10 kb of each feature (Lin et al. 2012). We compared interactions in control and cohesin-deficient cells for each feature and pair of features within each open compartment and combined these scores as a measure for the cohesin dependence of long-range interactions between specific features. (B) Impact of cohesin deficiency on homotypic cohesin-based and alternative interactions. SIMA was used to determine the enrichment of Hi-C reads in interactions connecting “like” features in control and cohesin-deficient thymocytes. The difference between normalized enrichment ratios in each condition was assessed by the Wilcoxon signed-rank test (see Methods). (C) Impact of cohesin deficiency on all pairwise feature-based interactions. SIMA results for homotypic interactions are shown together with those associated with interactions connecting different features (heterotypic interactions). Refer to B for details. Homotypic RAD21-RAD21 interactions are the most decreased, followed by CTCF-RAD21 interactions. Interactions between features associated with active transcription are strongly increased in cohesin-deficient thymocytes, as are interactions involving the repressive histone modification H3K27me3 (Zhang et al. 2012) with marks of active transcription. (D) Cytoscape representation of SIMA results in C. Edge color and width correspond to the Wilcoxon signed-rank test effect size and significance, respectively. (E) Length scale of lost and gained cohesin-dependent feature-based interactions. Boxplots are based on SIMA comparisons stratified into three classes according to compartment size (≤1 Mb, 1–3 Mb, >3 Mb). Effect sizes for decreased interactions (RAD21-RAD21, CTCF-RAD21, CTCF-CTCF) and increased interactions (remainder) were grouped and are indicated separately. The effect of reduced cohesin-based interactions is most pronounced within smaller compartments and decreases when larger compartments are considered (Pearson's correlation coefficient r = −0.73, P < 0.05), whereas alternative interactions increased with compartment size (r = 0.7, P < 0.001). Outliers are not depicted.
This approach revealed that interactions between RAD21 and CTCF sites were reduced in cohesin-deficient thymocytes (Fig. 5B), and the analysis of interactions between all pairs of features revealed that RAD21-RAD21 and RAD21-CTCF interactions were most strongly decreased (Fig. 5C). This result is consistent with previous 3C experiments that indicated reduced interactions between such sites in cohesin-depleted cells (Fig. 4B; Hadjur et al. 2009; Kagey et al. 2010; Seitan et al. 2011).
Interactions that remained—or even increased—in cohesin-deficient cells included features of active transcription such as NIPBL, H3K4me3, and RNA Pol II and extended to interactions between activation-associated features and repressive H3K27me3 marks (Fig. 5B,C). Upon cohesin depletion, promoters of genes that were silent in control cells also showed detectable interactions with a range of features linked to transcriptional activation (Fig. 5C).
To determine whether these interactions in cohesin-deficient cells were selective, we identified control sites that were at least 10 kb removed from other features. Interactions involving these “random” sites showed little cohesin dependence, suggesting that increased interactions preferentially involve sites that are marked by the features analyzed, and thereby provide a measure of selectivity (Supplemental Fig. S4).
Since SIMA analysis is based on averaged occurrences of the specific features we do not know whether the increased representation of alternative interactions in cohesin-deficient cells is absolute or relative to the loss of cohesin-based interactions. Either way, the data indicate a shift in the chromatin landscape in cohesin-deficient cells from cohesin-based to alternative interactions (Fig. 5D).
As a proxy for the scale of differential interactions with decreased and increased representation in our Hi-C data, we stratified SIMA results by compartment size. Interestingly, the reduction of cohesin-based interactions was most pronounced in compartments <1 Mb, suggesting an upper limit for cohesin-based interactions within compartments (Fig. 5E; Supplemental Fig. S5). The alternative interactions detected in cohesin-deficient cells increased with compartment sizes >1 Mb, suggesting that they preferentially occur over larger distances or in larger compartments, which may have a higher degree of complexity (Fig. 5E; Supplemental Fig. S5).
