SUMMARY
Embryonic stem cells (ESC) are apparently homogeneous self-renewing cells, but Stella, a marker of preimplantation embryos and germ cells showed heterogeneous expression in ESCs. Here we show that the Stella-positive ESCs were like the inner cell mass (ICM) while the Stella-negative cells were like the epiblast cells, although these populations exhibited interchangeability that reflected an adherence to an inherent program. Thus, ESCs are in a metastable state since they shift between the ICM- and epiblast-like phenotypes, which provides an insight into their instability, plasticity and pluripotency. This equilibrium was skewed reversibly by environmental and epigenetic cues; for example, the absence of signals from feeder cells caused a shift towards an epiblast-like state while trichostatin A, an inhibitor of histone deactelylase, restored Stella-positive population. Consistently, the two populations showed different histone modifications but with hypomethylated DNA in stella locus, as well as functional differences when induced to undergo differentiation. The Stella-negative cells were apparently more like the post-implantation epiblast-derived stem cells (EpiSCs), except that the stella locus was hypermethylated and repressed in the latter, which denotes a robust boundary between ESCs and EpiSCs.
Keywords: Embryonic stem cells, Inner cell mass, Epiblast, Stella, Self-renewal
INTRODUCTION
Mouse embryonic stem cells (ESCs) exhibit a degree of similarity with three different transient cell populations in the mouse embryo: the inner cell mass (ICM) of blastocysts, the epiblast cells of early postimplantation embryos and primordial germ cells (PGCs), which includes expression of pluripotency marker genes, such as Oct4 (Chambers and Smith, 2004; Pesce et al., 1998; Surani et al., 2007; Zwaka and Thomson, 2005). However, unlike the cells in vivo, ESCs retain pluripotency and exhibit the capacity for indefinite self-renewal, while the cells in vivo undergo differentiation according to a strict developmental program. As long as ESCs are cultured in an appropriate medium, such the one containing leukemia inhibitory factor (LIF) and with either serum or bone morphogenetic protein 4 (BMP4), they can undergo self-renewal without compromising pluripotency (Ying et al., 2003). For this reason, ESCs are generally regarded as a homogeneous group of cells in the majority of studies.
However, the precise provenance of ESC for which there is no strict in vivo equivalent remains to be fully elucidated. Based on some recent studies, it has been suggested that germ cells may be the closest in vivo equivalent of ESCs (Zwaka and Thomson, 2005), partly because expression of Stella has been reported in both mouse and human ESCs, albeit heterogeneously (Clark et al., 2004; Payer et al., 2006). Stella a definitive marker of the germ cell lineage, is however first observed in preimplantation embryos. Thereafter, Stella is repressed in the epiblast (Payer et al., 2006; Sato et al., 2002), and subsequently re-expressed only following specification of PGCs (Payer et al., 2006). Other proteins such as Pecam1, Nanog and SSEA1 also exhibit heterogeneous expression in undifferentiated ESCs (Chambers et al., 2007; Cui et al., 2004; Furusawa et al., 2004; Payer et al., 2006; Toyooka et al., 2008). Thus, while heterogeneity is a hallmark of ESCs, it remains to be fully elucidated how this is compatible with pluripotency and self-renewal.
In this study, we set out to investigate the nature of Stella-expressing cells in undifferentiated ESCs. We demonstrate that Stella-positive ESCs are closely related to the ICM and not to the epiblast or PGCs. Moreover, cultured under conditions that maintain pluripotency, the proportion of Stella-positive cells remained relatively constant, while they were able to reversibly convert to Stella-negative ESCs. Expression of stella is regulated by chromatin-based modifications in ESCs, which can respond to both the environmental and epigenetic cues. We propose that while undifferentiated ESCs undergo self-renewal, they are in a highly dynamic state as they continuously fluctuate between an ICM- and epiblast-like phenotype. By contrast, stella is robustly repressed by DNA methylation in pluripotent stem cells derived from postimplantation epiblast cells (EpiSC), which do not readily revert to an ICM-like state. Furthermore, stella is subsequently activated but only following specification of PGCs.
RESULTS
The population of Stella-GFP-positive cells remains relatively constant
We previously generated two stella-gfp transgenic ESC lines (SH10.10 and BAC9) with different lengths of the stella flanking sequences coupled to a gene for green fluorescent protein (GFP) as the reporter (Payer et al., 2006). In these ESCs, expression of Stella as well as of Stella-GFP was detectable only in a subset of ESCs (Figures 1A-C). Similar heterogeneous expression of Stella was also confirmed in non-transgenic ESCs by immunsostaining (data not shown). Notably, cells with expression of Stella-GFP coincided significantly with those observed for endogenous Stella expression. For example, this was the case in 126/141 (89%) randomly chosen ESC, although some Stella-positive cells were negative for Stella-GFP (5%), and some strongly Stella-GFP-positive cells (6%) expressed Stella only weakly. Fluorescence-activated cell sorting (FACS) consistently revealed that around 20-30% of ESCs were Stella-GFP-positive under our culture conditions, irrespective of the passage number. However the exact proportion of Stella-GFP positive cells occasionally deviated from this range, as they are susceptible to slight differences in culture conditions (Figure 1D and data not shown). These observations were made whether we used ESCs from stella-gfp BAC9 or from stella-gfp SH10.10 transgenic mice, except that the fluorescence activity of Stella-GFP in BAC9 ESCs was weaker (Figure 1D), which is most likely the result of the integration of different copy number of transgenes in the genome. For convenience, we used stella-gfp SH10.10 ESCs in the rest of the studies described here.
