Abstract
The DNA-uptake signal sequence (USS) of the bacterium Haemophilus influenzae is highly over-represented in its genome (1,471 copies of the core sequence AAGTGCGGT), and DNA fragments containing USS are preferentially taken up by competent cells. Because this bias favors uptake of conspecific DNA, USSs are often considered a kind of mate recognition system in bacteria, acting as species-specific barriers against uptake of unrelated DNA. However, the H. influenzae USS is highly over-represented in the genomes of three otherwise-divergent Pasteurellaceae species (Pasteurella multocida, Haemophilus somnus, and Actinobacillus actinomycetemcomitans, 927, 1,205, and 1,760 copies, respectively), suggesting that USSs do not always limit exchange. USSs in all these genomes are mainly in coding regions and show no orientation bias around the chromosome, weakening proposed USS functions in transcription termination and chromosome replication. Alignment of homologous genes was used to determine evolutionary relationships between individual USSs. Most H. influenzae USSs were found to have perfect or imperfect homologs (USS at the same location) in at least one other species, and most USSs in the other species had perfect or imperfect homologs in H. influenzae. These homologies suggest that the use of a common USS is due to inheritance of the USS-based uptake system from a common ancestor of the Pasteurellaceae, and it indicates that individual USSs can be evolutionarily stable elements of their genomes. The pattern is consistent with a molecular drive model of USS evolution, with new USSs arising by mutation and preferentially spread to new genomes by the biased DNA-uptake system.
Keywords: natural competence, transformation, Actinobacillus, Haemophilus, Pasteurella
Natural competence is the genetically specified ability of many bacteria to take up DNA from the surrounding environment. The distribution of natural competence is sporadic; although it is widespread it is not confined to a specific lineage in the bacterial phylogenetic tree, suggesting that its value to the cells varies with genetic and environmental conditions. It has been well characterized in a number of different bacteria, including Neisseria gonorrhoeae and other Neisseria species (1–3), Streptococcus pneumoniae (4, 5), Haemophilus influenzae (6), Bacillus subtilis (7), and Acinetobacter calcoaceticus (8) [for a general review see Dubnau (9)]. However, its main function is still controversial; although competence allows genetic exchange and recombination in bacteria (10), the nutritional benefits of taking up DNA may provide sufficient selective advantage to account for its evolutionary maintenance (11).
The self-specificity of the DNA uptake system in some bacteria poses additional questions about the evolution of DNA uptake. Although most competent bacteria will take up any DNA, the well characterized competent species of the Neisseriaceae and Pasteurellaceae families preferentially take up DNA from close relatives (12–17). Two factors are responsible for this bias. First, the DNA uptake machinery on the cell surface preferentially binds and takes up fragments containing a specific short sequence (9 or 10 bp) called the uptake signal sequence (USS). Although the cell-surface proteins that bind the USS have not yet been identified, sequence-specific binding at the cell surface is strongly supported (references in ref. 6), and Danner et al. (14) have shown that DNA uptake is specifically blocked by ethylation of bases in the USS. Second, the genomes of these species are highly enriched for their preferred sequence, so most fragments larger than 2 kb will contain one or more USSs (18–20). In the Neisseriaceae species N. gonorrhoeae and Neisseria meningitidis the highly conserved core of the USS is the 10-bp sequence GCCGTCTGAA, present in 1,891 copies in the 2.18-megabase (Mb) N. meningitidis Z2491 genome (18, 20). In H. influenzae Rd (Pasteurellaceae) the USS core is the 9-bp sequence AAGTGCGGT, with 1,471 copies in the 1.83-Mb genome (20).
The evolutionary forces responsible for the biased uptake system have not been investigated. Most researchers originally assumed that the system exists because uptake of self-DNA is more beneficial than uptake of DNA from other species (i.e., USSs are genomic identity tags). However, competent H. influenzae cells efficiently take up DNA from other members of the Pasteurellaceae (17, 21). Furthermore, DNA containing H. influenzae USSs is preferred by Actinobacillus actinomycetemcomitans over unrelated DNAs, and its genome is enriched with copies of the H. influenzae USS core (22). The finding that some species with moderately divergent genomes (70–75% DNA identity in homologous genes) may share a common USS suggests that the USS system may not be maintained by the benefit of excluding foreign DNA.
A satisfactory explanation for USS-biased systems must account for both the bias of a bacterium's DNA uptake machinery and the abundance of the preferred sequence in its own genome. The latter requirement is especially problematic because under the identity tag model USSs provide no direct benefits: they affect only DNA uptake by other cells, and only if the cell originally containing them releases its DNA. Thus under this model accumulation of USSs in the genome might be expected to require (at least) strong kin selection, if not the extreme altruism of programmed cell death (23). However the problem can be greatly simplified by recognizing that, provided the uptake machinery is biased, USSs may be maintained (14, 19) or even increase (24) without being beneficial. Instead USSs will accumulate as a direct consequence of the uptake bias, in a form of what is often called “molecular drive” (25). Provided the DNA that cells take up sometimes recombines with and replaces a chromosomal homolog, any bias in the fragments taken up by the uptake system will be introduced into the genome. Thus the AAGTGCGGT repeat may have become abundant in the H. influenzae genome simply because the DNA uptake system prefers fragments containing it.