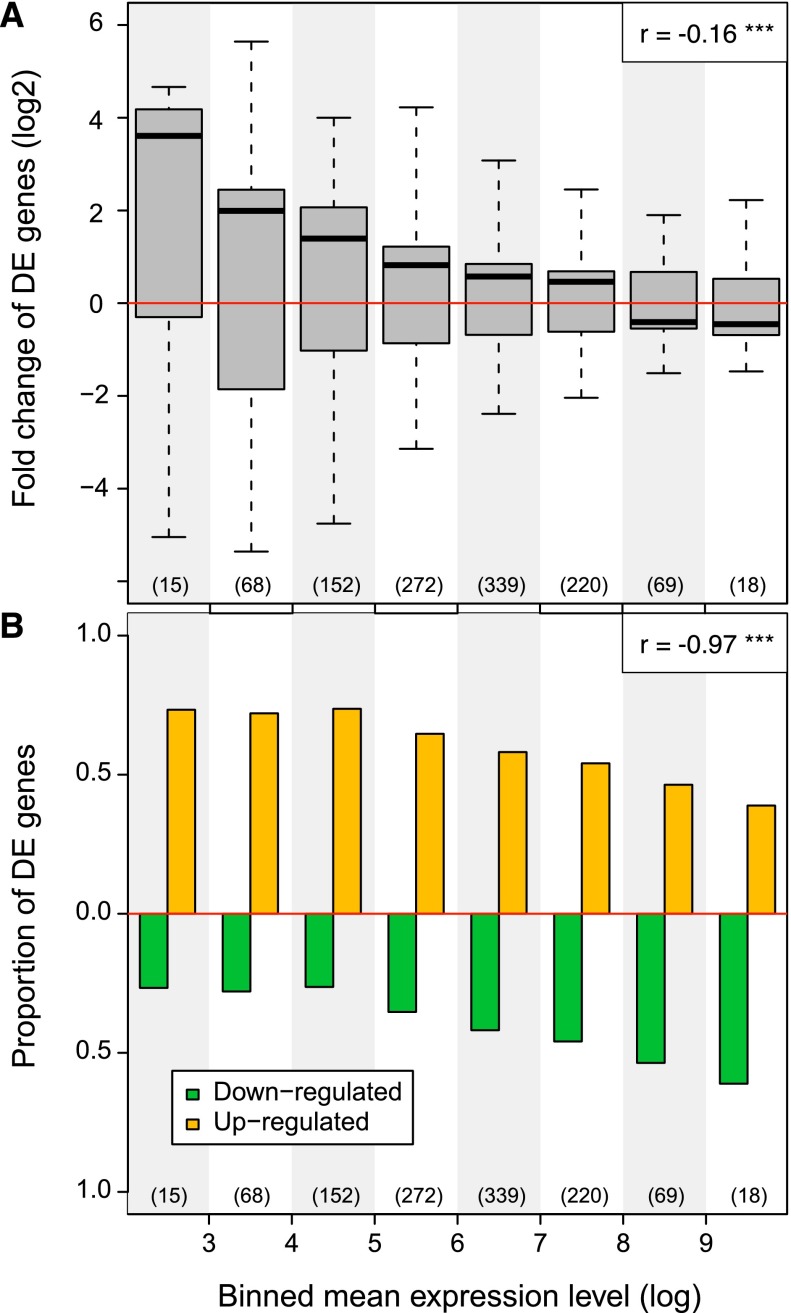
The prominence of interactions between features associated with both active gene expression and silencing in cohesin-deficient cells (Fig. 5C) resonates with models where cohesin not only facilitates specific interactions, but also provides separation between genes and regulatory elements (Wallace and Felsenfeld 2007; Phillips and Corces 2009; van Steensel 2011; Dixon et al. 2012). To address how the altered chromatin interactions in cohesin-deficient cells may impact on gene expression, we stratified genes according to their level of expression. Gene expression was perturbed across the entire range of the expression spectrum (Fig. 6A). In addition, genes with low expression were more often up-regulated, whereas genes with high expression were more often down-regulated (Fig. 6A). This resulted in a systematic skewing of gene expression away from the extremes of the dynamic range and toward average values (Pearson's correlation coefficient r = −0.16, P < 10−7) (Fig. 6A). Accordingly, the proportion of up-regulated genes was higher at the low end of the expression spectrum, whereas the proportion of down-regulated genes was higher at the high end of the expression spectrum (Pearson's correlation coefficient r = −0.97, P < 10−4) (Fig. 6B). Hence, genes showed a more uniform expression in cohesin-deficient cells.
