Abstract
The structures of the bacterial RNA polymerase holoenzyme have provided detailed information about the intersubunit interactions within the holoenzyme. Functional analysis indicates that one of these is critical in enabling the holoenzyme to recognize the major class of bacterial promoters. It has been suggested that this interaction, involving the flap domain of the β subunit and conserved region 4 of the σ subunit, is a potential target for regulation. Here we provide genetic and biochemical evidence that the σ region 4/β-flap interaction is targeted by the transcription factor AsiA. Specifically, we show that AsiA competes directly with the β-flap for binding to σ region 4, thereby inhibiting transcription initiation by disrupting the σ region 4/β-flap interaction.
The bacterial RNA polymerase (RNAP) holoenzyme consists of a catalytically proficient core enzyme (subunit structure α2ββ′ω) and a σ subunit that confers on the holoenzyme the ability to recognize specific promoter sequences (1). The primary σ factor in Escherichia coli is σ70, and the σ70-containing holoenzyme (Eσ70) typically recognizes promoters defined by two conserved sequence elements (the –10 and –35 hexamers) positioned roughly 10 and 35 bp upstream of the transcription start point (1). Most σ factors share four conserved regions (2), and of these, regions 2 and 4 interact with the –10 and –35 elements, respectively (1). Nevertheless, intact σ70 recognizes specific promoter sequences only in the context of the holoenzyme because of critical conformational changes in σ70 that take place when it associates with the core enzyme (3). These include an increase in the interdomain distance between regions 2 and 4 that is caused by an interaction between σ region 4 and the β-flap domain (4). The σ region 4/β-flap interaction is essential for the recognition of –10/–35 promoters because it positions regions 2 and 4 of σ70 for simultaneous interaction with the promoter –10 and –35 elements (4). This role of the σ region 4/β-flap interaction suggested that there may exist regulatory factors that inhibit transcription from –10/–35 promoters by disrupting the σ region 4/β-flap interaction (4, 5).
Here we examine the mechanism of action of the bacteriophage T4-encoded anti-σ factor AsiA. AsiA is known to bind tightly to region 4 of σ70 and inhibit transcription from –10/–35 promoters when complexed with the holoenzyme (6–10). The finding that AsiA binds to region 4 of σ70 and inhibits transcription specifically from –10/–35 promoters raises the possibility that AsiA works by disrupting the σ70 region 4/β-flap interaction, a model that is consistent with recent NMR analysis (11). We provide genetic evidence that AsiA and the β-flap interact with overlapping determinants on σ70 region 4. We then present complementary biochemical and biophysical evidence that AsiA works by a competitive binding mechanism, inhibiting transcription from –10/–35 promoters by disrupting the σ region 4/β-flap interaction in the context of the holoenzyme.
Materials and Methods
Mutant Screen. Mutagenic PCR was used to introduce random mutations into the appropriate fragment of plasmid pBRα-σ70 D581G as described (12). A pool of mutant α-σ70 D581G chimeras was transformed into strain F′93+62 (13) bearing the test promoter (plac2λop) and a linked lacZ reporter gene on an F′ episome and containing pACΔ–35λcI–AsiA K20A (a derivative of pACΔ–35λcI–AsiA; ref. 13). The resulting transformants were plated on indicator medium containing 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) (40 μg/ml) and the β-galactosidase inhibitor tPEG (250 μM), permitting the identification of colonies containing α-σ70 D581G mutants that caused a reduction in λcI–AsiA-stimulated transcription from the test promoter. Plasmid DNA from these colonies was isolated, and the mutant α-σ70 D581G chimeras were assayed for their abilities to stimulate transcription from placCons–35C.1 (identical in sequence to placCons–35C from the transcription start point to position –62; see ref. 12) in strain BG18. Mutants were sought that stimulated transcription from placCons–35C.1 as well or nearly as well as the parent chimera.
β-Galactosidase Assays. Cells were grown in LB supplemented with the appropriate antibiotics at the following concentrations: carbenicillin (100 μg/ml), chloramphenicol (25 μg/ml), and kanamycin (50 μg/ml). Isopropyl-β-d-thiogalactoside (IPTG) was provided at the indicated concentrations. SDS·CHCl3-permeabilized cells were assayed as described (13). Assays were performed at least three times in duplicate on separate occasions, with similar results. Values are the averages from one experiment; duplicate measurements differed by <5%.
Proteins. N-terminally his-tagged versions of wild-type σ70 and the σ70 mutants were purified as described after overproduction from the corresponding derivative of vector pLHN12-His (14). E. coli RNAP core enzyme was purchased from Epicentre, and holoenzymes were assembled by incubation of RNAP core (62.5 nM) with a 5-fold molar excess of the appropriate σ70 at 37°C for 10 min. C-terminally his-tagged AsiA was purified as described (15) and AsiA-containing RNAP holoenzymes were assembled by preincubating the appropriate σ70 (in 5-fold molar excess over RNAP core) with different concentrations of AsiA in σ storage buffer (14) at 4°C for 10 min. RNAP core (62.5 nM) was then added, and the mixtures were incubated at 37°C for 10 min.