Although biased uptake can explain the present abundance of USSs, identification of other functions could help clarify their origin and present maintenance. There is no evidence that USSs influence the fate of incoming DNA once it enters the cell, although such effects are difficult to rule out. USSs are often found in inverted-repeat pairs downstream from coding regions, where they are likely to act as transcription terminators (18, 20). Termination is clearly not the primary function of USSs, because 50% of the USSs in N. meningitidis and 65% of those in H. influenzae lie within ORFs and most of the rest are not in inverted-repeat pairs (20). Because stem-loops with conserved sequences are not characteristic of transcription terminators in other genomes, this role for USSs is likely to be a consequence of accumulation of USSs in the genome, with selection for efficient termination favoring dyad-pair USSs that arise at the ends of transcripts. USSs are unlikely to function as Chi sequences, interacting with the RecBCD complex and promoting production of recombinogenic strands, because H. influenzae has a Chi sequence unrelated to its USS (26). Because closely spaced USSs are less common in the H. influenzae genome than predicted for randomly located sequence elements, Karlin et al. (27) proposed that USSs have a structural function in the compaction of the chromosome or in DNA replication or repair. However, no intracellular USS-binding proteins are known (19), and the H. influenzae and N. meningitidis USSs show no orientation bias around the chromosome, such as would be expected for a sequence that interacts with DNA replication machinery. Furthermore, this nonrandom distribution can also be explained by biased uptake, because molecular drive will act less strongly on two closely spaced USSs than on two isolated ones.
Comparisons of related genomes could address many questions about USS evolution. What forces determine the density and distribution of USSs in the genome? Are individual USSs stable entities, or are they often gained and lost? Do USSs evolve faster or slower than the rest of the genome? Does a species' USS consensus change with time? Are imperfect USSs subject to the same forces as perfect USSs? Fortunately, suitable comparisons are now possible for the H. influenzae USSs, as the annotated Pasteurella multocida genome is available and several other pasteurellacean genomes are nearing completion. H. influenzae and P. multocida shared a common ancestor ≈270 million years ago (28), and a preliminary analysis found many copies of the H. influenzae USS core in the P. multocida genome.§ Below we use comparisons of these two sequences to investigate the origin, maintenance, and evolution of USSs. A similar analysis of the unfinished genome sequences of Haemophilus somnus and A. actinomycetemcomitans is then used to test whether the conclusions can be generalized to the Pasteurellaceae family.
Methods
H. influenzae, P. multocida, and Haemophilus ducreyi complete genome sequences were downloaded from The Institute for Genomic Research and National Center for Biotechnology Information websites [ftp://ftp.tigr.org/pub/data/(NC_000907), ftp://ftp.ncbi.nih.gov/genomes/Bacteria/(NC_002663), and ftp://ftp.ncbi.nih.gov/genomes/Bacteria/(NC_002940)]. The available genome sequences of H. somnus and A. actinomycetemcomitans were downloaded from http://genome.ornl.gov/microbial/hsom/ and www.genome.ou.edu/act.html, respectively. The perl programs n-mer_HI.pl and n-mer_PM.pl were used to search the H. influenzae and P. multocida sequences for over-represented oligonucleotides of 8–12 bases. These and other programs and supplementary files referred to below are available at http://pooh.phy.ncu.edu.tw/∼tychen/USSI/. The perl programs exp_HI.pl and exp_PM.pl were used to calculate the expected numbers of these oligonucleotides in these genomes. The numbers of H. influenzae USS cores in the available H. somnus and A. actinomycetemcomitans sequences were determined by using the search function of Microsoft word.
Interspecific comparisons of H. influenzae and P. multocida used the 719 homologous gene pairs identified as genes specifying the same named proteins. The USSs in the set of 154 H. influenzae:P. multocida homologs where both members of the pair contained at least one USS (gene set 1) were initially characterized by a position score, calculated as the proportional distance of the 5′ end of every USS from the 5′ end of its gene. USSs in the minus orientation with respect to the direction of transcription were given negative scores.
Pairwise alignment of coding genes used the program macclade 4.0, which allows simultaneous examination of nucleotide and amino acid sequences. Alignments were done first for 54 of the 107 H. influenzae:P. multocida gene pairs in gene set 1 having at least one perfect USS with no putative homolog (no USS in the homolog with a position score within 5% of its position score) (gene set 2), and then for 78 homologous gene pairs with at least one perfect USS in H. influenzae but none in their P. multocida homologs, and for 40 with at least one in P. multocida but none in H. influenzae (gene set 3). Alignments to H. influenzae genes were also done for homologs of 20 of these genes in H. somnus and A. actinomycetemcomitans. The homologs were identified by blast searches (www.ncbi.nlm.nih.gov/sutils/genom_table.cgi) because the genomes have not yet been annotated. A Microsoft Word search was used to locate H. ducreyi copies of the H. influenzae USS, and blast was used to search for H. influenzae homologs of those H. ducreyi genes with USS.