Figure 6.
Cohesin depletion compresses the dynamic range of gene expression. (A) Genes were stratified into 10 equally sized log intervals from low (0–1) to high (>9) based on the average gene expression of control and cohesin-deficient thymocytes. Boxplots indicate the distribution of gene expression fold changes in cohesin-deficient thymocytes. The number of genes in each bin is indicated (bins 1 and 2 are empty). Note that lowly expressed genes are frequently up-regulated, whereas highly transcribed genes tend to be down-regulated. Note that this pattern does not result from ascertainment bias in which lowly expressed genes can only be up-regulated and vice versa, because we stratified genes according to the mean of their expression in control and cohesin-depleted cells. Therefore, the direction of regulation is the inverse when control cells are compared to cohesin-deficient cells (not shown). P-values are based on one-sample Wilcoxon signed-rank tests and indicate significant difference from zero (no change). Genes with zero mean expression in both cohesin-deficient and control thymocytes are excluded and outliers are not depicted. (B) Bar plot indicating the proportion of up- and down-regulated genes in each gene expression interval (see A). The proportion of up-regulated genes is anti-correlated with interval rank (Pearson's correlation coefficient r = −0.97, P < 10−4).
Discussion
The cohesin complex provides physical linkage between sister chromatids from the time of chromosome duplication in S-phase until chromosomes segregate in cell division (Nasmyth and Haering 2009) and can form long-range interactions that link gene regulatory elements with their targets in interphase (Hadjur et al. 2009; Kagey et al. 2010; Seitan et al. 2011). Unexpectedly, our Hi-C analysis shows that the maintenance of compartments in nondividing mammalian cells was not affected by the depletion of cohesin, at least not at a level we can achieve in our experimental system. This reduction is far in excess of the 20%–30% reduction known to cause severe developmental abnormalities in model organisms and in human Cornelia de Lange syndrome (CdLS), suggesting that a breakdown of genome organization at the level of compartments may not be the cause for deregulated gene expression in CdLS (Strachan 2005; Kawauchi et al. 2009). In contrast to the preservation of architectural genome organization, we find that cohesin depletion alters long-range chromosomal interactions within compartments and results in a more uniform expression of genes affected by cohesin depletion.
We found that the density of cohesin binding sites correlated with differential interactions of 100-kb regions that showed both increased and decreased interactions in cohesin-deficient thymocytes. Analysis at the local level showed that cohesin depletion reduced Hi-C reads that mapped within 10 kb of RAD21 and CTCF binding sites. Other interactions remained or even increased in cohesin-deficient cells. These alternative interactions included features of transcriptionally active and repressed chromatin. Using compartment size as a proxy for the length scale of interactions, cohesin-based interactions and the alternative interactions detected in cohesin-deficient cells appeared to differ in scale. Consistent with correlative data (de Wit et al. 2013; Phillips-Cremins et al. 2013), cohesin-dependent interactions within compartments were mostly confined to <1 Mb, i.e., the scale of topologically associated domains (TADs). In contrast, the alternative interactions detected in cohesin-deficient cells increased with compartment sizes >1 Mb. This result suggests that TAD-scale cohesin-based interactions prevent the detection of longer-range alternative interactions. Hence, disrupting cohesin-based interactions may lead to a degree of mixing between chromosomal domains beyond the TAD scale (>1 Mb).