In Vitro Transcription Assays. Assays were performed by preincubating template DNA (10 nM) in transcription buffer (10 mM MgCl2/90 mM KCl/40 mM Tris·HCl, pH 8.0/125 μg/ml BSA/5 mM DTT/10% vol/vol glycerol). RNAP holoenzyme (with or without AsiA) was then added to a final concentration of 10 nM and the reactions incubated for 10 min at 37°C to allow open complex formation. Transcription was initiated by addition of 200 μM GTP (initiating nucleotide), 5 μM ATP, 5 μM CTP, 3 μM[α-32P]UTP at 2 mCi/ml, and 100 μg/ml heparin (1 Ci = 37 GBq). The total volume of each reaction was 25 μl. Reactions were incubated for 15 min at 37°C and quenched by the addition of 25 μl of stop solution (95% vol/vol formamide/20 mM EDTA/0.05% wt/vol bromophenol blue/0.05% wt/vol xylene cyanol). Samples were electrophoresed on 6% (wt/vol) polyacrylamide sequencing gels. Bands were visualized by PhosphorImager, and the data were analyzed by imagequant.
Fluorescence Resonance Energy Transfer (FRET) Analysis. AsiA/RNAP holoenzyme complexes. σ70 labeled at residue 581 by cpm (Molecular Probes) was prepared by using mutant σ70 containing a single reactive cysteine at position 581 as described (3). AsiA labeled at position 86 with fluorescein (Molecular Probes) was prepared by using mutant AsiA containing a single reactive cysteine at position 86 as described (15). To assess the binding of AsiA to RNAP holoenzyme containing wild-type or mutant σ70, equal amounts of labeled AsiA/holoenzyme complex and unlabeled holoenzyme were mixed together as follows. Core (12.5 pM) was combined with 11 pM σ70-cpm and 10 pM AsiA–FAM (fluorescein malemide) in 200 μl of buffer F containing 50 mM Tris·HCl (pH 8), 100 mM NaCl, and 0.5 mg/ml BSA in a quartz cuvette. The mixture was incubated for 20′ at 37°C to ensure that labeled holoenzyme–AsiA complex reached equilibrium. Under these conditions, at least 90% of labeled σ70 is bound to the core (data not shown; see also ref. 4). Then, 5 μl containing 12.5 pM core and 11 pM unlabeled wild-type or mutant σ70 were added to the labeled holoenzyme-AsiA complex. The mixture was incubated at 37°C for 5 min before measurements were taken. Fluorescence emission spectra with the excitation at 380 nm (cpm excitation) were recorded from 400 to 600 nm. Fluorescence spectra were recorded by using an Aminco Bowman Series 2 spectrofluorometer (Spectronic Instruments, Rochester, NY). Spectra were acquired with the scan rate of 2 nm/sec and with excitation and emission slits of 2 and 4 nm, correspondingly. The ratio of fluorescence intensity integrated between 500 and 600 nm (acceptor emission) to the intensity integrated between 400 and 500 nm (donor emission) was used as an apparent parameter describing the extent of FRET between the donor and acceptor. The value of this parameter is directly proportional to the amount of Eσ70-cpm–AsiA–FAM complex in solution. For these experiments, wild-type RNAP core enzyme was prepared by in vitro reconstitution as described (16), and unlabeled wild-type and mutant σ70 subunits were purified as described (14).
AsiA/σ70 complexes. Complexes between labeled σ70 and labeled AsiA were formed by mixing the proteins in 200 μl of buffer F in a quartz cuvette at a ratio of 1:1.5 (50 nM σ70-cpm and 75 nM AsiA–FAM). Relative binding affinities of σ70 mutants were assessed by adding various amounts of unlabeled protein to the mixture of labeled σ70 and AsiA. Addition of unlabeled σ70 (wild type or mutant) led to a substitution of labeled σ70 in the σ70-cpm–AsiA–FAM complex by the unlabeled competitor protein, resulting in diminished intensity of the FRET signal.
Results
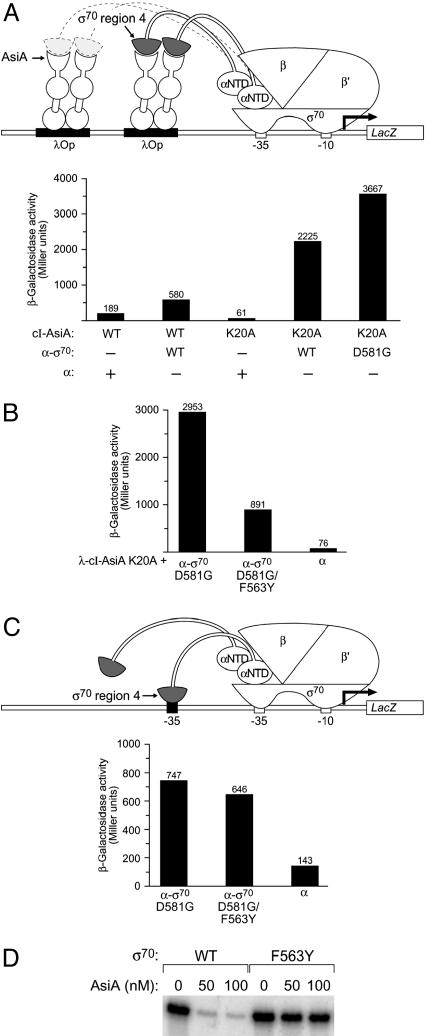
Design of Genetic Screen. To probe the mechanism of transcription inhibition by AsiA, we took a genetic approach. Previous work suggested that the interaction of AsiA with σ70 region 4 is required for AsiA-mediated transcription inhibition (8–10). Therefore, we sought to identify amino acid residues in region 4 of σ70 that participate directly in the interaction with AsiA through the use of a bacterial two-hybrid assay (17, 18). In this assay, contact between a protein domain fused to a component of RNAP and an interacting protein (or protein domain) fused to a DNA-bound protein (here, the cI protein of bacteriophage λ) activates transcription of a lacZ reporter gene (see Fig. 1A). In particular, we made use of a λcI–AsiA fusion protein and an α-σ70 chimera in which the C-terminal domain of α (the α-CTD) has been replaced by a fragment of σ70 (residues 528–613) encompassing region 4 (Fig. 1 A). The λcI–AsiA fusion protein activated transcription from test promoter plac2λop ≈3-fold specifically in cells containing the α-σ70 chimera (Fig. 1 A; see also ref. 13).