Results
USS Frequency. To confirm that the H. influenzae core USS was the only sequence highly over-represented in the H. influenzae and P. multocida genomes, we tabulated and examined all of the over-represented sequences of 8–12 bases. In both genomes, the most abundant 9-bp repeats are indeed identical to the H. influenzae USS. The most common shorter repeats are subsets of the USS, and the most common longer repeats contain it (see supplementary files HI_8-12_mer.zip and PM_8-12_mer.zip at http://pooh.phy.ncu.edu.tw/∼tychen/USSI/). The analysis confirmed the 1,471 USSs in the H. influenzae genome reported by Smith et al. (20) and identified 927 in the P. multocida genome. Only 8 (H. influenzae) or 12 (P. multocida) USS copies would be expected for arbitrary sequences of the same lengths and base compositions as these genomes. In P. multocida as in H. influenzae there is no significant difference between the numbers of USSs in the + and – orientations (468 + and 459 –, and 737 + and 734 –, respectively). Although most of the P. multocida USSs are in coding sequences (557), the proportion (60%) is lower than expected for the 89% coding sequences. This distribution is also similar to that in H. influenzae, where only 65% of the USSs are in the 86% coding sequences. Less than half of all genes of both species contain USSs (38% of H. influenzae genes and 22% of P. multocida genes). Overall the similarities of the H. influenzae and P. multocida USS distributions suggest that the USSs are created and maintained by the same types of evolutionary forces in both species. Although P. multocida has not been reported to take up DNA under laboratory conditions (29), its genome does contain the full complement of genes known to be needed for DNA uptake by H. influenzae (M.B. and R.J.R., unpublished results). The lower density of USSs in its genome (about half that of H. influenzae) may reflect a reduced frequency or importance of DNA uptake.
Initial Characterization of USS Homology by Position in the Gene. The high frequency of the H. influenzae USS in P. multocida may be best explained by inheritance of a USS-specific DNA uptake system from the common ancestor of both species. The analysis below confirms this explanation and further demonstrates that many copies of the USS are in homologous positions in the two genomes, and thus are likely to have been directly inherited from the ancestral genome.
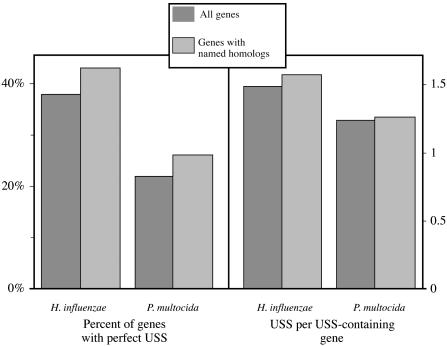
The H. influenzae and P. multocida genomes are too diverged to allow determination of homology of specific USS elements by nucleotide sequence comparisons alone. Instead the greater conservation of amino acid sequences in protein-coding genes was used to identify homologous USSs in the underlying DNA sequences. The analysis was limited to homologous genes assigned the same name, as this set contains genes of known function and reasonable full-length sequence similarity. The comparisons in Fig. 1 show that, with respect to USS content, these homologous genes are a representative sample of the total genes in both H. influenzae and P. multocida, allowing results for the homologous genes to be extrapolated to the total genes of these species. The frequency of USSs in the genes with named homologs (shaded bars) is slightly higher than in the total genes (solid bars), consistent with the larger average size of these genes (≈1,113 bp) than the average gene in either species (927 bp in H. influenzae and 997 bp in P. multocida).
Fig. 1.
Comparison of USSs in all H. influenzae and P. multocida genes (dark bars) with those in genes with named homologs (light bars).
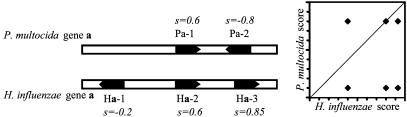
To identify homologies between USSs, the analysis was initially restricted to the 154 homologous gene pairs where both the H. influenzae and P. multocida homologs had at least one USS (gene set 1). A low-resolution positional criterion was first used to determine whether pairs of USSs were likely to be homologous. To avoid biasing the search by any assumption of homology, the positions and orientations of all the USSs in each of the pairs of homologous genes were compared, even though this analysis inevitably included many nonhomologous comparisons. The logic of this analysis is illustrated in Fig. 2. Each USS in each gene was assigned a score between –1 and +1, reflecting its position within the gene and its orientation relative to the direction of transcription. For example, a USS whose 5′ end (5′-AAGTGCGGT-3′) was at nucleotide 400 of a 1,000-bp gene would be assigned a score of 0.4, and one in the opposite orientation (5′-TCCGCACTT-3′), a score of –0.4. The position scores of all of the USSs in each of the homologous genes were then plotted against each other.