Figure 1. Heterogeneous expression of Stella in undifferentiated ESCs.
(A-C) Immunoflurescence analysis of Stella-GFP (A) and endogenous Stella (B) expression in transgenic stella-gfp SH10.10 ESCs, and their merged images with DAPI (C). Scale bar: 25 μm. (D) Percentage of Stella-GFP-positive cells in control ces3 ESCs (ESWT; left), stella-gfp SH10.10 (middle) and stella-gfp BAC9 (right) transgenic lines. Numbers above the gates indicate the percentage of the total cells plotted in each dotplot. Numbers described in upper left indicate the passage number.
Stella-GFP-positive population is enriched in ICM-specific markers
Next, we examined transcription profile of the Stella-GFP-positive and -negative populations, by quantitative reverse polymerase chain reaction (Q-PCR). Notably, we found that the two fractions clearly differed in the levels of expression of Fgf5, a marker of pluripotent epiblast cells of early postimplantation embryos, which is not expressed in the ICM (Pelton et al., 2002; Rathjen et al., 1999). Furthermore, Gbx2 was also detected at a higher level in the Stella-GFP-negative cells, and like Fgf5, Gbx2 is also up-regulated in the primitive ectoderm cells soon after implantation (Figure 2A) (Chapman et al., 1997; Kurimoto et al., 2006). By contrast, Stella-GFP-positive population was slightly enriched in Pecam1 and Zfp42/Rex1 transcripts which are preferentially expressed in ICM compared to the epiblast (Figure 2A) (Pelton et al., 2002; Robson et al., 2001). Genes, such as alphafetoprotein and collagen type IV that are up-regulated in differentiated ESCs were not detectable in either of the two populations (data not shown), indicating that Stella-GFP-negative population is not representative of differentiating ESCs.
Figure 2. Differential gene expression in Stella-positive and Stella-negative population of stella-gfp SH10.10 ESCs.
(A) Q-PCR analysis of each gene in FACS-sorted Stella-GFP-positive and Stella-GFP-negative populations. Relative levels of each gene expression are estimated by referring to the value of Gapdh. The mean values are calculated from three independent experiments. (B) Q-PCR analysis using randomly selected single cells from stella-gfp ESCs. Left graph shows the putative number (log10) of transcript in each single cell. Three populations, Stellahigh, Stellalow and Stellanegative were identified and are set apart by broken lines. Right graph shows the average number of transcripts in each population.
Single cell analysis
The above analysis was conducted on the two FACS-sorted cell populations, which would mask the extent of variations amongst individual cells. In addition, whereas Stella-GFP expression in ESCs showed extensive overlap with the endogenous expression of Stella, they do not coincide completely (Figures 1A-C and 2A). Therefore, we decided to further investigate the nature of heterogeneity amongst the ESCs, by examining gene expressions in 87 randomly selected single ESCs. Q-PCR analysis revealed variable stella transcripts in 63 (72.4%) cells (Figure 2B), amongst which we found 26 (29.9%), Gfp-expressing cells. Notably, Gfp was detected predominantly in cells with high expression of stella (Figure 2B), which is consistent with the 20-30% of Stella-positive cells we detected by immunostaining. Amongst these, 15/26 cells showed high stella expression (stella-high;57.8%), and 9/26 belonged to the stella-low group (34.6%) (Figure. 2B). The differences in the percentage of cells with the stella transcript and those positive for the protein might be caused by translational regulation of stella transcript. It is possible that translational regulation is an important general feature of ESCs (see below). Nevertheless, these results demonstrate that the number of stella transcripts in individual cells was variable, but virtually all the stella-Gfp-positive cells (92.3% 26/28) were present within the group with high stella transcripts (Figure 2B).
Next, we extended our single cell Q-PCR analysis to further assess expression of the other key marker genes. In agreement with the data from the FACS-sorted populations, we detected Fgf5 expression significantly amongst the stella-negative cells. By contrast, most of Pecam1 expressing cells were amongst those with expression of stella, which also showed higher expression of Zfp42/Rex1(Figure 2B). Furthermore, it is of interest that expression of Gbx2 was detected amongst the stella-low and stella-negative cells, which is consistent with the in vivo expression of Gbx2 that starts to increase in the E4.5 primitive ectoderm at the time when stella expression begins to decline (Kurimoto et al., 2006). It is striking that the average number of Nanog, and Sox2 transcripts were also significantly lower in stella-negative cells compared to the stella-positive cells (0.01<P<0.05) (Figure 2B), which may reflect a decrease in these transcripts in vivo in the E4.5 primitive ectoderm and epiblast cells of mid-streak embryos, respectively (Avilion et al., 2003; Chambers et al., 2003). By contrast, Oct4 trasncripts were detected uniformly in all cells, which is the case in vivo in ICM and epiblast cells. Notably, these results indicated that the developmentally regulated genes are not expressed stochastically amongst ESCs as their expression seems to reveal an adherence to the in vivo developmental program.