Fig. 1.
Bacterial two-hybrid assay used to screen for mutations in region 4 of σ70 that disrupt the interaction with AsiA. (A) The diagram depicts test promoter plac2λop (previously called placCOP-93+OL2–62, ref. 13), which bears a consensus λ operator sequence centered 93 bp upstream and the λ operator OL2 centered 62 bp upstream from the initiation point of the lac core promoter. In strain F′93+62 this test promoter is linked to lacZ on an F′ episome. Replacement of the RNAP α-CTD by a fragment of σ that harbors region 4 permits interaction with the AsiA moiety of either of two DNA-bound λcI–AsiA fusion proteins. The graph shows the effect of the λcI–AsiA fusion protein (with or without the K20A substitution) on transcription in vivo from plac2λop in the presence of the α-σ70 chimera (λcI–AsiA and λcI–AsiA K20A) or the α-σ70 D581G chimera (λcI–AsiA K20A only). F′93+62 cells harboring compatible plasmids directing the synthesis of the indicated proteins were grown in the presence of 50 μM IPTG and assayed for β-galactosidase activity. Plasmids pACΔ–35λcI–AsiA (13) and derivative pACΔ–35λcI–AsiA K20A directed the synthesis of the λcI–AsiA and λcI–AsiA K20A fusion proteins, and plasmids pBRα-σ70 (28), pBRα (28), and pBRα-σ70 D581G (12) directed the synthesis of the α-σ70 chimera, full-length α, and the α-σ70 D581G chimera. (B) Effect of F563Y substitution in σ moiety of α-σ70 D581G chimera on transcription in vivo from plac2λop in presence of the λcI–AsiA K20A fusion protein. F′93+62 cells harboring compatible plasmids directing the synthesis of the indicated proteins were grown in the presence of 50 μM IPTG and assayed for β-galactosidase activity. Plasmid pACΔ–35λcI–AsiA K20A directed the synthesis of the λcI–AsiA K20A fusion protein and plasmids pBRα-σ70 D581G, pBRα-σ70 D581G/F563Y, and pBRα directed the synthesis of the α-σ70 D581G chimera, the α-σ70 D581G/F563Y chimera, and full-length α. (C) Substitution F563Y in σ moiety of α-σ70 D581G chimera does not affect the ability of the tethered σ70 region 4 moiety to bind DNA. The diagram depicts test promoter placCons–35C.1 (see Materials and Methods), which contains a consensus –35 element centered 55 bp upstream from the transcription start site. In strain BG18, this test promoter is linked to lacZ on an F′ episome. Replacement of the RNAP α-CTD by a fragment of σ that harbors region 4 containing the D581G substitution permits interaction with the –35 element centered at position –55. The graph shows the effect of the F563Y substitution in the σ moiety of the α-σ70 D581G chimera on transcription in vivo from placCons–35C.1. BG18 cells harboring compatible plasmids directing the synthesis of the indicated proteins were grown in the presence of 50 μM IPTG and assayed for β-galactosidase activity. Plasmids pBRα-σ70 D581G, pBRα-σ70 D581G/F563Y, and pBRα directed the synthesis of the α-σ70 D581G chimera, the α-σ70 D581G/F563Y chimera, and full-length α. (D) Eσ70 F563Y is resistant to inhibitory effect of AsiA on transcription from a –10/–35 promoter. Shown are results of single round in vitro transcription assays performed on –10/–35 promoter T7A2 in the absence or presence of increasing concentrations of AsiA (50 or 100 nM) by using wild-type Eσ70 or mutant Eσ70 bearing the F563Y substitution in σ70.
To increase the sensitivity of our genetic screen, we sought to identify a λcI–AsiA variant that would activate transcription more strongly in our two-hybrid assay. Because AsiA forms stable homodimers that must first dissociate for AsiA to bind to σ70 region 4 (11, 19, 20), we hypothesized that the ability of the fused AsiA moiety to activate transcription might be limited by its tendency to dimerize in the context of the dimeric λcI–AsiA fusion protein (13). To test this hypothesis, we introduced into the AsiA moiety an amino acid substitution (K20A) that specifically disrupts the formation of AsiA homodimers without inhibiting the ability of AsiA to interact with σ70 region 4 (R.J.B.U. and J.U., unpublished data). The resulting λcI–AsiA K20A variant stimulated transcription from promoter plac2λop ≈36-fold in the presence of the α-σ70 chimera (Fig. 1 A), providing strong support for the hypothesis that the interaction of the fused AsiA moieties of the λcI–AsiA dimer competes with the interaction of the AsiA and σ70 region 4 moieties in the two-hybrid assay.