Fig. 2.
Schematic analysis of the H. influenzae and P. multocida homologs of a hypothetical gene a. H. influenzae gene a contains three USSs, with position scores s =–0.2, s = 0.6, and s = 0.85. P. multocida gene a contains two USSs, with scores s = 0.6 and s =–0.8 (position 0.8 in the negative orientation). Plotting all these scores against each other gives the six spots shown in the graph. One comparison falls on the diagonal and is thus between putative homologs (comparison Ha-2:Pa-1), and the remaining five comparisons fall off the diagonal and are likely between nonhomologous USSs.
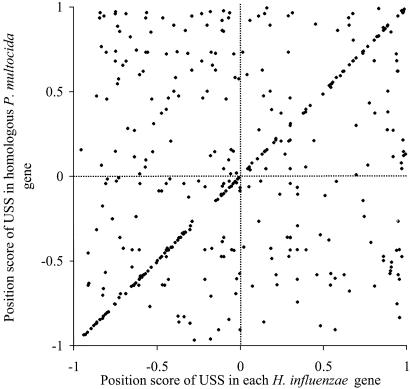
The results for all of the homologs are shown in Fig. 3. The area within 5% of the diagonal contains 136 spots, each of which represents a pair of USSs in the same orientations and approximate positions in a pair of homologous genes. Interpretation of the spots not on the diagonal requires consideration of the large number of nonhomologous comparisons inevitable with this analysis. For example, the pair of homologous genes shown in Fig. 2 could have given at most two USS comparisons between homologs (giving points on or near the diagonal in Fig. 3) and four nonhomologous comparisons (points off the diagonal), because one member of the pair has only two USSs. Table 1 summarizes the expectations for the actual pairs compared, showing that only 193 of the 408 USS comparisons (points in Fig. 3) could have been between homologous USSs. As this is only 57 more than the observed 136 putative homologs (spots falling within 5% of the diagonal), and these 136 pairs include almost half of the 281 H. influenzae USSs and 66% of the P. multocida USSs in gene set 1, we conclude that about half of USSs have likely homologs.
Fig. 3.
Relationship between positions and orientations of H. influenzae and P. multocida USSs within homologous genes (see text and Fig. 2 for detailed explanation of the analysis). Each point represents a single intragenic USS comparison. Positive and negative values reflect USSs orientation with respect to the direction of transcription.
Table 1. Number of possible H. influenzae (Hi):P. multocida (Pm) USS pairs and homologies in the 154 homologous genes with USSs in both species (gene set 1).
| No. of USSs in each homolog (Hi:Pm) | No. of Hi:Pm gene pairs of this type | Comparisons per gene pair of this type | Total comparisons of this type | Possibly homologous comparisons per gene pair | Possibly homologous comparisons of this type |
|---|---|---|---|---|---|
| 1:1 | 61 | 1 | 61 | 1 | 61 |
| 1:2 + 2:1 | 10 + 37 | 2 | 94 | 1 | 47 |
| 1:3 + 3:1 | 1 + 8 | 3 | 27 | 1 | 9 |
| 1:4 + 4:1 | 0 + 2 | 4 | 8 | 1 | 2 |
| 1:5 + 5:1 | 0 + 1 | 5 | 5 | 1 | 1 |
| 2:2 | 14 | 4 | 56 | 2 | 28 |
| 2:3 + 3:2 | 0 + 11 | 6 | 66 | 2 | 22 |
| 2:4 + 4:2 | 0 + 2 | 8 | 16 | 2 | 4 |
| 2:5 + 5:2 | 0 + 3 | 10 | 30 | 2 | 6 |
| 3:3 | 3 | 9 | 27 | 3 | 9 |
| 4:5 + 5:4 | 0 + 1 | 20 | 20 | 4 | 4 |
| Total | 154 | 408 | 193 |
The points in the upper left and lower right quadrants of Fig. 3 represent comparisons between USSs in different orientations in the same gene. The absence of any concentration of points on the negative-slope diagonal indicates that oppositely oriented USSs are not preferentially found at the same positions and thus are unlikely to be homologous. This finding implies that USSs do not invert orientations during their evolution. The even distribution of USSs with positive and negative scores shows that USS orientation is not influenced by their direction of transcription.