Consistent with the Q-PCR analyses of single cells, the entire population of Stella-GFP-positive ESCs was contained within the Pecam1-positive population by FACS analysis (Figure S1). Pecam1 is a developmentally regulated gene that is homogeneously expressed in the ICM and completely absent from the epiblast (Figure S1 and (Robson et al., 2001). This being the case, it is clear that the Stella-GFP-positive cells are contained within these ICM-like cells. By contrast, expression of SSEA1 (stage-specific embryonic antigen-1), which also shows heterogeneous expression in ESCs (Cui et al., 2004; Furusawa et al., 2004), was detected in both the Stella-GFP-positive and Stella-GFP-negative cells (Figure S1). However, unlike Pecam1, there is heterogeneous expression of SSEA1 in both the ICM and epiblast cells (Cui et al., 2004; Furusawa et al., 2004). We observed that the Stella-GFP-positive cells were detected within the SSEA1-positive as well as the SSEA1-negative population at comparable levels (17.4% vs 11.0%) (Figure S1). Immunofluorescence analysis confirmed that Stella-GFP-positive cells did not entirely overlap with the SSEA1-positive cells (Figure S1). These results again emphasise that heterogeneity amongst ESCs has a basis in the inherent developmental program in vivo.
The sub-populations of ESCs are interchangeable
Next we asked how the relatively stable population of 20-30% Stella-GFP-positive cells are maintained amongst the ESCs, for which there are at least two possibilities: either they are in a state of flux and interchange continuously between positive and negative states while maintaining a constant ratio, or they give rise only to descendants of the same type. To distinguish between these possibilities, FACS sorted Stella-positive and Stella-negative ESCs populations were cultured and analysed separately. To avoid contamination of cells that express endogenous stella but no Gfp transcripts, we used Stella-GFP-negative/Pecam1-negative population since single cell analysis revealed that cells that do not express endogenous stella were enriched within the double negative population. Remarkably, after 3 days of culture, approximately 50% of the cells in both the individual fractions were found to be Stella-GFP-negative/Pecam1-positive (Figure 3A). We consider these cells to be phenotypically closest to the two original isolated FACS-sorted populations described above. Indeed, these individual cultures were eventually fully restored to the original state of about 20-30% Stella-GFP-positive cells we first encountered amongst the stella-gfp ESCs. Interestingly, the Stella-GFP-positive/Pecam1-positive population reverted to the original state faster than the Stella-GFP-negative/Pecam1-negative cells, which occurred within 5 and 7 days of culture, respectively (Figures 3A and B). These results show that the FACS-sorted populations of ESCs representing the two extreme populations are interchangeable. Moreover, the two individual groups of cells first generated a large proportion of Stella-GFP-negative/Pecam1-positive cells, which were in turn followed by the generation of cell types that progressively departed further away from the original FACS-sorted Stella-GFP-positive/Pecam1-positive and Stella-GFP-negative/Pecam1-negative cell types, respectively. These observations suggest an adherence to an intrinsic program during their inter-conversion, rather than a stochastic change in gene expression.
Figure 3. ESCs display a state of dynamic equilibrium.
(A) Stella-GFP-positive/Pecam1-positive (green) and Stella-GFP-negative/Pecam1-negative (yellow) FACS-sorted stella-gfp ESCs were cultured for 7 days. The percentage of cells with Stella-GFP expression was analyzed on day 0, 3, 5 and 7. Gating was determined independently at each time point using unsorted stella-gfp ESCs analysed in parallel as a reference. The percentage of gated cells is given in the upper corner of each dotplot. (B) Summary of phenotypic changes in Stella-GFP-positive/Pecam1-positive and Stella-GFP-negative/Pecam1-negative cells. Note that the Stella-GFP-positive/Pecam1-positive cells (green cells) revert to the original proportion more quickly than the Stella-GFP-negative/Pecam1-negative (yellow) cells. Cells depicted with white cytoplasm indicate Stella-GFP-negative/Pcam1-positive cells. (C) Clonal analysis of Stella-GFP-positive/Pecam1-positive and Stella-GFP-negative/Pecam1-negative FACS-sorted stella-gfp ESCs. Cells were expanded for 9 days and the percentage of Stella-GFP-positive cells in each independent clone (n=64) determined by FACS analysis. The averaged percentage is shown in each graph. (D) Different rate of colony formation of Stella-GFP-positive/Pecam1-positive and Stella-GFP-negative/Pecam1-negative stella-gfp ESCs. The averaged numbers of colonies from Stella-GFP-positive/Pecam1-positive and Stella-GFP-negative/Pecam1-negative FACS-sorted stella-gfp ESCs are shown. The mean values are calculated from three independent experiments.