To identify amino acid substitutions in σ70 region 4 that specifically affected its interaction with AsiA and eliminate from consideration those that disrupted the structural integrity of the σ70 moiety, we took advantage of a related genetic assay that reports on the ability of the tethered σ70 region 4 moiety to bind to a –35 element (12). In this assay, the interaction of the region 4 moiety with an ectopic –35 element positioned upstream of the core promoter elements activates transcription from the test promoter depicted in Fig. 1C (promoter placCons-35C.1). We showed previously that although the wild-type σ70 moiety does not bind detectably to the ectopic –35 element of placCons–35C, a variant bearing amino acid substitution D581G does (12). Thus, we used the D581G variant of the α-σ70 chimera in our genetic screen, having first verified that the D581G substitution did not compromise the ability of the AsiA moiety of the λcI–AsiA fusion protein to interact with σ70 region 4 in our two-hybrid assay (Fig. 1 A).
Identification and Characterization of a σ70 Mutant That Is Specifically Defective in the Interaction with AsiA. We introduced random mutations into the gene fragment encoding the σ70 moiety of an α-σ70 D581G chimera. After screening the mutagenized library of α-σ70 D581G chimeras, we identified an amino acid substitution, F563Y, that reduced λcI–AsiA K20A-stimulated transcription from test promoter plac2λop, but had only a slight effect on the ability of the tethered σ70 moiety to activate transcription from test promoter placCons-35C.1 (Fig. 1 B and C). We conclude that substitution F563Y specifically weakens (but does not abolish) the interaction of σ70 region 4 with AsiA.
We then wished to determine whether or not introduction of the F563Y substitution into full-length σ70 would affect the ability of AsiA to inhibit transcription from a –10/–35 promoter (T7A2) in vitro. Reconstituted holoenzymes Eσ70 and Eσ70 F563Y were equally proficient in initiating transcription from this promoter in the absence of AsiA (Fig. 1D). As expected, AsiA inhibited transcription by the wild-type holoenzyme (Fig. 1D, lanes 1–3). In contrast, the mutant holoenzyme containing σ70 F563Y was almost completely refractory to the inhibitory effect of AsiA (Fig. 1D, compare lanes 4–6 with lanes 1–3). Thus, amino acid substitution F563Y in σ70 both weakened the σ70 region 4/AsiA interaction in the two-hybrid assay and disrupted AsiA-mediated transcription inhibition in vitro, indicating that the σ70 region 4/AsiA interaction detected in the two-hybrid assay is required for AsiA to function as an inhibitor of transcription.
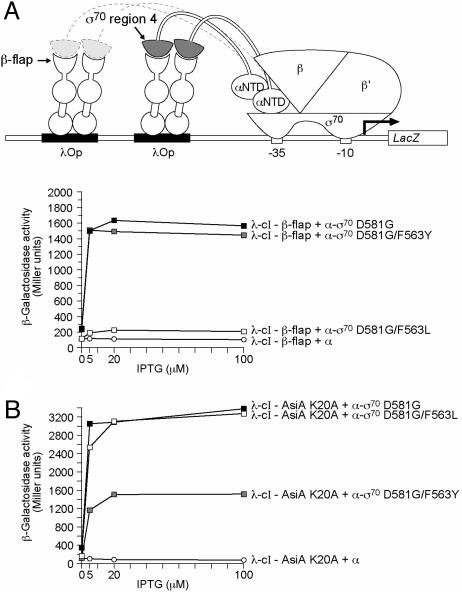
Substitutions of σ70 Residue 563 Affect both the σ70/AsiA and the σ70/β-Flap Interactions. Two independent lines of evidence implicate residue F563 of σ70 in the interaction of σ70 region 4 with the β-flap. First, the RNAP holoenzyme structures indicate that the residue corresponding to F563 of σ70 lies at the region 4/β-flap interface (21, 22). Second, we found that a substitution (F278L) affecting the corresponding residue of σ38 (the stationary phase-specific σ factor in E. coli) specifically disrupted the interaction of σ38 region 4 with the β-flap (B.E.N., unpublished data). We therefore wished to compare the effects of the F563Y and F563L substitutions in σ70 on both the σ70 region 4/β-flap and the σ70 region 4/AsiA interactions. Accordingly, we introduced the F563L substitution into the σ70 moiety of the α-σ70 D581G chimera. Whereas the F563L substitution in σ70 region 4 nearly abolished the stimulatory effect of the λcI–β-flap fusion protein from test promoter plac2λop (Fig. 2A), it had no effect on the ability of the λcI–AsiA K20A fusion protein to stimulate transcription from the same promoter (Fig. 2B). Conversely, we found that amino acid substitution F563Y in σ70 region 4, which reduced the stimulatory effect of the λcI–AsiA K20A fusion protein, had no effect on the ability of the λcI-β-flap fusion protein to stimulate transcription (Fig. 2 A and B). We conclude, therefore, that the F563L substitution specifically affects the interaction of σ70 region 4 with the β-flap, whereas the F563Y substitution specifically affects its interaction with AsiA, suggesting that the molecular details of the two interactions differ.