Confirmation of USS Homology by Sequence Alignments. Nucleotide and amino acid sequence alignments were used to confirm that the putatively homologous USSs identified above are in genuinely homologous positions and to characterize the USSs apparently lacking homologs. The results are summarized in Table 2. Because the alignments were laborious, the analysis was initially limited to half of the 107 pairs of homologs in gene set 1, where at least one USS lacked a putative homolog (gene set 2). Alignments showed that 36 of the 38 putatively homologous USSs in the 54 homologous gene pairs were truly homologous, confirming that the 5% cut-off used in Fig. 3 was conservative. None of the other 85 H. influenzae USSs and 39 P. multocida USSs acquired perfect homologs once their sequences were correctly aligned, confirming that the 5% cut-off was not excessively conservative.
Table 2. USS homology in homologous H. influenzae (Hi) and P. multocida (Pm) genes.
| Putative USS homology
|
Authentic USS homology after alignment
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Imperfect homologs‡
|
|||||||||||
| Gene set | Species | No. of genes | Total USSs | Homologs* | Nonhomologs† | Homologs‡ | 8 matches | 7 matches | 6 matches | 5 matches | Nonhomologs‡ |
| 2 | Hi | 54 | 123 | 85 | 13 | 10 | 13 | 17 | 34 | ||
| 38 | 36 | ||||||||||
| Pm | 54 | 77 | 39 | 4 | 2 | 3 | 7 | 25 | |||
| 3 | Hi | 78 | 93 | 0 | 12 | 13 | 15 | 16 | 37 | ||
| Pm | 40 | 41 | 0 | 15 | 2 | 3 | 5 | 16 | |||
≤5% difference in position scores of perfect USS.
>5% difference in position scores of perfect USS.
Determined by nucleotide sequence alignment.
Because the H. influenzae genome also contains many copies of sequences imperfectly matched to the USS consensus, the alignments of USSs without putative homologs were also examined for imperfect matches. Most were found to have homologously positioned imperfect matches to the USS core. The right-hand columns of Table 2 break this down, showing the numbers of USSs matching imperfect homologs at 5, 6, 7, and 8 of the 9 positions. Chance matches at 5 of 9 bases are unlikely (P = 0.034), and chance matches at 6 or more positions are very unlikely (P ≤ 0.008). In gene set 2, 16 of the 41 P. multocida USSs with no putative homologs aligned with imperfect USSs in H. influenzae, and 53 of the 87 H. influenzae USSs with no putative homologs aligned with imperfect USSs in P. multocida.
Analysis of imperfectly matched USSs was then extended to a subset of the original 719 gene pairs that had not previously been examined, those where only one member of the pair has any perfect USSs (gene set 3, Table 2). We analyzed 78 USS-containing H. influenzae genes whose P. multocida homologs lacked USSs, and 40 USS-containing P. multocida genes whose H. influenzae homologs lacked USSs. Fifty-six of the 93 H. influenzae USSs (60%) and 25 of the 41 P. multocida USSs (61%) had imperfect homologs. Overall, most USSs examined by alignment were found to have homologous perfect or imperfect USS in the other species (145/216 = 67% in H. influenzae and 77/118 = 65% in P. multocida). These then represent perfect or imperfect USSs present in the common ancestor and maintained over 270 million years of evolution.
USS Homology in Other Species. To find out whether USS stability is general across the Pasteurellaceae, the analysis was extended to other members of this family. The fully annotated genome sequence of H. ducreyi is available on line, but a description has not yet been published. We found it to carry only 41 USSs, more than the 8 predicted for a genome of this size and composition (1.7 megabases, 38% G+C) but far fewer than found in H. influenzae or P. multocida. Most of these USSs cluster in two small islands containing bacteriophage genes. Despite its genus assignment, H. ducreyi is thought to be only distantly related to other Pasteurellaceae (30, 31), and this USS distribution may reflect the presence of USSs in horizontally transferred sequences, as has been found for the H. influenzae phage HP1 (32).
Nonannotated genome sequences are available for two other Pasteurellaceae, H. somnus and A. actinomycetemcomitans, and these sequences were used to extend the analysis. The genome sequence of H. somnus is ≈90% complete and has been assembled into 64 contigs; the 1.9 megabases available are 37% G+C. The A. actinomycetemcomitans sequence is complete, with 2,105,332 bp and 42% G+C. Although only 8 and 12 perfect copies of the H. influenzae USS are expected by chance in these genomes, 1,205 were identified in H. somnus (606 in the + orientation and 599 in the –), and 1,760 in A. actinomycetemcomitans (881 + and 879 –). These densities and lack of orientation bias resemble those seen in H. influenzae and P. multocida.
To examine the divergence of individual USSs, blast searches were used to identify H. somnus and A. actinomycetemcomitans homologs of 20 of the 54 H. influenzae genes analyzed in gene set 2. Table 3 shows the USS homologies identified in the aligned sequences. Most of the perfect H. influenzae USSs examined aligned with perfect or imperfect A. actinomycetemcomitans and H. somnus USSs, and most of the perfect A. actinomycetemcomitans and H. somnus USSs examined aligned with perfect or imperfect H. influenzae USSs. Overall, in these genes, 66% of the perfect USSs in the H. influenzae:P. multocida comparisons have perfect or imperfect homologs, as do 68% of the USSs in the H. influenzae:H. somnus comparisons and 75% of the USSs in the H. influenzae:A. actinomycetemcomitans comparisons.