Since SIMA analysis is based on averaged occurrences of the specific features, we do not know whether the increases are absolute or relative to the loss of cohesin-based interactions. Either way, the data indicate a shift in the chromatin landscape in cohesin-deficient cells from cohesin-based to alternative interactions. Importantly, the homogenization of interactions that results from this shift is consistent with the pattern of deregulated gene expression in cohesin-deficient thymocytes. Similar to previous observations in Drosophila (Schaaf et al. 2009), genes at the low end of the expression spectrum were preferentially up-regulated, whereas genes at the high end of the expression spectrum were preferentially down-regulated. Perhaps this is the genome-scale equivalent of the ability of CTCF and CTCF-associated cohesin to mediate insulator and boundary functions. Although such functions were predicted based on correlative analysis of histone modifications (Cuddapah et al. 2009), lamin-associated domains (Guelen et al. 2008; McCord et al. 2013; Meuleman et al. 2013) and long-range interactions maps (Dixon et al. 2012), their impact on gene expression had been previously documented only in reporter assays (Wallace and Felsenfeld 2007; Phillips and Corces 2009) and at individual loci, based on the manipulation of individual CTCF sites (Guo et al. 2011) or the deletion of CTCF (Ribeiro de Almeida et al. 2011). We suggest that cohesin-based interactions limit the extent of alternative interactions to enable discrete gene expression states.
Our conclusion that cohesin contributes to functional interactions within preexisting chromosomal compartments contrasts with cohesin's structural role in providing stable cohesion between sister chromatids (Nasmyth and Haering 2009). On closer inspection, however, this conclusion is consistent with a substantial body of existing knowledge on how cohesin interactions with chromatin are regulated during the cell cycle. In the G1 phase of the cell cycle, chromatin-associated cohesin shows relatively rapid turnover and has a half-life of minutes (Gerlich et al. 2006; McNairn and Gerton 2009; Gause et al. 2010; Onn and Koshland 2011). It is only during S-phase that a subset of cohesin complexes is stabilized by the acetylation of specific lysine residues in the SMC3 subunit, which displace the cohesin unloading factor WAPL and allow association of the cohesin-stabilizing factor sororin to extend the half-life of a subset of cohesin complexes (Gerlich et al. 2006; Kueng et al. 2006; Gause et al. 2010). This subset of long-lived cohesin complexes is thought to have a structural role by mediating cohesin between sister chromatids from S-phase until the transition between metaphase and anaphase, which can be hours or even decades in the case of human oocytes (Jessberger 2012). However, a population of cohesin complexes with high turnover remains even after DNA replication, and perhaps it is those complexes that continue to contribute to the regulation of gene expression during the S- and G2-phases of the cell cycle. The depletion of the cohesin removal factor WAPL demonstrates the consequences of rendering all cohesin complexes stable (Kueng et al. 2006; Tedeschi et al. 2013). Interestingly, loss of WAPL causes dramatic changes in the structure of interphase chromatin and perturbs the regulation of gene expression (Kueng et al. 2006; Tedeschi et al. 2013). It therefore appears that dynamic cohesin turnover is essential for regulated gene expression, and we speculate that this behavior is reflected in the contribution of cohesin to genome organization in interphase that is described by our data.
Methods
Experimental procedures
The conditional Rad21 allele crossed to CD4Cre and methods for RT- and genomic PCR, chromosome conformation capture and ChIP-seq have been described (Seitan et al. 2011). ChIP was performed using Abcam ab992 rabbit polyclonal antibody to RAD21 (Seitan et al. 2011), Bethyl Laboratories A300-793A rabbit polyclonal antibody to MED1 (Kagey et al. 2010), and Bethyl Laboratories A301-779A rabbit polyclonal antibody to NIPBL (Kagey et al. 2010). ChIP primers (5′–3′) and amplicon positions (mm9) were: X:100692 (ChrX:100691761–100691894): fwd: TGGCTTGTACTTCCAGATCAT, rev: AATGTATAAGCAAGCTAGTACGCA; 17:13025 (mm9 Chr17:13025211–13025311): fwd: GGTCTCGCCAGCTTGCTATTTC, rev: TGGCTAGATGTCATTGTGGTGG; 17:17783 (Chr17:17782844–17782944): fwd: AGGACATAGTCGCTTGAGTGATGG, rev: TTGGGCCAGGCTGGTACTTT; 11:53442 (Chr11:53441557–53441657): fwd: CAGGATGAGGATATGACTAGCTGTGG, rev: TCCAGTCCAGTTCCAAAAGGAGC. RNA-seq was done as described (Seitan et al. 2011) except that we used TruSeq kits according to the manufacturers' instructions (Illumina). Hi-C libraries were prepared as described (Lieberman-Aiden et al. 2009; Belton et al. 2012).