Fig. 2.
Effects of substitutions at position 563 of σ70 on the interactions with AsiA K20A and the β-flap moiety of RNAP. (A) The effects of two different substitutions at position 563 of σ70 on the σ70 region 4/β-flap interaction. The diagram depicts test promoter plac2λop. The graph shows the effects of the λcI-β-flap fusion protein on transcription in vivo from plac2λop in the presence of α-σ70 chimeras bearing different amino acids at position 563 of the tethered σ70 region 4 moiety. F′93+62 cells harboring compatible plasmids directing the synthesis of the indicated proteins were grown in the presence of different concentrations of IPTG and assayed for β-galactosidase activity. Plasmid pACλcI-β-flap (4) directed the synthesis of the of λcI-β-flap fusion protein, and plasmids pBRα-σ70 D581G (12), pBRα-σ70 D581G/F563L, pBRα-σ70 D581G/F563Y, and pBRα directed the synthesis of the α-σ70 D581G chimera, the α-σ70 D581G/F563L chimera, the α-σ70 D581G/F563Y chimera, and full-length α.(B) The effect of two different substitutions at position 563 of σ70 on the σ70 region 4/AsiA K20A interaction. The graph shows the effects of the λcI–AsiA K20A fusion protein on transcription in vivo from plac2λop in the presence of α-σ70 chimeras bearing different amino acids at position 563 of the tethered σ70 region 4 moiety. F′93+62 cells harboring compatible plasmids directing the synthesis of the indicated proteins were grown in the presence of different concentrations of IPTG and assayed for β-galactosidase activity. Plasmid pACΔ–35λcI–AsiA K20A directed the synthesis of the λcI–AsiA K20A fusion protein, and plasmids pBRα-σ70 D581G, pBRα-σ70 D581G/F563L, pBRα-σ70 D581G/F563Y, and pBRα directed the synthesis of the α-σ70 D581G chimera, the α-σ70 D581G/F563L chimera, the α-σ70 D581G/F563Y chimera, and full-length α.
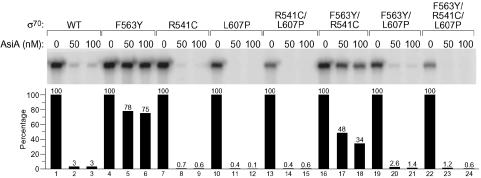
Evidence That AsiA Competes with the β-Flap for Binding to σ70 Region 4. The identification of an amino acid residue (F563) that apparently participates in both the σ70 region 4/β-flap and the σ70 region 4/AsiA interactions suggested that AsiA and the β-flap interact with overlapping determinants of σ70 region 4. Therefore, AsiA and the β-flap would be expected to compete for binding to σ70 region 4 in the context of the RNAP holoenzyme. This competitive binding model for AsiA-mediated transcription inhibition implies that the relative affinities of AsiA and the β-flap for σ70 region 4 should dictate whether AsiA can effectively inhibit transcription from –10/–35 promoters. To test this competitive binding model, we asked whether weakening the interaction of σ70 F563Y with the β-flap could restore the susceptibility of Eσ70 F563Y to AsiA. To do this, we took advantage of two amino acid substitutions in σ70 (R541C and L607P) that specifically disrupt the interaction between σ70 region 4 and the β-flap (B.E.N., unpublished data). We first used the two-hybrid assay to test whether these substitutions had any effect on the interaction of σ70 region 4 and AsiA. When introduced into σ70 region 4 either singly or in combination, neither the R541C substitution nor the L607P substitution significantly affected the σ70 region 4/AsiA K20A interaction (data not shown).
We then introduced substitutions R541C and L607P, singly and in combination, into full-length σ70 already containing the F563Y substitution. We used these mutant σ70 proteins as well as σ70 R541C, σ70 L607P, σ70 R541C/L607P, σ70 F563Y, and σ70 WT to reconstitute a set of holoenzymes and examined the abilities of these holoenzymes to initiate transcription from the T7A2 promoter in the absence or presence of AsiA (Fig. 3; see also Table 1). As in the experiment of Fig. 1D, we found that AsiA efficiently inhibited transcription by Eσ70 WT (Fig. 3, lanes 1–3) but was unable to inhibit transcription by Eσ70 F563Y (Fig. 3, lanes 4–6). Furthermore, AsiA inhibited transcription by Eσ70 R541C, Eσ70 L607P, and Eσ70 R541C/L607P even more efficiently than it inhibited transcription by Eσ70 WT (Fig. 3, compare lanes 7–9, 10–12, and 13–15, with lanes 1–3). Finally, we found that introduction of substitutions R541C and/or L607P into σ70 F563Y enabled AsiA to inhibit transcription by the corresponding doubly and triply substituted holoenzymes (Fig. 3, lanes 16–18, 19–21, and 22–24). Moreover, the efficiency with which AsiA inhibited transcription by each of these holoenzymes correlated with the severity of the β-flap-binding defect of the corresponding mutant σ70. That is, σ70 substitutions R541C, L607P, and R541C/L607P resulted in increasingly severe β-flap-binding defects (B.E.N., unpublished data), and correspondingly, Eσ70 F563Y/R541C, Eσ70 F563Y/L607P, and Eσ70 F563Y/R541C/L607P were increasingly susceptible to inhibition by AsiA (Fig. 3, lanes 16–24). These findings demonstrate that the effect of weakening the σ70 region 4/AsiA interaction can be suppressed by weakening a competing interaction between σ70 region 4 and the β-flap.