Table 3. Numbers of homologous USSs identical at different numbers of positions in 20 homologous genes of H. influenzae (Hi), P. multocida (Pm), H. somnus (Hs), and A. actinomycetemcomitans (Aa).
| Identity to perfect 9-bp USS core
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Comparison | 9:9 | 9:8 | 8:9 | 9:7 | 7:9 | 9:6 | 6:9 | 9:5 | 5:9 | 9:≤4 | ≤4:9 | Total 9s |
| Hi:Pm | 12 | 4 | 1 | 1 | 1 | 7 | 2 | 4 | 2 | 15 | 8 | 43:26 |
| (Cumulative total) | (24) | (29) | (31) | (40) | (46) | (69) | ||||||
| Hi:Hs | 16 | 4 | 3 | 1 | 1 | 0 | 3 | 7 | 2 | 15 | 10 | 43:35 |
| (Cumulative total) | (32) | (39) | (41) | (44) | (53) | (78) | ||||||
| Hi:Aa | 23 | 1 | 2 | 1 | 1 | 2 | 2 | 5 | 3 | 11 | 10 | 43:41
|
| (Cumulative total) | (46) | (49) | (51) | (55) | (63) | (84) | ||||||
Discussion
Why are the DNA uptake mechanisms of some naturally competent bacteria biased toward a repeated sequence highly over-represented in their own genomes, whereas other bacteria treat all DNAs equally? Until now USS-biased uptake systems have been viewed as adaptations to prevent uptake of DNA from cells of other species. In the conceptual framework that viewed competence as a process evolved because of the benefits of genetic exchange, USSs were thought to function as identity tags to allow recognition of DNA from suitable mates (24). This hypothesis predicted that distinctive USS systems (different uptake biases and corresponding abundant repeats) would be found in species likely to encounter each other's DNA, and also that USSs would evolve rapidly, as is the case for known mate-recognition systems (33).
However the presence of the same USS sequence in the genomes of three different genera of human pathogens shows this view to be mistaken. The explanation is not simply that closely related bacteria have been incorrectly assigned to different genera. Although the phylogeny of the Pasteurellaceae is not well resolved, the average DNA sequence identity between identifiable homologs of H. influenzae and P. multocida is comparable to that between Escherichia coli and Salmonella typhimurium [71% (34)], low enough to substantially limit recombination with incoming foreign DNA (21). Not only are the USS consensus sequences in different genera identical but also most individual USS elements are shared, indicating they have been inherited from the common ancestor of these species. USSs are thus now shown to be evolutionarily stable entities, lacking the features typical of mate-recognition systems (33).
Although rarely questioned, the mate-recognition function originally proposed for USSs lacks a solid evolutionary foundation. Beneficial new combinations can be produced by genetic exchange, but there is little reason to expect the long-term benefits of exchange to outweigh its short-term costs, in part because recombination also breaks up coadapted sets of genes. This break-up is one of the reasons we still lack a satisfactory explanation for eukaryote sexual reproduction (35). Genes acquired by DNA uptake are especially likely to be deleterious because most if not all of the DNA in the environment comes from dead cells and will be disproportionately burdened with deleterious mutations (36, 37). Identification of adaptive horizontally transferred segments in bacterial genomes does not address these genetic costs (38), because we cannot know how many deleterious acquisitions have had to be eliminated by natural selection over the same time period. Other costs of genetic exchange by transformation include the reorganization of the cell surface required for DNA uptake and the risk of cell death when induction of the SOS response by incoming DNA activates resident prophages (39). Evolution of USSs as identity tags remains problematic even if genetic exchange is beneficial, because the USSs in a cell's genome play no role in transformation until after they have been released from the cell. Because the effect of each USS is to increase the chance that the DNA fragment containing it will be taken up by a neighboring cell, the direct benefit is experienced mainly by the USS itself, and to a lesser extent by flanking sequences. From this perspective USSs appear to function more as selfish elements than as factors conferring evolvability on the population or species. This distinction between direct (selfish) consequences and indirect population-level ones is key, because natural selection acts far more strongly on the former than the latter.
Thus USSs may not have evolved by selection for the benefits of restricting DNA uptake to closely related sequences. Some alternative hypotheses explain the abundance of USSs by invoking an intracellular function independent of DNA uptake, for example as transcriptional terminators (18). Although such functions may have influenced the initial choice of the specific sequence of the USS, and may play secondary roles today, they are unable to account for the majority of USSs in extant organisms' genomes and also fail to explain why there should be such a strong correspondence between the bias of the DNA uptake machinery and the abundance of its preferred sequence.