Computational methods
ChIP-seq read mapping and peak calling
Raw read alignment, filtering, and peak-calling for RAD21, MED1, NIPBL CTCF, and definition of CNCs was done as previously described (Faure et al. 2012). ChIP-seq data for H3K27me3 (Zhang et al. 2012) was similarly processed, except CCAT version 3.0 (Xu et al. 2010) was used to identify the relatively broad regions occupied by this mark (precompiled histone modification configuration). Single base pair summit positions for H3K4me3 and RNAP2 obtained from the Mouse ENCODE Project (Shen et al. 2012) were extended to a width of 200 bp to define peak regions.
RNA-seq data analysis
Raw reads for each condition and replicate were independently aligned to mouse transcript sequences (cDNA sequences from Ensembl version 66, NCBI37/mm9) (Flicek et al. 2013) using Bowtie version 0.12.8 (Langmead et al. 2009) with default parameters. Gene expression estimates and normalized count equivalents were obtained using MMSEQ version 0.11.2 (Turro et al. 2011). We used the Bioconductor R package DESeq version 1.6.1 (Anders and Huber 2010) to determine significantly differentially expressed genes in cohesin-deficient thymocytes versus control cells (FDR = 0.05). Empirical gene expression dispersion values were estimated in a condition-specific fashion (method = “per-condition”) and used to fit a dispersion-mean relationship, where only the fitted values were used (sharingMode = “fit-only”). T cell receptor gene segments, pseudogenes, ribosomal genes, and genes with an aggregate exon mapability score in the lowest ten percentile (UCSC Genome Browser track “wgEncodeCrgMapabilityAlign50mer”) were excluded from the analysis. Expressed genes were defined as those having log(expression_level + 1) ≥ 1 in control cells; otherwise genes were considered silent.
Hi-C data analysis
Iterative error correction of Hi-C data was performed as described (Imakaev et al. 2012). The HOMER Hi-C software analysis pipeline (http://biowhat.ucsd.edu/homer/interactions/) was used to determine significant interactions, differential interactions and to perform Structured Interaction Matrix Analysis (SIMA) (Lin et al. 2012). Briefly, paired-end reads were trimmed to remove sequence following the canonical HindIII ligation junction sequence (1bp mismatch allowed to account for potential star activity). Trimmed reads were aligned independently to the mouse reference genome assembly (NCBI37/mm9) using BWA (Li and Durbin 2009). Paired-end reads were merged and filtered to remove duplicate read pairs (“-tbp 1”), paired-end reads likely representing continuous genomic fragments or religation events (“-removePEbg”), self-ligations (“-removeSelfLigation”), and reads originating from regions with unusually high tag density (“-removeSpikes 10000 5”). Additionally, only read-pairs where both ends mapped near restriction sites were retained (“-both”).
To identify differential interactions, we first determined a “universe” of interactions on which to focus the analysis, defined as significant interactions between 100-kb genomic regions in either control or cohesin-deficient thymocytes (replicates pooled; FDR = 0.01). HOMER uses the binomial distribution to determine significant deviations above the expected number of Hi-C reads occurring between two loci, where the background model takes into account linear genomic distance and sequencing depth. For each replicate, high scoring differential interactions (P < 0.05) were then determined by comparing Hi-C read levels between control and cohesin-deficient thymocytes within this subset of all possible interactions. Only differential interactions consistently identified in both Hi-C replicates were retained for downstream analysis.
