Abstract
Comprehensive identification of sequences that regulate transcription is one of the major goals of genome biology. Focal alteration in chromatin structure in vivo, detectable through hypersensitivity to DNaseI and other nucleases, is the sine qua non of a diverse cast of transcriptional regulatory elements including enhancers, promoters, insulators, and locus control regions. We developed an approach for genome-scale identification of DNaseI hypersensitive sites (HSs) via isolation and cloning of in vivo DNaseI cleavage sites to create libraries of active chromatin sequences (ACSs). Here, we describe analysis of >61,000 ACSs derived from erythroid cells. We observed peaks in the density of ACSs at the transcriptional start sites of known genes at non-gene-associated CpG islands, and, to a lesser degree, at evolutionarily conserved noncoding sequences. Peaks in ACS density paralleled the distribution of DNaseI HSs. ACSs and DNaseI HSs were distributed between both expressed and nonexpressed genes, suggesting that a large proportion of genes reside within open chromatin domains. The results permit a quantitative approximation of the distribution of HSs and classical cis-regulatory sequences in the human genome.
Keywords: DNaseI hypersensitive sites, cis-regulatory elements, subtraction
Understanding the human genome and those of other complex organisms will require comprehensive delineation of the functional elements that regulate transcription and other chromosomal processes. In vivo, regulatory sequences are found to coincide with focal alterations in chromatin structure (1–4). Chromatin architecture plays a defining role in the control of eukaryotic genes in vivo because it determines the accessibility of critical genomic sequences to the regulatory and transcriptional machineries (1, 2). Active regulatory foci within genomic sequences are detectable experimentally on the basis of pronounced sensitivity to cleavage when intact nuclei are exposed to DNA-modifying agents, canonically, the nonspecific endonuclease DNaseI (3–5). The colocalization of DNaseI hypersensitive sites (HSs) with cis-active elements spans the spectrum of known transcriptional and chromosomal regulatory activities, including transcriptional enhancers, promoters, and silencers, insulators, locus control regions, and domain boundary elements (1, 3, 6). It is therefore expected that a comprehensive library of DNaseI hypersensitive sites from the human genome would contain many (if not all) of these classical cis-regulatory sequences.
We sought to exploit in vivo DNaseI hypersensitivity as the basis of a powerful and generic approach for de novo identification of functional noncoding sequences on a genome-wide level. We developed a method for isolating and cloning sequences flanking DNaseI cut sites introduced in the context of intact nuclei, and for enriching sequences associated with DNaseI hypersensitive sites using a subtractive procedure. Sequencing and genomic mapping of the resulting collection of active chromatin sequences (ACSs) provide the basis for genome-wide localization of DNaseI hypersensitive sites and for global analysis of the relationship between chromatin structure and gene expression.
Methods
Cell Culture. We cultured K562 [American Type Culture Collection (ATCC)] cells in humidified incubators at 37°C and 5% CO2 in air. Cells were grown in RPMI medium 1640 (Invitrogen) supplemented with 10% FBS. Cultures were harvested at a density of 5 × 105 cells/ml.
DNaseI Digestion and DNA Purification. We performed DNaseI digestions according to a standard protocol (7). After DNaseI treatments, DNA was purified by using the Puregene system (Gentra Systems) and resuspended in 10 mM Tris·Cl (pH 8.0). Samples were quantitated in triplicate by using a Spectramax 384 Plus UV spectrophotometer (Molecular Devices).
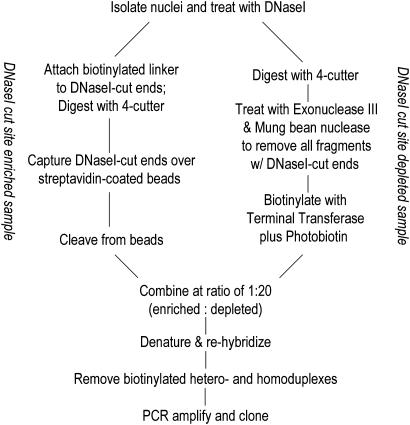
Creation of Genomic DNA Libraries Comprising ACSs. Under limiting conditions, DNaseI preferentially introduces cuts into open or “active” chromatin. We isolated DNaseI cut genomic DNA ends directly by using a linker-adapter strategy, and then used a subtractive procedure to remove background and further enrich for sequences from DNaseI hypersensitive sites. A schematic of the procedure appears in Fig. 1. Detailed protocol information is provided as Supporting Methods, which is published as supporting information on the PNAS web site.
Fig. 1.