We suggest that the best explanation for USS abundance is the molecular drive created by the biased uptake system, which can produce an excess of its preferred sequence in the genome. This drive arises from the interaction of four processes: random mutation produces variant versions of the USSs (more-favored and less-favored by the uptake machinery); new and old versions are released into the environment, usually by cell death; biased uptake preferentially brings the favored version into the cytoplasm; and recombination replaces resident versions with those brought from outside. Thus mutations creating more-favored USSs are more likely to spread by transformation than other mutations, and mutations creating less-favored USSs are more likely to be preferentially lost by transformation with DNA from wild-type cells. Finally, selection eliminates those USS variants that interfere with gene function. From the perspective of the USSs, biased uptake creates the selective environment in which they can evolve as selfish elements.
This model requires only that bacteria sometimes take up DNA from members of their own or a closely related species, and that this DNA sometimes replaces a chromosomal homolog. It does not require USSs to have originally arisen by mutation. However, a mutational origin is likely, because USS-encoded amino acids in H. influenzae and N. meningitidis genes align with homologous amino acids in proteins from organisms lacking USS, such as E. coli and B. subtilis (unpublished data). An uptake system that strongly discriminates between perfect and singly mismatched USSs would explain the otherwise-surprising low frequency of the latter in the H. influenzae genome (27).
How could biased uptake have arisen in the first place? All known DNA-binding proteins have some intrinsic sequence bias, so we must expect that any DNA uptake system could produce a weak drive. However, in most bacteria any weak initial bias has not evolved into the extreme uptake bias and USS accumulation seen in the Pasteurellaceae and Neisseriaceae. We suggest that these Gram-negative bacteria have evolved USS-dependent DNA uptake in response to the barrier posed by their outer membranes. DNA is taken up across the Gram-positive membrane and the inner membrane of Gram-negative cells as a linear single strand, which can be threaded through a narrow channel. But H. influenzae and Neisseria spp. transport double-stranded DNA across their outer membranes, and in H. influenzae this transport is known to not require a free end (i.e., closed circular molecules are efficiently transported without nicking or cleavage) (40). At the scale of the cell surface, double-stranded DNA is a stiff and very hydrophilic rod [persistence length ≈50 nm (41)], and transport across the outer membrane likely requires both tight binding to a transport protein and the ability to create a kink in the DNA rod, perhaps by inducing local strand separation. Binding affinity is tightly linked to sequence specificity (42), so selection for effective transport across the outer membrane may have entailed selection for increased uptake bias and may have indirectly led to accumulation of increasingly precise sequences in the genome. Testing this hypothesis will require understanding the mechanism of sequence-specific uptake.
Acknowledgments
Trevor Cowley and Philip Chen contributed to early versions of this work. We thank the National Center for Biotechnology Information, the Institute for Genomic Research, and the Haemophilus ducreyi Sequencing Project (collaborative project between the Institute for Systems Biology, Seattle, and the laboratory of Dr. Robert Munson at the Children's Research Institute and the Ohio State University) for providing the genome sequences. We thank the editor and referees for helpful suggestions. This work was supported by National Institutes of Health Grant RO1 GM60715 (to R.J.R.) and Republic of China National Science Council Grant 91-2119-M-008-12 (to H.C.L.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: USS, uptake signal sequence.
Footnotes
Hong, B. K. & Dewhirst, F. E., IADR/AADR/CADR 80th General Session, March 6–9, 2002, San Diego, abstr. 1448.
References
- 1.Biswas, G. D., Thompson, S. A. & Sparling, P. F. (1989) Clin. Microbiol. Rev. 2, Suppl., S24–S28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jyssum, S. & Jyssum, K. (1970) Acta Pathol. Microbiol. Scand. [B] 78, 140–148. [DOI] [PubMed] [Google Scholar]