To determine genomic features associated with chromatin interactions, we used a method that pools Hi-C information associated with a given set of genomic regions within a specified set of domains (SIMA) (Lin et al. 2012). We used default resolution (“-res 2500”) and optimal Hi-C interaction search space parameters (“-superRes 10000”). Domains of interest were defined as merged adjacent 140-kb regions within open compartments at least 500 kb (“-minDsize 500000”) and not more than 5 Mb in length. Within-domain associations were assessed independently in control and cohesin-deficient thymocytes for all peak sets (RAD21, MED1, NIPBL, CTCF, H3K4me3, H3K27me3, RNAP2) as well as all canonical TSSs (excluding pseudogenes; Ensembl version 66; Flicek et al. 2013), promoters of silent genes, expressed genes, and significantly differentially expressed genes. Normalizing by the number of expected Hi-C reads under the background model and comparing to the randomized average (after shuffling feature positions 10,000 times), we obtained an enrichment ratio for each genomic region, indicating the association of each feature (or feature pair) with interactions contained within that region. To determine the impact of cohesin on these interaction associations, we compared observed/randomized enrichment ratios in control and cohesin-deficient cells for each feature (or feature pair) within each compartment. The size and direction of change in these ratios in cohesin-depleted thymocytes were compared using a paired statistical test (Wilcoxon signed-rank test) to provide a measure for the cohesin dependence of long-range interactions between specific features. See Supplemental Figure S5 for SIMA results stratified by compartment size.
Multinomial logistic regression model
We used gene features derived from ChIP-seq, RNA-seq, Hi-C, genomic sequence, and annotation to predict gene expression changes in cohesin-deficient thymocytes using a multinomial logistic regression model (nnet R package version 7.3-1) (Venables and Ripley 2002). Binary variables used included RAD21 (overlapping CTCF), cohesin-non-CTCF (CNC), CTCF, NIPBL, MED1, H3K4me3 and RNAP2 peak, and CpG island (CGI) (Illingworth et al. 2010) overlap within the gene promoter (TSS ± 2.5kb), paused promoter RNAP2 (stalling index ≥4) (Hendrix et al. 2008), gene body overlap with 100-kb differentially interacting (DI) regions (Up, increased; Down, decreased) as well as a randomly selected “control” set of interacting regions. We also included total gene length as a continuous variable. The three-class categorical response variable was encoded as follows: “0” nondifferentially expressed, “−1” significantly down-regulated, “1” significantly up-regulated.
Data access
ChIP-seq, RNA-seq, and Hi-C data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE48763.
Acknowledgments
We thank Drs. L. Game and M. Jones for sequencing; G. Dharmalingam, S. Khadayate, Drs. T. Carroll, C. Berthelot, and J. Marioni for help with data analysis; and Drs. J. Elliott and M. Opanowicz for cell sorting. This work was supported by the Medical Research Council, UK (A.G.F., M.M.); the Wellcome Trust (M.M.); the European Molecular Biology Laboratory (A.J.F., P.F.); the National Institutes of Health (J.D.); and the National Human Genome Research Institute (HG003143 and HG003143-06S1) (J.D.). E.H. was supported by the Fondation pour la Recherche Médicale, the Agence Nationale de la Recherche, EpiGeneSys FP7 no. 257082 Network of Excellence, a European Research Council (ERC) Advanced Investigator Award no. 250367, and EU FP7 SYBOSS Grant no. 242129.
Author contributions: V.S., A.J.F., E.H., A.G.F., P.F., J.D., and M.M. devised the study; V.S., Y.Z., L.G., and M.M. performed experiments; A.J.F., R.P.M., B.R.L., and J.D. analyzed and managed data; and V.S., A.J.F., A.G.F., and M.M. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.161620.113.