Cloning of active chromatin sequences. We developed a strategy to create genomic DNA libraries containing sequences flanking DNaseI cut sites introduced into nuclear chromatin under limiting (hypersensitive) conditions. After DNA purification, free DNA ends are enzymatically repaired and ligated to a biotinylated linker adaptor. The DNA sample is then fragmented further with a four-cutter enzyme (NlaIII). At this stage, the genome has been partitioned into two predominant fragment populations: NlaIII–NlaIII fragments (derived from the non-DNaseI cut background) and NlaIII-adaptor fragments (carrying for DNaseI cut sites). Adapted DNA is efficiently isolated on paramagnetic streptavidin-coated beads, whereas NlaIII–NlaIII background fragments are cleansed. A second linker adaptor is then appended to the NlaIII end of captured DNA, and the product is released from the beads. This DNaseI cut site-enriched population is enriched and is retained for the subsequent subtraction step. A DNaseI cut site-depleted population is prepared by further fragmenting DNaseI-treated genomic DNA with a four-cutter that leaves a 3′ overhang (e.g., NlaIII). Further digestion of this sample with Exonuclease III followed by mung bean nuclease will preserve the NlaIII–NlaIII fragments (which are resistant to processive degradation), whereas fragments with DNaseI cut ends will be efficiently eliminated. The residual remaining population of DNaseI cut site-depleted DNA is then heavily biotinylated. An excess of this population is mixed with the DNaseI cut site-enriched population, and the sample is denatured and is slowly reannealed. Nonbiotinylated fragments generated by repeated DNaseI cleavage events at or around the same genomic coordinate (i.e., a hypersensitive site) will be more likely to self-anneal than find a partner in the DNaseI cut site-depleted population. Sites that have only been cut once (i.e., due to non-HS-specific cutting or to genomic shear) will form heteroduplexes. Extraction of the mixture with paramagnetic beads isolates the nonbiotinylated homoduplexes that are now further enriched in DNaseI hypersensitive sites. This population is PCR-amplified and cloned to make the genomic ACS libraries.
Creation of DNaseI hypersensitive site-enriched DNA. Isolated intact nuclei were digested with DNaseI to preferentially introduce double-stranded breaks into DNaseI hypersensitive sites. These ends were repaired and ligated to a common biotinylated adaptor (see Supporting Methods). Genomic DNA was then fractionated by digestion with NlaIII, and the biotinylated DNA was purified on paramagnetic streptavidin-coated beads (Dynal, Great Neck, NY). The isolated DNaseI cut site-enriched DNA was then ligated to a second adaptor (see Supporting Methods) and recovered from the beads by NotI-digestion.
Creation of DNaseI HS-depleted DNA. DNA was isolated from DNaseI-digested nuclei and was further digested with NlaIII to generate ends with four nucleotide 3′ overhangs, rendering them resistant to digestion with exonuclease III. The sample was then digested with exonuclease III followed by mung bean nuclease to eliminate single-stranded products (see Supporting Methods). Exonuclease III digests processively from double-stranded breaks introduced by DNaseI to render the fragments single stranded. Intact NlaIII–NlaIII fragments (which hence contain no DNaseI cut sites) will not be digested. These remaining fragments were biotinylated by the dual action of Terminal Transferase (Sigma) in the presence of Biotin-ddUTP, followed by chemical labeling with photobiotin. The resultant population was heavily biotinylated and depleted in hypersensitive sites.
Creation of HS-enriched library by using subtraction. DNaseI hypersensitive site-enriched DNA was mixed with an excess of DNaseI HS-depleted DNA, denatured, and slowly rehybridized. Biotinylated DNA (nonhypersensitive) was extracted by using streptavidin beads (Dynal), and the remaining nonbiotinylated DNA was amplified by using PCR and cloned (see Supporting Methods).
Primer Selection. We designed primers to amplify ≈250-bp genomic segments spanning candidate HS sequences with Primer3 (8).
Analysis of DNaseI Hypersensitivity by Hypersensitivity Quantitative PCR (HSqPCR). We used a real-time quantitative PCR-based method to quantify DNaseI hypersensitivity (9). Quantitative PCR reactions were performed on an ABI 7900HT Sequence Detection System (Applied Biosystems), and all of the reactions were assembled robotically by using a Biomek FX (Beckman). Melting curve analysis was performed for each amplicon, and those yielding multiple products were discarded. Efficiency-corrected Ct values were used to compute relative copy number ratios (DNaseI-treated vs. untreated samples) for each amplicon. Relative DNaseI sensitivity ratios were thus obtained. Ratios <1 are indicative of relative copy loss due to sequence-specific preferential cleavage of chromatin by DNaseI under limiting conditions.