To exclude the possibility that the results could be influenced by the reciprocal contamination of the two cell populations, ESC sub-clones (n=64) derived from FACS-sorted single Stella-GFP-positive/Pecam1-positive or Stella-GFP-negative/Pecam1-negative cells were analyzed at 9 days after FACS-sorting. Clonal analysis revealed that both sets of cells could generate the reciprocal set of cells as in the parental cell lines (Figure 3C), except that there was a lower average percentage of Stella-GFP-positive cells detected from the latter sub-clones, which is consistent with the relatively less efficient generation of the reciprocal cell types as described previously. Moreover, the clonal analysis also showed less efficient plating of Stella-GFP-negative/Pecam1-negative ESCs, which formed fewer colonies compared to the cells of the opposite phenotype (Figure 3D). Thus, while the overall population of Stella-positive ESCs is maintained through mutual conversion of positive and negative cells, the efficiency of the process clearly differs. We reason that the conversion of Stella-positive/Pecam1-positive cells into Stella-negative/Pecam1-negative cells follows an inherent developmental program form ICM towards epiblast, whereas the reciprocal conversion is contrary to the normal developmental program in vivo. This could account for the slower conversion rate of the latter.
Epigenetic regulation of metastable Stella expression in ESCs
The intermittent expression of Stella in ESCs indicates that these pluripotent stem cells are probably in a metastable state, which should be reflected in their epigenetic states. We therefore examined histone modifications and DNA methylation in the endogenous genomic DNA sequences surrounding the start codon of Stella. We found that the Stella-GFP-positive ESCs displayed a relative enrichment of acetylated histone H3 lysine 9 (H3K9ac) and tri-methylated H3K4 (H3K4me3), which are a hallmark of an active gene, compared to the levels in Stella GFP-negative/Pecam1-negative ESCs (Figure 4A). The levels of tri-methylation of H3K27 (H3K27me3), a repressive gene marker was however comparable in the two populations (Figure 4A). Furthermore, the DNA sequence, which has 10 CpGs, was hypomethylated in both the Stella-GFP-positive and Stella-GFP-negative/Pecam1-negative FACS-sorted cells (Figure4B), except for a slight increase in some of the latter cells. These results indicate that the metastable expression of Stella is regulated by chromatin-based epigenetic changes, which is independent of DNA methylation.
Figure 4. Epigenetic regulaton of the stella locus and modulation of Stella-GFP-positive cells.
(A) ChIP analysis of histone modifications in the Stella locus. Genomic DNAs from FACS-sorted Stella-GFP-positive/Pecam1-positive or Stella-GFP-negative/Pecam1-negative cells were immunoprecipitated with the antibodies as indicated, and were then subjected to Q-PCR using a primer set specific to the endogenous genomic locus encoding the start codon of Stella. Levels of histone modifications were estimated by dividing with the input value (see mateials and methods). (B) Bisulfite sequencing profiles of DNA methylation of the stella locus. CpG sequences are shown with filled (methylated) and open (unmethylated) circles. Gaps in the methylation profiles represent mutated or missing CpG sites. The numbers under the bisulfite sequencing profiles show the percentages of methylated CpG. (C) FACS analysis of Stella-GFP-positive cells under various conditions. Shown are the percentages of Stella-GFP-positive cells after culturing stella-gfp ESCs without MEFs (upper left), followed by re-culturing them on MEFs in chemically defined medium (upper right), or exposing them to TSA (lower left) or 5-aza (lower right) in the absence of MEFs. (D) Morphology of the colonies of stella-gfp ESCs cultured with TSA. Images show the change in morphology of the stella-gfp ESC colonies cultured without MEF (left), and than following addition of TSA (right). Note the relatively more compact ESC colonies in the latter. Windows in each image represent Stella-GFP in a colony. Scale bar: 50μm
As the histone modifications can be relatively plastic, we next tested the responsiveness of Stella expression under a variety of conditions. First, we found that the culture of ESCs in a chemically defined medium without embryonic fibroblast feeder cells (MEFs), which are a source of signaling molecules that affect ESCs (Chambers and Smith, 2004), resulted in almost complete loss of Stella-GFP expression (Figure 4C). Notably, Stella-GFP expression was however restored, as before, in over 20% of the ESCs when they were returned to culture with MEFs (Figure 4C). This clearly shows that the metastable state of Stella-GFP expression is influenced by environmental cues, one of which is signals from MEF. Notably, Stella-GFP expression was also restored in response to 10 nM Trichostatin A (TSA), an inhibitor of histone deacetylase, but DNA5-azacytidine, an inhibitor DNA methyltransferase had no effect. Indeed, the proportion of Stella-GFP-positive cells increased to 37.5% in response to TSA. These ESCs also showed a striking phenotypic change as they formed compact colonies, which is characteristic of ESCs grown on MEFs (Figure 4D). These results demonstrate that histone acetylation, but not DNA methylation of Stella locus is one of the epigenetic modifications regulating expression of Stella.