References
- Akalin A, Fredman D, Arner E, Dong X, Bryne JC, Suzuki H, Daub CO, Hayashizaki Y, Lenhard B 2009. Transcriptional features of genomic regulatory blocks. Genome Biol 10: R38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon L, Martinez-Perez E, Merkenschlager M 2013. Condensin, cohesin and the control of chromatin states. Curr Opin Genet Dev 23: 204–211 [DOI] [PubMed] [Google Scholar]
- Belton JM, McCord RP, Gibcus JH, Naumova N, Zhan Y, Dekker J 2012. Hi-C: A comprehensive technique to capture the conformation of genomes. Methods 58: 268–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K 2009. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 19: 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Bouwman BAM, Zhu Y, Klous P, Splinter E, Verstegen MJAM, Krijger PHL, Festuccia N, Nora EP, Welling M, et al. 2013. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature 501: 227–231 [DOI] [PubMed] [Google Scholar]
- Dimitrieva S, Bucher P 2013. UCNEbase—a database of ultraconserved non-coding elements and genomic regulatory blocks. Nucleic Acids Res 41: D101–D109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Merkenschlager M 2013. Cohesin at active genes: A unifying theme for cohesin and gene expression from model organisms to humans. Curr Opin Cell Biol 25: 327–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure AJ, Schmidt D, Watt S, Schwalie PC, Wilson MD, Xu H, Ramsay RG, Odom DT, Flicek P 2012. Cohesin regulates tissue-specific expression by stabilizing highly occupied cis-regulatory modules. Genome Res 22: 2163–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, et al. 2013. Ensembl 2013. Nucleic Acids Res 41: D48–D55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P, Bickmore W 2007. Nuclear organization of the genome and the potential for gene regulation. Nature 447: 413–417 [DOI] [PubMed] [Google Scholar]
- Fudenberg G, Mirny LA 2012. Higher-order chromatin structure: Bridging physics and biology. Curr Opin Genet Dev 22: 115–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause M, Misulovin Z, Bilyeu A, Dorsett D 2010. Dosage-sensitive regulation of cohesin chromosome binding and dynamics by Nipped-B, Pds5, and Wapl. Mol Cell Biol 30: 4940–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J 2006. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr Biol 16: 1571–1578 [DOI] [PubMed] [Google Scholar]
- Gibcus JH, Dekker J 2013. The hierarchy of the 3D genome. Mol Cell 49: 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951 [DOI] [PubMed] [Google Scholar]
- Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng HL, Hansen E, Despo O, Bossen C, Vettermann C, et al. 2011. CTCF-binding elements mediate control of V(D)J recombination. Nature 477: 424–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M 2009. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460: 410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix DA, Hong J-W, Zeitlinger J, Rokhsar DS, Levine MS 2008. Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc Natl Acad Sci 105: 7762–7767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth RS, Gruenewald-Schneider U, Webb S, Kerr ARW, James KD, Turner DJ, James KD, Turner DJ, Smith C, Harrison DJ, et al. 2010. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet 6: e1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imakaev M, Fudenberg G, McCord RP, Naumova N, Goloborodko A, Lajoie BR, Dekker J, Mirny LA 2012. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods 9: 999–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessberger R 2012. Age-related aneuploidy through cohesion exhaustion. EMBO Rep 13: 539–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi S, Calof AL, Santos R, Lopez-Burks ME, Young CM, Hoang MP, Chua A, Lao T, Lechner MS, Daniel JA, et al. 2009. Multiple organ system defects and transcriptional dysregulation in the Nipbl+/− mouse, a model of Cornelia de Lange Syndrome. PLoS Genet 5: e1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA 2009. Circos: An information aesthetic for comparative genomics. Genome Res 19: 1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM 2006. Wapl controls the dynamic association of cohesin with chromatin. Cell 127: 955–967 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Benner C, Mansson R, Heinz S, Miyazaki K, Miyazaki M, Chandra V, Bossen C, Glass CK, Murre C 2012. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat Immunol 13: 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdinier F, Yusufzai TM, Felsenfeld G 2004. Both CTCF-dependent and -independent insulators are found between the mouse T cell receptor α and Dad1 genes. J Biol Chem 279: 25381–25389 [DOI] [PubMed] [Google Scholar]