Determination of DNaseI Hypersensitivity Threshold. DNaseI sensitivity ratios vary as a continuous function of genomic position. We therefore used a classifier approach to establish a threshold DNaseI sensitivity ratio for rigorous discrimination of candidate sequences as HSs. Subject to this criterion, HSs should appear as statistical outliers relative to background variability in the DNaseI HS ratio. To establish 95% confidence bounds on background variability, we took advantage of the fact that the DNaseI HS status of the entire alpha- and beta-globin gene domains has been extensively analyzed in K562 cells (10, 11). We selected 125 kb of nonhypersensitive sequence from these regions and designed 550 ≈225-bp nonoverlapping amplicons. We then obtained DNaseI sensitivity ratios for these amplicons as described above. Nine independent data points were collected for each amplicon (total 4,950 measurements). We then used the same approach to analyze 19 previously validated DNaseI HSs from these regions, which spanned a functional spectrum including enhancers, promoters, and locus-control region elements and insulator elements (10–12). To separate DNaseI hypersensitive amplicons and the genomic background, we considered the full distribution of HS ratios and used robust outlier methods (13) to identify replicate clusters that deviated significantly from the average background sensitivity. Further, only low variance replicates were accepted by selecting a cutoff data quality value equal to the median of the of the 569 variance measurements obtained. In this way, the data enabled us to obtain conservative global 95% outlier confidence values for hypersensitive sites and a corresponding cut-off hypersensitivity ratio, below which candidate sequences could be classified as “hypersensitive” with >95% confidence. We further generalized this model by incorporating data from 479 additional amplicons (4,311 measurements) spanning a total of 172 kb selected from five noncontiguous gene loci (ADA, TCR-α, c-Myc, CD2, and OPN1LW). The threshold HS criteria are conservative because they require both high data quality (low replicate variance) and a hypersensitivity profile that corresponds to a group of prominent HSs, leaving open the possibility that less intense “minor” hypersensitive sites may be misclassified as “nonhypersensitive.”
Microarray Expression Analysis. Gene expression analysis was performed on Agilent Human 1A oligo microarrays. Total RNA was isolated from 5 × 107 K562 cells with RNeasy total RNA isolation kit (Qiagen). cDNA and cRNA for expression analysis were generated from 5 μg of total RNA by using the Agilent Fluorescent Linear Amplification Kit (Agilent Technologies, Palo Alto, CA), and 4 μg of nucleic acid was used in each hybridization.
Results
Active Chromatin Sequences Are G + C-Rich. We produced a library (K5008) of ACSs from K562 cells, which display an erythroid phenotype. The motivation for selection of this tissue was the prior extensive experience with it for chromatin and DNaseI hypersensitivity studies (10, 11).
We cloned and sequenced a total of 92,115 ACSs from the K5008 library. After filtering to remove ACSs that fell within repeated sequences or did not map to the human genome build, we recovered a total of 61,561 ACSs. The mean length of filtered sequences was 86 bp, which permitted unique localization within the human genome sequence. The mean G + C content of K5008 library sequences was 51%, significantly higher than the genomic average of 38–41% (14, 15). This difference cannot be accounted for by statistical overrepresentation of classical CpG islands because the absolute number of such sequences within the overall K5008 set was small (see below). Its significance is further emphasized by the fact that the subtractive technique used to produce the ACS library will tend to select against G + C-rich sequences; on average these fragments have higher annealing temperatures and will cross-anneal (and thus be eliminated) more readily under the low-stringency hybridization conditions used to create the library.
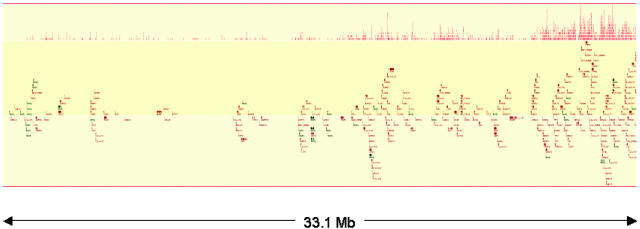
ACSs Derive Predominantly from Genic Regions. The distribution and density of 1,174 ACSs from the K5008 library that mapped to chromosome 21 are shown in Fig. 2. Striking correspondence in both parameters is evident between chromosome 21 ACSs and genes.
Fig. 2.
Genomic distribution of ACSs parallels genes. Distribution of ACSs (small vertical bars, top) and genes (ensembl) are shown along 33.1 Mb of human chromosome 21. Vertical stacking of ACSs and genes is due to compactness of the horizontal axis.
The overall distribution of ACSs relative to known genomic sequence classes differed from expectation in several categories. To quantify expectation, we programmed a simulation that selected random genomic sequences of the same average length as K5008 sequences and filtered them in an identical manner; several iterations were run, with each producing 2,000 unique mapping events that were then characterized with respect to the closest genomic feature. Results showed that 19.9% of ACSs fell within introns of known genes compared with an average of 27.5% from the simulation runs (P < 0.001). We found that 1.85% of ACS fell within exons of known genes (predominantly first exons), significantly greater than expectation (simulation 1.2%; P < 0.01). Also, 13.4% of ACSs either partially overlapped or fell within a known CpG island, compared with 1.3% expected by chance (P < 0.01).
Approximately 17.5% of ACSs mapped >50 kb away from any known genic feature. Well-demarcated multigene domains of open chromatin have been proposed to be a regular feature of the higher-order organization complex genomes (16). Aside from global correspondence with genic regions, we found no evidence for large-scale well circumscribed domains. However, it is unclear whether more subtly demarcated regions would have been detected given the current ACS sample size.