Fig. 3.
Mutations that weaken the σ70 region 4/β-flap interaction restore sensitivity of Eσ70 F563Y to AsiA. Shown are the results of single round in vitro transcription assays performed on T7A2 promoter in the absence or presence of increasing concentrations of AsiA (50 or 100 nM) by using wild-type or mutant RNAP holoenzymes. For each RNAP, the transcripts were quantified (by imagequant) to determine the magnitude of the inhibitory effect of AsiA. Thus, the amount of transcript produced in the absence of AsiA was set at 100% for each holoenzyme. Note that all of the mutant holoenzymes except those containing σ70 with both the R541C and L607P substitutions initiated transcription from T7A2 approximately as well as Eσ70 WT (compare lanes 4, 7, 10, 16, and 19 with lane 1). Eσ70 R541C/L607P and Eσ70 F563Y/R541C/L607P initiated transcription from T7A2 only ≈50% as efficiently as Eσ70 WT. Disruption of the interaction of σ70 with the β-flap is expected to cause defects in the recognition of –10/–35 promoters, and analysis of the effects of the R541C and L607P substitutions indicated that it was necessary to introduce both of these substitutions into σ70 to detect a significant defect in the recognition of strong –10/–35 promoters in vitro (B.E.N., unpublished data). No defects in the recognition of a control extended –10 promoter were detected with σ70 mutants bearing substitutions R541C and L607P either singly or in combination (B.E.N., unpublished data).
Table 1. Sensitivity of mutant RNAP holoenzymes to AsiA.
| σ70 mutant | Sensitivity of σ70-containing holoenzyme to AsiA in vitro |
|---|---|
| Wild type | + + + + |
| F563Y* | + |
| R541C† | ≥ + + + + |
| L607P | ≥ + + + + |
| R541C/L607P | ≥ + + + + |
| F563Y/R541C | + + |
| F563Y/L607P | ≥ + + + + |
| F563Y/R541C/L607P | ≥ + + + + |
Boldface indicates that the F563Y substitution disrupts the interaction of σ70 region 4 with AsiA (but not with the β-flap).
Italics indicate that the designated substitutions disrupt the interaction of σ70 region 4 with the β-flap (but not with AsiA).
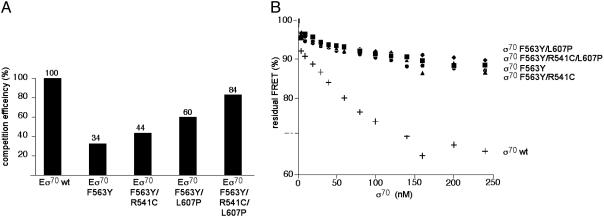
To assay directly the binding of AsiA to the wild-type and mutant holoenzymes in solution, we used a FRET-based binding assay. FRET donor and acceptor probes were incorporated into AsiA and σ70 by using single-cysteine mutants of the proteins. The labeled polypeptides were mixed with RNAP core to form the AsiA-containing holoenzyme. Association of acceptor-labeled AsiA with donor-labeled holoenzyme produced a FRET signal. The labeled holoenzyme–AsiA complex was then mixed with an equal amount of unlabeled holoenzyme (containing either wild-type or mutant σ70), and the decrease in FRET was quantified. The decrease in FRET reflects the ability of the unlabeled holoenzyme species to compete with the labeled wild-type holoenzyme for the binding of labeled AsiA. Compared to the wild-type holoenzyme, the holoenzyme containing the σ70 F563Y mutant competed poorly for the binding of AsiA, as expected (Fig. 4A). Furthermore, this defect in the ability of the σ70 F563Y-containing holoenzyme to bind AsiA was suppressed by the introduction into σ70 F563Y of substitutions R541C and/or L607P, which weaken the σ70 region 4/β-flap interaction (Fig. 4A). The results of the FRET analysis thus paralleled the results of the in vitro transcription assays (Fig. 3), indicating that the ability of AsiA to inhibit transcription correlates with its ability to bind the holoenzyme.
Fig. 4.
Effects of substitutions in σ70 on AsiA binding in the context of RNAP holoenzyme (A) and free σ70 (B). (A) The indicated RNAP holoenzymes were added to a ternary complex containing RNAP core and fluorescently labeled σ70 and AsiA. The decrease in FRET signal was determined and converted to competition efficiency (assumed to be 100% for wild-type RNAP holoenzyme). (B) A complex between fluorescently labeled σ70 and AsiA was formed and the effect of the addition of increasing concentrations of unlabeled wild-type or mutant σ70 subunits on the FRET signal was determined (plus signs, wild-type σ70; circles, σ70 F563Y; triangles, σ70 F563Y/R541C; diamonds, σ70 F563Y/L607P; squares, σ70 F563Y/R541C/L607P). The decrease in FRET signal reflects the ability of each added σ70 to compete for labeled AsiA.