- 3.Saez-Nieto, J. A., Lujan, R., Martinez-Suarez, J. V., Berron, S., Vazquez, J. A., Vinas, M. & Campos, J. (1990) Antimicrob. Agents Chemother. 34, 2269–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lacks, S. (1979) J. Bacteriol. 138, 404–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lacks, S. & Greenberg, B. (1973) J. Bacteriol. 114, 152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodgal, S. H. (1982) Annu. Rev. Genet. 16, 169–192. [DOI] [PubMed] [Google Scholar]
- 7.Dubnau, D. (1991) Microbiol. Rev. 55, 395–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorenz, M. G., Reipschlager, K. & Wackernagel, W. (1992) Arch. Microbiol. 157, 355–360. [DOI] [PubMed] [Google Scholar]
- 9.Dubnau, D. (1999) Annu. Rev. Microbiol. 53, 217–244. [DOI] [PubMed] [Google Scholar]
- 10.Griffith, F. (1928) J. Hyg. 27, 113–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Redfield, R. J. (1993) J. Hered. 84, 400–404. [DOI] [PubMed] [Google Scholar]
- 12.Scocca, J. J., Poland, R. L. & Zoon, K. C. (1974) J. Bacteriol. 118, 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sisco, K. L. & Smith, H. O. (1979) Proc. Natl. Acad. Sci. USA 76, 972–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danner, D. B., Deich, R. A., Sisco, K. L. & Smith, H. O. (1980) Gene 11, 311–318. [DOI] [PubMed] [Google Scholar]
- 15.Graves, J. F., Biswas, G. D. & Sparling, P. F. (1982) J. Bacteriol. 152, 1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathis, L. S. & Scocca, J. J. (1982) J. Gen. Microbiol. 128, 1159–1161. [DOI] [PubMed] [Google Scholar]
- 17.Albritton, W. L., Setlow, J. K., Thomas, M., Sottnek, F. & Steigerwalt, A. G. (1984) Mol. Gen. Genet. 193, 358–363. [DOI] [PubMed] [Google Scholar]
- 18.Goodman, S. D. & Scocca, J. J. (1988) Proc. Natl. Acad. Sci. USA 85, 6982–6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith, H. O., Tomb, J.-F., Dougherty, B. A., Fleischmann, R. D. & Venter, J. C. (1995) Science 269, 538–540. [DOI] [PubMed] [Google Scholar]
- 20.Smith, H. O., Gwinn, M. L. & Salzberg, S. L. (1999) Res. Microbiol. 150, 603–616. [DOI] [PubMed] [Google Scholar]
- 21.Albritton, W., Setlow, J., Thomas, M. & Sottnek, F. (1986) Int. J. Syst. Bacteriol. 36, 103–106. [Google Scholar]
- 22.Wang, Y., Goodman, S. D., Redfield, R. J. & Chen, C. (2002) J. Bacteriol. 184, 3442–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bayles, K. W. (2003) Trends Microbiol. 11, 306–311. [DOI] [PubMed] [Google Scholar]
- 24.Redfield, R. J. (1991) Nature 352, 25–26.2062373 [Google Scholar]
- 25.Dover, G. (1982) Nature 299, 111–117. [DOI] [PubMed] [Google Scholar]
- 26.Sourice, S., Biaudet, V., El Karoui, M., Ehrlich, S. D. & Gruss, A. (1998) Mol. Microbiol. 27, 1021–1029. [DOI] [PubMed] [Google Scholar]
- 27.Karlin, S., Mrazek, J. & Campbell, A. M. (1996) Nucleic Acids Res. 24, 4263–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.May, B. J., Zhang, Q., Li, L. L., Paustian, M. L., Whittam, T. S. & Kapur, V. (2001) Proc. Natl. Acad. Sci. USA 98, 3460–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jablonski, L., Sriranganathan, N., Boyle, S. M. & Carter, G. R. (1992) Microb. Pathog. 12, 63–68. [DOI] [PubMed] [Google Scholar]
- 30.Casin, I., Grimont, F., Grimont, P. A. D. & Sanson-Le Pros, M.-J. (1985) Int. J. Syst. Bacteriol. 35, 23–25. [Google Scholar]
- 31.Hedegaard, J., Okkels, H., Bruun, B., Kilian, M., Mortensen, K. K. & Norskov-Lauritsen, N. (2001) Microbiology 147, 2599–2609. [DOI] [PubMed] [Google Scholar]
- 32.Esposito, D., Fitzmaurice, W. P., Benjamin, R. C., Goodman, S. D., Waldman, A. S. & Scocca, J. J. (1996) Nucleic Acids Res. 24, 2360–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blows, M. W. (1999) Proc. R. Soc. London Ser. B 266, 2169–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McClelland, M., Sanderson, K. E., Spieth, J., Clifton, S. W., Latreille, P., Courtney, L., Porwollik, S., Ali, J., Dante, M., Du, F., et al. (2001) Nature 413, 852–856. [DOI] [PubMed] [Google Scholar]
- 35.Kondrashov, A. S. (1993) J. Hered. 84, 372–387. [DOI] [PubMed] [Google Scholar]
- 36.Redfield, R. J. (1988) Genetics 119, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Redfield, R., Schrag, M. & Dean, A. (1997) Genetics 146, 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawrence, J. G. & Ochman, H. (1998) Proc. Natl. Acad. Sci. USA 95, 9413–9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Setlow, J. K., Boling, M. E., Beattie, K. L. & Kimball, R. F. (1972) J. Mol. Biol. 68, 361–378. [DOI] [PubMed] [Google Scholar]
- 40.Barany, F., Kahn, M. E. & Smith, H. O. (1983) Proc. Natl. Acad. Sci. USA 80, 7274–7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hagerman, P. J. (1988) Annu. Rev. Biophys. Biochem. 17, 265–286. [DOI] [PubMed] [Google Scholar]
- 42.Schildbach, J. F., Karzai, A. W., Raumann, B. E. & Sauer, R. T. (1999) Proc. Natl. Acad. Sci. USA 96, 811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]