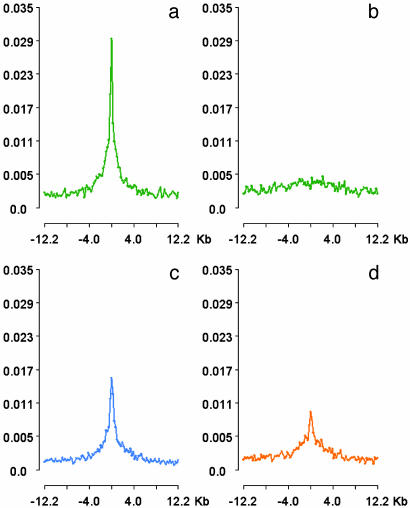
High Relative Density of ACSs at TSSs and CpG Islands. Regulatory sequences identified by DNaseI hypersensitive sites are expected to be found with higher density in certain genomic locales such as transcriptional start sites (TSSs). To examine the correspondence between ACSs and specific genomic landmarks, we computed the normalized density of ACSs relative to (i) the annotated 5′ TSSs of known genes, (ii) the 3′ transcription termini of known genes, and (iii) CpG islands (Fig. 3). Viewed on a 25-kb (± 12.5 kb) (Fig. 3a) horizon, a clear and symmetrical peak in the relative density of ACSs was observed around TSSs. No significant peak was observed at 3′ termini (Fig. 3b), confirming the specificity of this finding for TSSs. We also observed a prominent peak in ACS density relative to CpG islands (Fig. 3c). Because CpG islands are a regular (although by no means universal) feature of human promoters, we performed a second analysis that included only CpG islands located >2.5 kb distant (5′ or 3′) from a known TSS (Fig. 3d). This analysis also revealed a prominent peak, suggesting that a proportion of intergenic CpG islands lie within active chromatin domains. We also considered what component of the observed ACS peak around TSSs could be explained by CpG islands. We found that the strong peak in ACS density at TSSs persisted even when only non-CpG island-associated TSSs were analyzed (not shown), further confirming that this peak is due to a chromatin feature intrinsic to TSSs.
Fig. 3.
Density of ACS peaks at TSSs and CpG islands. y axes show the average number of ACSs per 100 bp bin; x axes show normalized distance (kb) relative to TSSs (a) and 3′ transcription termini of 16,169 RefSeq genes (b), and to promoter-associated (c) and non-promoter-associated (d) CpG islands. Peaks in ACS density at TSSs and at CpG islands are evident whereas no peak is found at 3′ transcript termini. ACS density peaks at CpG islands are evident even when non-promoter-associated CpGs are considered (d). Centered distances of the ACSs from each genomic feature set were computed by using a fractional counting technique to avoid the problem of multiply assigned ACSs. The number of times an ACS was assigned to a genomic feature was recorded, and a histogram corresponding with equal subdivisions was constructed wherein the number of ACSs assigned to each class was scaled by the fractional multiple assignment count. Thus, if an ACS was assigned to two distinct TSSs, a value of 1/2 was assigned to each histogram class. Finally, normalizing the classes by the total number of assigned tags gives the average tag density in the class as depicted.
One explanation for the observed distribution of ACSs around TSSs is that it reflects a large-scale chromatin disruption. An alternative and readily testable hypothesis is that this finding instead signifies continuous (although nonlinear) averaged distribution of DNaseI HSs both 5′ and 3′ of TSSs (see below).
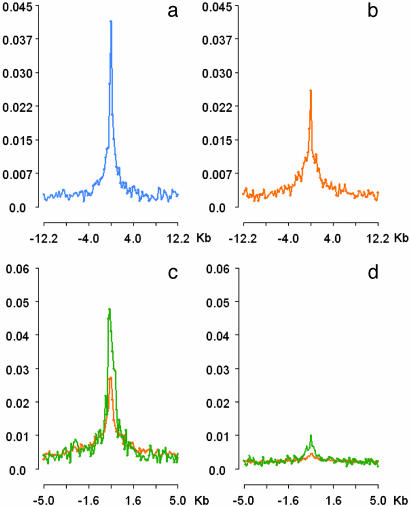
ACSs Show a Preference for Expressed Genes. We next asked whether the peak in ACS density at TSSs was confined to expressed genes. To evaluate this question, we assayed expression of 17,976 genes in K562 cells with a standard microarray platform (see Methods). We defined genes to be expressed (irrespective of relative magnitude) if their mean intensity exceeded the mean intensity of the background by 1 SD, a widely accepted empirical threshold (17, 18). We found that 8,333 genes (46.3%) met these criteria; 9,654 genes (53.6%) were considered nonexpressed. Genes for which no data were available because of lack of inclusion on the array or to technical issues were not included in the analysis. We then plotted ACS density relative to the TSSs of the expressed gene set (Fig. 4a), and also relative to the TSSs of nonexpressed genes (Fig. 4b). This analysis showed a prominent increase in the density of ACSs relative to expressed genes compared with nonexpressed genes. However, the absolute difference was surprisingly modest, suggesting that a large number of nonexpressed genes reside within open chromatin domains.