Functional differences between the subpopulations of ESCs
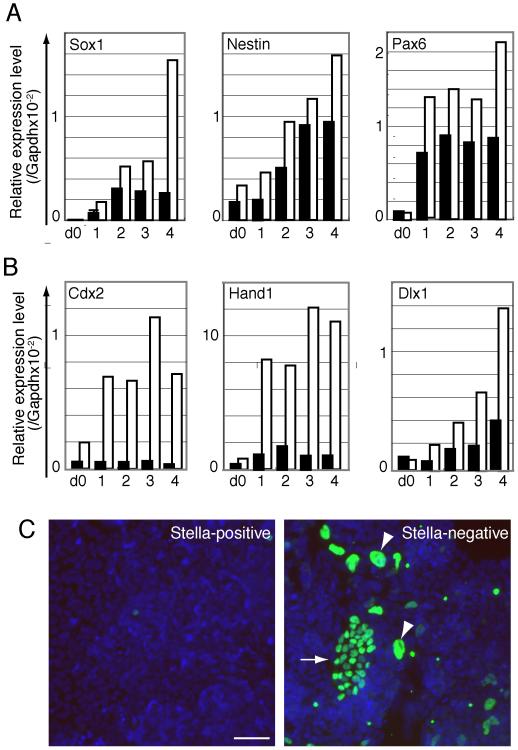
Our studies above show that the subpopulations of ESCs exhibit distinct gene expression profiles, while being in a metastable epigenetic state. Since the subpopulations were constantly interchangeable under conditions that maintain them in an undifferentiated state, we decided to investigate if they had distinct properties if they were forced to undergo differentiation. First we attempted to check their differentiation potential in embryoid bodies (EB). However, the Stella-negative/Pecam-negative cells seldom formed EBs, whereas Stella-GFP-positive/Pecam1-positive cells could so as expected and as judged by the detection of differentiation marker genes (data not shown). This observation shows a difference in the properties of the two populations. Next, we cultured each cell population with retinoic acid (RA) that induces mainly neuronal differentiation. Q-PCR analysis showed that expression of early neuronal lineage-marker gene, Sox1, Nestin and Pax6 was enhanced in Stella-GFP-negative/Pecam1-negative cells, compared to the levels of expression detected with Stella-GFP-positive/Pecam1-positive cells (Figure 5A). This is not due to a difference in the basal levels of expression of these genes in each population, since Sox1 and Pax6 transcripts were rather lower in non-stimulated Stella-GFP-negative/Pecam1-negative cells (Figure 5A). These results demonstrate that Stella-GFP-negative/Pecam1-negative cells were more sensitive to the signal to undergo differentiation, at least with respect to the response to RA-induced differentiation.
Figure 5. Differentiation potential of Stella-positive and Stella-negative populations.
(A) Response to retinoic acid-induced differentiation and expression of neuronal markers. Graphs show a representative Q-PCR analysis of Sox1, Nestin and Pax6 expression in FACS-sorted Stella-GFP-positive/Pecam1-positive (black) and Stella-GFP-negative/Pecam1-negative (white) ESCs cultured with RA as for days (d) as indicated. Samples at d0 were prepared immediately after FACS-sorting followed by RA-induction. (B) Expression of trophoectoderm marker genes. Graph shows representative Q-PCR analysis of Cdx2, Hand and Dlx1 expression in each subpopulation, as described in (A) when cultured under TS cell condition. Repeated Q-PCR analysis using different sets of sorted samples showed similar results as shown in both (A) and (B). (C) Enhanced expression of Cdx2 protein. Images are immunostaining of Cdx2 (green) and DAPI (blue) in each subpopulation cultured for 3 days under TS condition. Arrow and arrowhead indicate representative Cdx2-positive dense colony and cells having large nuclei, respectively. Scale bar: 50μm
Next we checked their response when each population was cultured under trophoectoderm stem (TS) cell culture condition. Surprisingly, the Stella-GFP-negative/Pecam1-negative population exhibited remarkable up regulaton of Cdx2 gene expression, a marker of trophoectoerm in pre- and peri-implantation embryos., as well as of other markers of this lineage, such as Hand1 and Dlx1 (Figure 5B). Immunofluorescence analysis showed that Cdx2 was clearly detectable in nuclei of cells forming dense colonies, as well as in large nuclei of presumptive trophoblast giant cells located around the periphery of differentiating colonies (Figure 5C). Recently, evidence from pluripotent stem cells derived from post-implantation epiblast, cells (EpiSCs), show that these are also capable of differentiation into trophoectoderm (Brons et al., 2007; Tesar et al., 2007). This is consistent with our data that that the Stella-negative/Pecam1-negative ES cells are more like the epiblast cells. By contrast, the Stella-positive/Pecam1-positive cells failed to show this response. This is consistent with the fact that ES cells show a small number of differentiating trophectoderm cells when cultured under TS cell condition (in preparation). Taken together, these results demonstrate that Stella-negative cells possess functionally distinctive properties, one of which is a permissive state for trophoectodermal differentiation that is also observed with EpiSCs.