We performed a control FRET experiment to confirm that the introduction of substitutions R541C and L607P into the σ70 F563Y mutant did not affect the affinity of AsiA for free σ70. In this experiment, donor/acceptor complexes were formed with labeled AsiA and labeled σ70. Increasing concentrations of unlabeled wild-type or mutant σ70 were then added, and the decrease in FRET was quantified. The decrease in FRET signal reflects the ability of each added σ70 to compete with the labeled σ70 for the binding of labeled AsiA (Fig. 4B). The results confirmed that the binding of AsiA to free σ70 was weakened significantly by the F563Y substitution and that this binding defect was not reversed by either the R541C or the L607P substitution (or the combination of the two) (Fig. 4B).
Discussion
Our findings provide strong support for the idea that AsiA targets the σ70/β-flap interaction, inhibiting transcription from –10/–35 promoters by displacing σ70 region 4 from the β-flap in the context of the holoenzyme. We note that this competitive binding model for AsiA-mediated transcription inhibition may provide an explanation for the previous demonstration that AsiA binds more readily to free σ70 than to the σ70-containing holoenzyme (23). Our data are consistent with the results of a recent analysis of the σ70 region 4/AsiA interaction by NMR (11). This analysis identified amino acid side chains in σ70 region 4 that are contacted by AsiA in the complex that forms when AsiA is combined with a pair of σ70 peptides comprising subregions 4.1 and 4.2. The identified side chains (which include F563) are distinct from those that participate in contacts with the –35 element as seen in the crystal structure of a σ region 4–DNAcomplex (24), but map instead to a hydrophobic pocket that binds the β-flap in the context of the holoenzyme (21, 22). Additional evidence indicated that the binding of AsiA to the σ70-containing holoenzyme resulted in a decrease in the interdomain distance between regions 2 and 4 of σ70, consistent with the expected disruption of the σ70 region 4/β-flap interaction (11).
We found that the ability of AsiA to inhibit transcription by the σ70-containing holoenzyme depends on the relative affinities of AsiA and the β-flap for σ70 region 4, supporting a competitive binding mechanism for AsiA-mediated transcription inhibition. Specifically, we showed that, when the interaction between σ70 region 4 and AsiA is weakened by substitution F563Y in σ70, AsiA fails to inhibit transcription unless the interaction between σ70 region 4 and the β-flap is also weakened by the introduction of one or more additional substitutions into σ70 (see Table 1). A prediction of the competitive binding model for AsiA action is that AsiA's ability to inhibit transcription would be compromised not only by amino acid substitutions that weaken the interaction of AsiA with σ70 region 4, but also by those that strengthen the interaction of σ70 region 4 with the β-flap. We have recently identified amino acid substitutions in σ70 region 4 that strengthen its interaction with the β-flap in the two-hybrid assay (ref. 4 and S.J.G., unpublished data). As predicted by the competitive binding model, we found that these substitutions, which did notweaken the interaction of σ70 region 4 with AsiA, blocked the ability of AsiA to inhibit transcription (B.D.G., unpublished data).
In addition to its role as an inhibitor of σ70-dependent transcription from –10/–35 promoters, AsiA functions as a coactivator of T4 middle gene transcription (25, 26). T4 middle promoters (which are recognized by the σ70-containing holoenzyme) bear a binding site for the phage-encoded regulator MotA that is located at approximately the position of the –35 element of a standard –10/–35 promoter (25). Activation of these promoters depends on a protein–protein interaction between DNA-bound MotA and residues near the C terminus of σ70 (27). Because this portion of σ70 has also been implicated in the β-flap interaction (21, 22), we speculate that, by disrupting the σ70 region 4/β-flap interaction, AsiA may facilitate the MotA/σ70 region 4 interaction.
The demonstration that AsiA works by disrupting the σ region 4/β-flap interaction suggests that it will be worthwhile to search for other regulators that target this interaction in the context of the σ70-containing holoenzyme as well as other holoenzyme species. Moreover, because evidence indicates that the strength of the σ region 4/β-flap interaction varies with different σ factors (4), different holoenzyme species are likely to be differentially susceptible to such regulators.
Acknowledgments
We thank Y. Xia for technical assistance and R. Hellmiss for artwork. This work was supported by National Institutes of Health Grants GM44025 (to A.H.), GM59295 and GM64530 (to K.S.), GM54998 (to J.L.U.), and GM50514 (to T.H.).
Abbreviations: RNAP, RNA polymerase; IPTG, isopropyl-β-d-thiogalactoside; FRET, fluorescence resonance energy transfer.