Fig. 4.
Distribution of ACSs as a function of gene expression (a and b) and distribution of ACS clusters relative to TSSs and CNGs (c and d). For explanation of the y axes, see Fig. 3. The x axes show normalized distance (kb) relative to TSS (a–c) and to CNGs (d). (a and b) Distribution of ACSs vs. expressed (a) and nonexpressed (b) genes. Genes (RefSeq) were categorized according to whether or not they were expressed in K562 cells. The average density of ACSs within 25-kb windows around TSSs of expressed and nonexpressed genes was computed. ACSs show a clear preference for expressed genes. However, a prominent peak in ACS density is still evident at nonexpressed genes, suggesting that many of these lie within open chromatin domains. (c and d) ACS clusters provide more powerful discrimination. We identified 3,293 ACS clusters comprising 2–8 ACSs distributed within a 1-kb window. ACS clusters (green) are better predictors of DNaseI hypersensitivity than ACSs (orange) (see text) and show more prominent aggregation around known or suspected functional genomic landmarks including TSSs (c), CpG islands (not shown), and evolutionarily conserved nongenic sequences (d). Note the difference in y axis scale vs. Fig. 3 and a and b. Relative densities were calculated as described in Fig. 3.
Correspondence Between ACSs and DNaseI Hypersensitive Sites. To determine the approximate percentage of ACSs that overlapped a DNaseI HS, we randomly selected 48 ACSs and assayed them for hypersensitivity in K562 cells with HSqPCR (see Methods). Of these ACSs, 3 (6.25%) overlapped a hypersensitive site. This finding suggests that in total the K5008 library contained at least ≈3,800 HSs. However, the fact that ACSs displayed marked distributional preferences for certain genomic features suggested that the correspondence between ACSs and DNaseI HSs would likewise depended on genomic context. We hypothesized specifically that the proportion of ACSs coinciding with DNaseI HSs would parallel the distribution of ACSs; namely, it would be maximal at the TSS and would diminish rapidly and symmetrically in both 5′ and 3′ directions.
To test this hypothesis, we randomly sampled three classes of ACS: (i) ACSs mapping between 0 and 250 bp upstream of the TSS; (ii) ACSs mapping 1,000 ± 100 bp upstream of the TSS; and (iii) ACSs mapping 1,000 ± 100 bp downstream of the TSS. Primers to 48 randomly selected members of each class were designed and assayed for hypersensitivity in K562 cells with HSqPCR. We found that 23/48 (47.9%) of ACSs mapping 0–250 bp upstream of the TSS coincided with HSs. However, promoter sequences are expected to contain DNaseI HSs although their position relative to the TSS may vary. To assess the significance of this finding, we therefore determined the background prevalence of DNaseI hypersensitivity at annotated TSSs. Because we observed high ACS densities at both expressed and nonexpressed genes, our gene selection was stratified accordingly. Primers were designed to encompass the first 250 bp upstream of a total of 192 genes: 92 randomly selected genes from the top quartile of K562-expressed genes, and 92 nonexpressed genes (see above). Quantitative determination of DNaseI hypersensitivity was then performed as described (see Methods). We found that 17/92 (18.5%) high expressing and 18/92 (19.6%) of nonexpressed genes harbored DNaseI HSs immediately upstream of their annotated transcriptional start sites. Although all expressing genes are expected to display hypersensitivity over their promoter regions, the precise location of promoter elements has been determined functionally for only a fraction of genes. In many cases, promoter elements may be situated several hundred bases or even up to >10 kb distant from the annotated TSS (19, 20). Moreover, for many genes, a promoter region may be located downstream from the annotated TSS within the first intron (21). The finding that a comparable proportion of nonexpressed genes also have hypersensitive sites immediately upstream of their annotated TSS is not altogether surprising, and is compatible both with previous observations (22–24) and with the distribution of ACSs (Fig. 4), suggesting that a significant number of human genes may be “poised” for transcription.
Of ACSs mapping –1,000 bp and +1,000 bp relative to the TSS, 2/47 (4.2%) and 2/46 (4.3%), respectively, were hypersensitive sites. Taken together with the TSS-proximal results above, these findings parallel the distribution of ACSs relative to the TSS (including their symmetry) and suggest further that this distribution primarily reflects the averaged distribution of DNaseI hypersensitive sites relative to the TSS.