Comparisons between the state of Stella in ESCs and EpiSCs
Based on our analysis, ESCs consist of a population of Stella-GFP-negative/Pecam1-negative cells that are relatively closer to the epiblast cells and share some of the differentiation potential with EpiSCs, a stem cell line from epiblast cells of postimplantation embryos. We therefore further characterized the Stella-GFP-negative/Pecam1-negative ESCs by comparing marker gene expression and epigenetic status of stella in EpiSCs. As expected, we detected Fgf5 expression in EpiSCs, but the levels were significantly higher compared to those in the epiblast-like cells in ESCs (Figure 6A). By contrast, expressions of stella, Pecam1 and Zfp42/Rex1 were negligible in EpiSCs, compared to the ESC population (Figures 6A and B). However, unlike a lack of Pecam1 expression, we detected heterogeneous expression of SSEA1 in EpiSCs (Figure 6B) consistent with similar expression in the epiblast cells in vivo. Levels of Nanog and Sox2 in EpiSCs are comparable to those in the epiblast-like cells in ESCs (Figure 6A). These observations suggest that the Stella-GFP-negative/Pecam1-negative population in ESCs represent an intermediate state between ICM-like and the EpiSCs. This was also supported by microarray analysis using single cell gene expression profile, which revealed that ESCs with low expression of stella had an intermediate gene expression profile when compared with ESCs with high expression of stella and EpiSCs (Figure S2). In other words, cells with low expression of stella were closer to the EpiSCs compared to the cells with high expression of stella.
Figure 6. Distinctive status of ESCs and EpiSCs.
(A) Q-PCR analysis of gene expression in FACS-sorted EpiSCs populations of Stella-GFP-positive and Stella-GFP-negative/Pecam1-negative ESC. Relative levels of gene expression in EpiSCs were estimated by reference to the value of Gapdh. (B) ChIP analysis of histone modifications in the stella locus of EpiSCs. Genomic DNAs were immunoprecipitated from FACS-sorted EpiSCs with antibodies as indicated, followed by Q-PCR analysis. Levels of histone modifications were estimated as indicated in Figure 2A. (C) FACS analysis of Pecam1 and SSEA1 in EpiSCs. Histograms show expression of Pecam1 (left) and SSEA1 (right). (D) Bisulfite sequencing profiles of DNA methylation in the Stella locus in EpiSCs. CpG sequences are shown with filled (methylated) and open (unmethylated) circles. Gaps in the methylation profiles represent mutated or missing CpG sites. The numbers under the bisulfite sequencing profiles show the percentages of methylated CpG. (E) Bisulfite sequencing profiles of LINE-1. Sequences are shown with filled (methylated) and open (unmethylated) circles. The numbers under the bisulfite sequencing profiles show the percentages of methylated CpG.
Next we compared the epigenetic status of stella locus in EpiSCs to compare it with the situation in ESCs. Compared to epiblast-like cells in ESCs, the levels of H3K9ac and H3K4me3 in EpiSCs were considerably diminished (Figure 6C), whereas that of H3K27 showed an increase. Most strikingly, the stella locus showed extensive DNA methylation in EpiSCs (Figure 6D). These results demonstrate that the epigenetic status and regulation of stella in the epiblast-like ESC subpopulations and EpiSCs differ significantly. Apart from the stella locus, LINE1 sequences, a retrotransposable element that is widely distributed in the genomic DNA, were also hypermethylated in EpiSCs, whereas they were hypomethylated in both populations from ESCs (Figure 6E), which demonstrates that DNA methylation takes place not only in the stella locus but also in genome-wide regions. These observations are consistent with the fact that de novo DNA methylation takes place in the pluripotent cell population in vivo during peri-implantation development (Kafri et al., 1992; Monk et al., 1987), which is essential for further development (Li et al., 1992; Okano et al., 1999). DNA methylation at this stage is linked to the incipient differentiation of pluripotent cells involving developmentally regulated genes, one of which is stella.
DISCUSSION
Single cell analysis demonstrates that ESCs exhibit heterogeneity that spans across ICM to epiblast-like phenotypes. Whereas expression of stella in ESCs represents one extreme of ICM-like cells, the mutually exclusive expression of Fgf5 amongst Stella-GFP-negative/Pecam1-negative cells represents the most epiblast-like cells. Thus, ESCs are not a homogeneous group of self-renewing cells as they constantly fluctuate between ICM and epiblast like states with an apparent adherence to an inherent program. Since the number of cells with stella transcripts exceeds those which exhibit endogenous Stella and Stella-GFP, it is possible that translational regulation mechanisms may be important during self-renewal and pluripotency. The expression of stella in ESCs is regulated by histone-based modifications, which is responsive to environmental and epigenetic factors. By contrast, stella in EpiSCs derived from the postimplantation epiblast cells is repressed robustly by DNA methylation, which denotes a barrier between the two types of pluripotent stem cells (Figure 7).