- McCord RP, Nazario-Toole A, Zhang H, Chines PS, Zhan Y, Erdos MR, Collins FS, Dekker J, Cao K 2013. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res 23: 260–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNairn AJ, Gerton JL 2009. Intersection of ChIP and FLIP, genomic methods to study the dynamics of the cohesin proteins. Chromosome Res 17: 155–163 [DOI] [PubMed] [Google Scholar]
- Merkenschlager M, Odom DT 2013. CTCF and cohesin: Linking gene regulatory elements with their targets. Cell 152: 1285–1297 [DOI] [PubMed] [Google Scholar]
- Meuleman W, Peric-Hupkes D, Kind J, Beaudry JB, Pagie L, Kellis M, Reinders M, Wessels L, van Steensel B 2013. Constitutive nuclear lamina–genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res 23: 270–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misulovin Z, Schwartz YB, Li XY, Kahn TG, Gause M, MacArthur S, Fay JC, Eisen MB, Pirrotta V, Biggin MD, et al. 2008. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma 117: 89–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon T, Duboule D 2012. Landscapes and archipelagos: Spatial organization of gene regulation in vertebrates. Trends Cell Biol 22: 347–354 [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH 2009. Cohesin: Its roles and mechanisms. Annu Rev Genet 43: 525–558 [DOI] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onn I, Koshland D 2011. In vitro assembly of physiological cohesin/DNA complexes. Proc Natl Acad Sci 108: 12198–12205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. 2008. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132: 422–433 [DOI] [PubMed] [Google Scholar]
- Phillips JE, Corces VG 2009. CTCF: Master weaver of the genome. Cell 137: 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, et al. 2013. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153: 1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro de Almeida C, Stadhouders R, De Bruijn MJ, Bergen IM, Thongjuea S, Lenhard B, van Ijcken W, Grosveld F, Galjart N, Soler E, et al. 2011. The DNA-binding protein CTCF limits proximal Vκ recombination and restricts κ enhancer interactions to the immunoglobulin κ light chain locus. Immunity 35: 501–513 [DOI] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A 2008. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci 105: 8309–8314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CA, Misulovin Z, Sahota G, Siddiqui AM, Schwartz YB, Kahn TG, Pirrotta V, Gause M, Dorsett D 2009. Regulation of the Drosophila enhancer of split and invected-engrailed gene complexes by sister chromatid cohesion proteins. PLoS ONE 4: e6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Schwalie PC, Ross-Innes CS, Hurtado A, Brown GD, Carroll JS, Flicek P, Odom DT 2010. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res 20: 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, et al. 2011. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature 476: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HY, Verma-Gaur J, Torkamani A, Feeney AJ, Galjart N, Krangel MS 2012. Tcra gene recombination is supported by a Tcra enhancer- and CTCF-dependent chromatin hub. Proc Natl Acad Sci 109: E3493–E3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM 2008. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J 27: 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan T 2005. Cornelia de Lange syndrome and the link between chromosomal function, DNA repair and developmental gene regulation. Curr Opin Genet Dev 15: 258–264 [DOI] [PubMed] [Google Scholar]
- Tedeschi A, Wutz G, Huet S, Jaritz M, Wuensche A, Schirghuber E, Davidson IF, Tang W, Cisneros DA, Bhaskara V, et al. 2013. Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature 501: 564–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turro E, Su S-Y, Gonçalves Â, Coin LJ, Richardson S, Lewin A 2011. Haplotype and isoform specific expression estimation using multi-mapping RNA-seq reads. Genome Biol 12: R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B 2011. Chromatin: Constructing the big picture. EMBO J 30: 1885–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables WN, Ripley BD 2002. Modern applied statistics with S, 4th ed. Springer, New York [Google Scholar]
- Wallace JA, Felsenfeld G 2007. We gather together: Insulators and genome organization. Curr Opin Genet Dev 17: 400–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451: 796–801 [DOI] [PubMed] [Google Scholar]
- Xu H, Handoko L, Wei X, Ye C, Sheng J, Wei C-L, Sung WK 2010. A signal-noise model for significance analysis of ChIP-seq with negative control. Bioinformatics 26: 1199–1204 [DOI] [PubMed] [Google Scholar]
- Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV 2012. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell 149: 467–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong XP, Krangel MS 1999. Enhancer-blocking activity within the DNAse I hypersensitivity site 2 to 6 region between the TCR α and Dad1 genes. J Immunol 163: 295–300 [PubMed] [Google Scholar]