ACSs in Distal Intergenic Regions. Regulatory sequences and associated DNaseI HSs have been reported to occur many tens or up to hundreds of kilobases distant from their cognate genes (25, 26). To test the utility of ACSs for identification of distal DNaseI HSs, we randomly selected 48 ACSs mapping >15 kb (but <100 kb) distant from the nearest gene (in either the 5′ or the 3′ direction) and tested for hypersensitivity in K562 cells with HSqPCR. Of these, 0/48 (0%) were found to be hypersensitive. In light of the overall prevalence of HS-overlapping ACSs in the K5008 library (6.25%), this finding suggests relative depletion of DNaseI HSs in the population of ACSs mapping within distal intergenic regions. However, given the size of the intergenic space and the a priori expected low density of DNaseI HSs within it, the relative proportion of DNaseI cut sites within true HS sites vs. nonspecific background cutting is expected to be low, resulting in low relative enrichment of HSs within ACSs from these regions.
ACS Clusters Provide More Powerful Discrimination. Poor correspondence between ACSs and DNaseI HSs in intergenic regions prompted us to search for metafeatures that might be more predictive of HSs. DNaseI HSs are defined by a high frequency of DNaseI cut sites over a given genomic interval. However, hypersensitivity becomes manifest only when cutting is averaged across a large population of individual chromosomes, each of which is cut in a stochastic fashion. We therefore hypothesized that, in the context of a library of ACSs where each member represents a unique cutting event, DNaseI hypersensitive sites would ultimately appear as clusters of ACSs mapping over small genomic intervals as larger numbers of clones were analyzed. To test this hypothesis, we clustered DNaseI cut sites along chromosomes to identify statistically significant groups of cut sites subject to the null hypothesis of uniformly distributed sites against the genomic background. This analysis revealed that, subject to the null hypothesis, clusters of size 2 contained within a 1,000-bp interval were statistically significant for our ACS library size.
We therefore defined an ACS cluster to be two or more DNaseI cut sites contained within a 1,000-bp window. One potential source of false-positive clustering is the presence of segmental duplications within the human genome sequence (27), which are not formally incorporated into current genome builds. We therefore filtered our preliminary cluster results against a library of known segmental duplications (27) and rejected any cluster occurring within these duplications. Applying these criteria, we identified a total of 3,293 clusters comprising 2–8 members. Plotting the distribution and density of ACS clusters relative to TSSs of known genes (Fig. 4c) produces a significantly more prominent peak than ACSs alone, suggesting that clusters are considerably more enriched in DNaseI hypersensitive sites. To test this idea directly, we designed primers to encompass the centroids (defined as the genomic coordinate mean of cluster members) of clusters with 4 or more members (irrespective of genomic position) and assayed these regions for DNaseI hypersensitivity in K562 cells. We identified DNaseI hypersensitive sites in 27/95 (28.4%) 4-member clusters, 12/29 (41.4%) 5-member clusters, 7/9 6-member clusters (77.8%), 1/2 (50%) 7-member clusters, and 2/5 (40%) 8-member clusters. Because these 140 clusters were widely distributed across the across the genome, the results confirm a substantial overall enrichment for DNaseI HSs within ACS clusters.
Coincidence Between ACSs and Evolutionarily Conserved Nongenic Sequences. Evolutionarily conserved nongenic sequences (CNGs) have been proposed to mark functional elements such as regulatory sequences (28). We analyzed the correspondence between ACSs and a global set of mouse-human conserved sequences described previously (29). For additional stringency, we did not consider sequences with <70% sequence conservation irrespective of length. A total of 420,431 such CNGs (mean length of 157 bp) were analyzed. Overall, we found that 4.1% of the DNaseI cut ends of ACSs fell within CNGs, significantly greater than expected by chance. This finding suggests that a measurable proportion of CNGs may harbor DNaseI hypersensitive sites and is further supported by a modest peak in the density of ACSs at CNGs, which is substantially more prominent when ACS clusters are considered (Fig. 4d). When CNGs proximal to the TSS are excluded from this analysis, the effect diminishes moderately, suggesting that CNGs in intergenic regions or distal introns are enriched in DNaseI HSs, although not dramatically so. For this reason, CNGs were not formally evaluated for hypersensitivity as a separate class. However, of all randomly selected ACSs described above, 22 overlapped CNGs. Of these, 6/22 (27%) coincided with hypersensitive sites. However, all HS-positive CNGs were located within the first 137 bp 5′ of TSSs. Of 9 CNGs located >200 bp from the TSS region, none were found to be hypersensitive.
Quantitative Approximation of the Distribution of HSs in the Human Genome. The distribution of DNaseI HSs in the human genome is of considerable interest given the close correspondence between HSs and classical cis-regulatory sequences. However, quantitative data that permit even preliminary estimation of this distribution have been lacking. ACSs and particularly ACS clusters may provide reliable surrogate markers for the distribution of HSs. We calculated binned (1,000 bp) percentages of ACSs and ACS clusters within a 10-kb window centered on the TSSs of known genes (Fig. 6, which is published as supporting information on the PNAS web site). Although this interval (±5 kb from the TSS) encompasses the peak ACS density region (Fig. 3), it contains only 22.4% of ACSs and 41.6% of ACS clusters. However, DNaseI HSs are markedly more prevalent in the vicinity of TSSs. We therefore used the results for DNaseI prevalence at the TSS and at ±1,000 bp described above to approximate the absolute distribution of DNaseI HSs within ±5 kb of the TSS by assuming that the prevalence of HSs paralleled the density of ACS clusters scaled for the observed proportion of HSs. This analysis suggested that ≈30% of all DNaseI HSs are expected to lie within this interval and implies that a majority of DNaseI HSs and, by extension, cis-regulatory sequences, are located >5 kb from the TSS.