Figure 7. Proposed model for the maintenance of dynamic equilibrium of distinct cell types in ESCs.
ESCs represented as consisting of distinct subpopulations of cells, each of which has a different combination of developmentally regulated genes spanning between the ICM to epiblast-like phenotype. The mechanism that drives ESCs from ICM to epiblast-like cells may be similar to that involved in the differentiation of ICM to epiblast in vivo, but the mechanism involved in the reversion of epiblast-like cells back to ICM-like cells suggests a dedifferentiation step; the mechanism regulating the latter is unknown. The self-renewal ‘region’ of ESCs is distinct from that in EpiSCs. Notably, DNA methylation of the stella locus is one criteria that distinguishing ESCs from EpiSCs (see text for details).
It was striking that the proportion of Stella-GFP-positive cells in ESCs remained relatively constant, even after the culture of Stella-GFP-positive/Pecam1-positive and Stella-GFP-negative/Pecam1-negative cells separately, as they regenerated the appropriate number of reciprocal cell types. However, the latter were slower than the former in converting into the reciprocal cell types. This is presumably because in the first case, the cells are progressing along an inherent developmental program form ICM- towards epiblast-like cells, while the converse represents a ‘dedifferentiation’ step that is contrary to the normal developmental program. Our single cell analysis also shows that cells lacking in stella transcripts, had exceedingly low levels of Pecam1 expression as well as lower levels of nanog expression. These observations are broadly in agreement with the differential expression of Pecam1 in ESCs, where the Pecam1-negative cells are predominantly epiblast-like cells (Furusawa et al., 2006; Kemp et al., 2005). Furthermore, Pecam1-positive or Nanog-positive cells constitute a relatively stable population of ESCs (Chambers et al., 2007; Furusawa et al., 2006; Furusawa et al., 2004), while the Pecam1-negative and Nanog-negative cells are harder to convert back into their reciprocal counterparts.
Consistent with the interchangeability of ESCs, we found that the proportion of Stella-GFP-positive cells was reversibly responsive to MEFs, which are a source of signaling molecules that may in turn influence the intrinsic epigenetic state of responding cells (Ansel et al., 2003; Kang et al., 2005; van Grunsven et al., 2005). Multiple signaling molecules, together with the transcription and epigenetic factors play an essential role in maintaining pluripotency in ESCs (Chambers and Smith, 2004; Surani et al., 2007). We stress that the proportion of Stella-GFP-positive cells could significantly exceed those we report here, provided appropriate conditions exist that promote the ICM-like phenotype. Indeed, ESCs cultured without MEFs that were almost completely in an epiblast-like state, showed a very significant increase in Stella-GFP-positive cells in response to TSA, an inhibitor of HDACs. Thus, TSA induced ‘reprogramming’ of ESCs caused a significant shift towards an ICM-like state from a virtual epiblast-like state prior to the treatment. Notably, the effect of TSA was striking because it also induced a marked phenotypic change in the ESCs colonies, which acquired an appearance that is reminiscent of the colonies grown on MEFs. Consistently, it was reported that the same concentration (10nM) of TSA inhibited differentiation of ESCs when cultured without LIF (Lee et al., 2004), although a significantly higher dosage of TSA induced ESC differentiation (McCool et al., 2007). Thus, the balance between histone acetylation and deacetylation may be at least one factor that may determine whether ESCs are in an ICM-like or epiblast-like state.
The gene expression profile indicated that the stella-negative ESCs are situated between ICM and fully developed epiblast. Under the culture condition that maintain ESCs, the stella-negative cells might not be able to progress to the EpiSCs owing to the extrinsic signals that force them back to the ICM-like state. However, when Stella-GFP-negative/Pecam1-negative ESCs were cultured under EpiSC condition, Fgf5 expression showed a further increase (data not shown). More importantly, we demonstrate that the Stella-positive and Stella-negative cells show clear differences with respect to their potential for differentiation. Notably, the Stella-negative cells can differentiate into trophectoderm cells (see Figures 5B and C), a property they share with EpiSCs. Analysis of epigenetic status however clearly showed that DNA methylation of the stella locus represents a boundary between ESCs and EpiSCs. EpiSCs despite showing expression of pluripotent marker genes, including Oct4, Sox2 and Nanog, are epigenetically quite distinct from ESCs, as they rarely contribute to chimeras, and unlike ESCs, they exhibit an inactive X chromosome (Tesar et al., 2007) Notably, the EpiSCs do not exhibit an ICM-like state unlike ESCs, which can fluctuate freely within the ‘self-renewal parameters’ encompassed by the ICM and epiblast-like states, and where stella locus is hypomethylated and its expression is regulated by chromatin-based modifications (Figure 7). Furthermore, this is consistent with the evidence that de novo DNA methylation is not essential for self-renewal of ESCs but it is essential for their differentiation (Chen et al., 2003; Lei et al., 1996).