References
- 1.Gross, C. A., Chan, C., Dombroski, A., Gruber, T., Sharp, M., Tupy, J. & Young, B. (1998) Cold Spring Harbor Symp. Quant. Biol. 63, 141–155. [DOI] [PubMed] [Google Scholar]
- 2.Lonetto, M., Gribskov, M. & Gross, C. A. (1992) J. Bacteriol. 174, 3843–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Callaci, S., Heyduk, E. & Heyduk, T. (1999) Mol. Cell 3, 229–238. [DOI] [PubMed] [Google Scholar]
- 4.Kuznedelov, K., Minakhin, L., Niedziela-Majka, A., Dove, S. L., Rogulja, D., Nickels, B. E., Hochschild, A., Heyduk, T. & Severinov, K. (2002) Science 295, 855–857. [DOI] [PubMed] [Google Scholar]
- 5.Colland, F., Rain, J.-C., Gounon, P., Labigne, A., Legrain, P. & De Reuse, H. (2001) Mol. Microbiol. 41, 477–487. [DOI] [PubMed] [Google Scholar]
- 6.Severinova, E., Severinov, K., Fenyo, D., Marr, M., Brody, E. N., Roberts, J. W., Chait, B. T. & Darst, S. A. (1996) J. Mol. Biol. 263, 637–647. [DOI] [PubMed] [Google Scholar]
- 7.Adelman, K., Orsini, G., Kolb, A., Graziani, L. & Brody, E. N. (1997) J. Biol. Chem. 272, 27435–27443. [DOI] [PubMed] [Google Scholar]
- 8.Severinova, E., Severinov, K. & Darst, S. A. (1998) J. Mol. Biol. 279, 9–18. [DOI] [PubMed] [Google Scholar]
- 9.Colland, F., Orsini, G., Brody, E. N., Buc, H. & Kolb, A. (1998) Mol. Microbiol. 27, 819–829. [DOI] [PubMed] [Google Scholar]
- 10.Minakhin, L., Camarero, J. A., Holford, M., Parker, C., Muir, T. W. & Severinov, K. (2001) J. Mol. Biol. 306, 631–642. [DOI] [PubMed] [Google Scholar]
- 11.Simeonov, M. F., Bieber Urbauer, R. J., Gilmore, J. M., Adelman, K., Brody, E. N., Niedziela-Majka, A., Minakhin, L., Heyduk, T. & Urbauer, J. L. (2003) Biochemistry 42, 7717–7726. [DOI] [PubMed] [Google Scholar]
- 12.Nickels, B. E., Dove, S. L., Murakami, K. S., Darst, S. A. & Hochschild, A. (2002) J. Mol. Biol. 324, 17–34. [DOI] [PubMed] [Google Scholar]
- 13.Dove, S. L. & Hochschild, A. (2001) J. Bacteriol. 183, 6413–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panaghie, G., Aiyar, S. E., Bobb, K. L., Hayward, R. S. & de Haseth, P. L. (2000) J. Mol. Biol. 299, 1217–1230. [DOI] [PubMed] [Google Scholar]
- 15.Minakhin, L., Niedziela-Majka, A., Kuznedelov, K., Adelman, K., Heyduk, T. & Severinov, K. (2003) J. Mol. Biol. 326, 679–690. [DOI] [PubMed] [Google Scholar]
- 16.Borukhov, S. & Goldfarb, A. (1993) Protein Expr. Purif. 4, 503–511. [DOI] [PubMed] [Google Scholar]
- 17.Dove, S. L., Joung, J. K. & Hochschild, A. (1997) Nature 386, 627–630. [DOI] [PubMed] [Google Scholar]
- 18.Dove, S. L. & Hochschild, A. (1998) Genes Dev. 12, 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urbauer, J. L., Adelman, K., Bieber Urbauer, R. J., Simeonov, M. F., Gilmore, J. M., Zolkiewski, M. & Brody, E. N. (2001) J. Biol. Chem. 276, 41128–41132. [DOI] [PubMed] [Google Scholar]
- 20.Lambert, L. J., Schirg, V., Demeler, B., Cadene, M. & Werner, M. H. (2001) EMBO J. 20, 7149–7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murakami, K. S., Masuda, S. & Darst, S. A. (2002) Science 296, 1280–1284. [DOI] [PubMed] [Google Scholar]
- 22.Vassylyev, D. G., Sekine, S., Laptenko, O., Lee, J., Vassylyeva, M. N., Borukhov, S. & Yokoyama, S. (2002) Nature 417, 712–719. [DOI] [PubMed] [Google Scholar]
- 23.Hinton, D. M. & Vuthoori, S. (2000) J. Mol. Biol. 304, 731–739. [DOI] [PubMed] [Google Scholar]
- 24.Campbell, E. A., Muzzin, O., Chlenov, M., Sun, J. L., Olson, C. A., Weinman, O., Trester-Zedlitz, M. L. & Darst, S. A. (2002) Mol. Cell 9, 527–539. [DOI] [PubMed] [Google Scholar]
- 25.Stitt, B. & Hinton, D. (1994) in Molecular Biology of Bacteriophage T4, eds. Karam, J. D., Drake, J. W., Mosig, G., Hall, D. H., Eiserling, F. A., Black, L. W., Spicer, E. K., Kuttner, E., Carlson, K. & Miller, E. S. (Am. Soc. Microbiol. Press, Washington, DC), pp. 142–160.
- 26.Brody, E. N., Kassavetis, G. A., Ouhammouch, M., Sanders, G. M., Tinker, R. L. & Geiduschek, E. P. (1985) FEMS Microbiol. Lett. 128, 1–8. [DOI] [PubMed] [Google Scholar]
- 27.Pande, S., Makela, A., Dove, S. L., Nickels, B. E., Hochschild, A. & Hinton, D. M. (2002) J. Bacteriol. 184, 3957–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dove, S. L., Huang, F. W. & Hochschild, A. (2000) Proc. Natl. Acad. Sci. USA 97, 13215–13220. [DOI] [PMC free article] [PubMed] [Google Scholar]