Discussion
The study of gene regulation in complex organisms has been severely constrained by the lack of functionally based methodologies for large-scale identification of cis-regulatory sequences. A variety of computational (30–32) and phylogenetic (33) approaches have been developed to address this deficit, but their utility has been limited by poor sensitivity and specificity for functional elements. By contrast, the use of DNaseI hypersensitivity studies for identification of in vivo-functional regulatory sequences is well established and has underpinned the discovery of hundreds of regulatory sequences controlling human genes and those of other eukaryotes.
We have described an approach for extending the DNaseI hypersensitivity paradigm to a genomic level through large-scale cloning and mapping of individual in vivo DNaseI cutting events. Although demonstrated in human tissue, cloning and analysis of ACSs should be applicable to any eukaryotic cell type, providing the basis for the accumulation of comprehensive databases of cis-regulatory sequences.
Analysis of a large library of active chromatin sequences has provided several insights into the relationship between chromatin structure and gene expression. We observed peaks in the density of ACSs at the transcriptional start sites of known genes, at non-gene-associated CpG islands, and, to a lesser degree, at evolutionarily conserved noncoding sequences. A remarkable feature of the distribution of ACSs around TSSs was its symmetry. This finding suggests that proximal intron regions may be a rich reservoir of regulatory sequences (34–37). Another surprising finding was the strong representation of both expressed and nonexpressed genes. This result suggests that a majority of genes reside within open chromatin domains. The fact that a large proportion of ACSs and ACS clusters are found within a 10-kb interval centered on transcription start sites of known genes is perhaps not surprising. However, the prediction that 70% of DNaseI hypersensitive sites (and, by extension, cis-regulatory sequences) lie outside this interval highlights the need for approaches such as the one described here for efficient culling of such sequences from the vastness of the genome.
All of the ACS cloning results described above used a single round of subtraction. However, this procedure may be applied iteratively to produce a population highly enriched for DNaseI hypersensitive sites. Additionally, generation of larger numbers of ACS library sequences will permit more full exploitation of the clustering effect. In combination, these techniques may permit definitive HS probability thresholds to be associated with clusters of different sizes, eliminating the need for direct hypersensitivity testing of large numbers of candidate sequences.
Supplementary Material
Acknowledgments
We thank Tony Shafer, Janelle Kawamoto, Josh Mack, and Rob Hall for outstanding technical assistance and expertise.
Abbreviations: ACS, active chromatin sequence; HS, hypersensitive site; TSS, transcriptional start site; CNG, conserved nongenic sequence; HSqPCR, hypersensitivity quantitative PCR.
References
- 1.Felsenfeld, G. (1996) Cell 86, 13–19. [DOI] [PubMed] [Google Scholar]
- 2.Felsenfeld, G. & Groudine, M. (2003) Nature 421, 448–453. [DOI] [PubMed] [Google Scholar]
- 3.Gross, D. S. & Garrard, W. T. (1988) Annu. Rev. Biochem. 57, 159–197. [DOI] [PubMed] [Google Scholar]
- 4.Elgin, S. C. (1984) Nature 309, 213–214. [DOI] [PubMed] [Google Scholar]
- 5.Wu, C. (1980) Nature 286, 854–860. [DOI] [PubMed] [Google Scholar]
- 6.Burgess-Beusse, B., Farrell, C., Gaszner, M., Litt, M., Mutskov, V., Recillas-Targa, F., Simpson, M., West, A. & Felsenfeld, G. (2002) Proc. Natl. Acad. Sci. USA 99, 16433–16437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reitman, M., Lee, E., Westphal, H. & Felsenfeld, G. (1993) Mol. Cell. Biol. 13, 3990–3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rozen, S. & Skaletsky, H. J. (2000) in Bioinformatics Methods and Protocols: Methods in Molecular Biology, eds. Krawetz, S. & Misener, S. (Humana, Totowa, NJ).
- 9.McArthur, M., Gerum, S. & Stamatoyannopoulos, G. (2001) J. Mol. Biol. 313, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuan, D., Solomon, W., Li, Q. & London, I. M. (1985) Proc. Natl. Acad. Sci. USA 82, 6384–6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgs, D. R., Wood, W. G., Jarman, A. P., Sharpe, J., Lida, J., Pretorius, I. M. & Ayyub, H. (1990) Genes Dev. 4, 1588–1601. [DOI] [PubMed] [Google Scholar]
- 12.Stamatoyannopoulos, G. & Grosveld, F. (2001) in Molecular Basis of Blood Diseases, eds. Stamatoyannopoulos, G., Majerus P., Perlmutter, R. & Varmus H. (Saunders, Philadelphia).
- 13.Rousseeuw, P. J. & van Zomeren, B. C. (1990) J. Am. Stat. Assoc. 85, 633–639. [Google Scholar]
- 14.International Human Genome Sequencing Consortium (2001) Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 15.Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural. R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 16.van Driel, R., Fransz, P. F. & Verschure, P. J. (2003) J. Cell. Sci. 116, 4067–4075. [DOI] [PubMed] [Google Scholar]
- 17.Wodicka, L., Dong, H., Mittmann, M., Ho, M. H. & Lockhart, D. J. (1997) Nat. Biotechnol. 15, 1359–1367. [DOI] [PubMed] [Google Scholar]
- 18.Epstein, C. B., Hale, W., IV, & Butow, R. A. (2001) Methods Cell Biol. 65, 439–452. [DOI] [PubMed] [Google Scholar]
- 19.Davuluri R. V., Grosse I. & Zhang M. Q. (2001) Nat. Genet. 29, 412–417. [DOI] [PubMed] [Google Scholar]
- 20.Shiraki, T., Kondo, S., Katayama, S., Waki, K., Kasukawa, T., Kawaji, H., Kodzius, R., Watahiki, A., Nakamura, M., Arakawa, T., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 15776–15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reisman, D., Greenberg, M. & Rotter, V. (1988) Proc. Natl. Acad. Sci. USA 85, 5146–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groudine, M., Kohwi-Shigematsu, T., Gelinas, R., Stamatoyannopoulos, G. & Papayannopoulou, T. (1983) Proc. Natl. Acad. Sci. USA 80, 7551–7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radomska, H. S., Satterthwaite, A. B., Burn, T. C., Oliff, I. A., Huetnnet, C. S. & Tenen, D. G. (1998) Gene 222, 305–318. [DOI] [PubMed] [Google Scholar]
- 24.Fraser, P. & Grosveld, F. (1998) Curr. Opin. Cell Biol. 10, 361–365. [DOI] [PubMed] [Google Scholar]
- 25.Antoniv, T. T., De Val, S., Wells, D., Denton, C. P., Rabe, C., de Crombrugghe, B., Ramirez, F. & Bou-Gharios, G. (2001) J. Biol. Chem. 276, 21754–21764. [DOI] [PubMed] [Google Scholar]
- 26.Spitz, F., Gonzalez, F. & Duboule, D. (2003) Cell 113, 405–417. [DOI] [PubMed] [Google Scholar]
- 27.Bailey, J. A., Gu, Z., Clark, R. A., Reinert, K., Samonte, R. V., Schwartz, S., Adams, M. D., Myers, E. W., Li, P. W. & Eichler, E. E. (2002) Science 297, 1003–1007. [DOI] [PubMed] [Google Scholar]
- 28.Ureta-Vidal, A., Ettwiller, L. & Birney, E. (2003) Nat. Rev. Genet. 4, 251–262. [DOI] [PubMed] [Google Scholar]
- 29.Alexandersson, M., Cawley, S. & Pachter, L. (2003) Genome Res. 13, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markstein, M., Markstein, P., Markstein, V. & Levine, M. S. (2002) Proc. Natl. Acad. Sci. USA 99, 763–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berman, B. P., Nibu, Y., Pfeiffer, B. D., Tomancak, P., Celniker, S. E., Levine, M., Rubin, G. M. & Eisen, M. B. (2002) Proc. Natl. Acad. Sci. USA 99, 757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stathopoulos, A., Van Drenth, M., Erives, A., Markstein, M. & Levine, M. (2002) Cell 111, 687–701. [DOI] [PubMed] [Google Scholar]
- 33.Pennacchio, L. A. & Rubin, E. M. (2001) Nat. Rev. Genet. 2, 100–109. [DOI] [PubMed] [Google Scholar]
- 34.Aronow, B., Lattier, D., Silbiger, R., Dusing, M., Hutton, J., Jones, G., Stock, J., McNeish, J., Potter, S., Witte, D., et al. (1989) Genes Dev. 3, 1384–1400. [DOI] [PubMed] [Google Scholar]
- 35.Bates, N. P. & Hurst H. C. (1997) Oncogene 15, 473–481. [DOI] [PubMed] [Google Scholar]
- 36.Rowntree, R. K., Vassaux, G., McDowell, T. L., Howe, S., McGuigan, A., Phylactides, M., Huxley, C. & Harris, A. (2001) Hum. Mol. Genet. 10, 1455–1464. [DOI] [PubMed] [Google Scholar]
- 37.Tone, M., Diamond, L. E., Walsh, L. A., Tone, Y., Thompson, S. A., Shanahan, E. M., Logan, J. S. & Waldmann, H. (1999) J. Biol. Chem. 274, 710–716. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.