Our preliminary evidence shows that the stella locus in EpiSCs is subject to DNA demethylation only in the cells that undergo specification into PGCs (K. Hayashi and M. A. Surani, in preparation), consistent with the fact that Stella is only expressed in the germ cell lineage after implantation (Saitou et al., 2002; Sato et al., 2002). This epigenetic change in the stella locus exclusively in PGCs is probably an important hallmark of other significant epigenetic reprogramming events in PGCs. Strikingly, these epigenetic changes in PGCs, confers the ability on early germ cells to undergo dedifferentiation into pluripotent embryonic germ cells, which resemble ESCs. EpiSCs themselves may lack the ability to convert directly into ESCs with a population of ICM-like cells.
In view of our findings, it is striking to note a virtual loss of stella expression in ESCs with a null mutation in the gene encoding methyl-CpG binding domain protein3 (Mbd3), which is a member of the NuRD complex (Kaji et al., 2006). Reciprocally, the Mbd3-null ESCs show high levels of Fgf5 expression, possibly as they acquire a more epiblast-like character and grow independently of LIF, although they are unable to differentiate. Mbd3 is also essential for peri-implantaton development in vivo. Similarly, Dicer-null ESCs can be maintained without LIF but they are impaired in their ability to undergo differentiation (Kanellopoulou et al., 2005; Murchison et al., 2005). We predict that these mutant cells may probably be more epiblast-like and devoid of Stella expression, and their differentiation is impaired because of attenuated DNA methylation due to misexpression of de novo DNA methyltransferases (Sinkkonen et al., 2008). Thus, the exit from ESC and loss of ICM-like stella-positive phenotype is coupled with DNA methylation as represented by methylation of the stella locus.
In conclusion, our study shows that ESCs are not a group of uniform self-renewing cells, since they appear to be in a metastable state, and shift between ICM-and epiblast-like states while retaining pluripotency. This equilibrium can shift in either direction in response to a variety of factors, including epigenetic regulators. Analysis of Stella expression may be used to determine the precise phenotypic state of different ESC lines, and of the factors, including epigenetic regulators, which affects its expression. The proportion of Stella-GFP-positive, and therefore ICM-like cells in ESC cultures, is a sensitive indicator of optimal environmental or epigenetic modulators that promote this phenotypic state. Further studies may also reveal the precise role of Stella in the derivation and maintenance of ESCs.
EXPERIMENTAL PROCEDURES
Cell culture
ESCs were grown in ESC medium (DMEM:F12, Invitrogen; supplemeted with 15% fetal calf serum, 1 mM glutamine, 100 u/ml penicillin/streptomycin, 0.1mM b-mercaptoethanol, and 1,000 u/ml LIF) on mitomycin C-treated MEFs, unless specifically mentioned. For clonal culture analysis, 500 FACS-sorted single ESCs were spread on MEFs seeded on 6cm tissue culture dish. After 5 days of culture, the number of colonies on the dish was counted, and then the colonies were picked up, trypsinised and transferred into 24-well dishes. At 4 days after the picking colonies up, Stella-GFP-positive cells were analysed by FACS. To establish MEF-independent stella-gfp ESCs, the ESC line was cultured in Clonal Grade Medium (Chemicon) for 10-14 days and then maintained in Knockout DMEM (Invitrigen) containing 20% of Knockout serum replacement, 1 mM glutamine, 100 u/ml penicillin/streptomycin, 0.1mM ß-mercaptoethanol and 1,000 u/ml LIF. The KSR-containing medium was used as the chemically defined medium. For differentiation analysis, FACS-sorted cells were cultured in the chemically defined medium with RA (1μM) or in TS medium with supernatant from medium cultured with MEFs (Tanaka et al., 1998). EpiSCs were cultured in N2B27 supplemented with 20%KSR, 2ng/ml of recombinant human Activin A (Peprotech) and 12ng/ml of bFGF (Invitrogen). Both ESCs and EpiSCs were cultured in 5% CO2/95% air at 37°C.
FACS and immunofluorescence analysis
For FACS analysis, single cell suspension was stained with R-Phycoerythrin-conjugated rat anti-Pecam1 (BD Pharmingen). The stained cells were analysed and sorted by FACSort with CELLQUEST software (BD Bioscience) and FACSAria (BD Bioscience), respectively. Immunofluorescence analysis of cells was performed essentially as described (de Sousa Lopes et al., 2004; Ohinata et al., 2005). The antibodies used were rabbit anti-PGC7/Stella, mouse anti-CDX2 (Cdx2-88, BioGenex) and rat anti-GFP (Nacalai tesque). All secondary antibodies used were Alexa Fluor® highly crossed adsorbed (Molecular Probes). Samples were mounted in Vectashield with Dapi (Vector). Immunofluorescence signals were detected by an Olympus IX71 inverted microscope or by a BioRad (Hercules, CA) Radiance 2000 confocal microscope.
RT-PCR analysis
For Q-PCR analysis using fractionated cells, total RNA was extracted from sorted ESCs and EpiSCs by using Trizol (Sigma). cDNA was synthesized using Superscript II and oligo d(T) primer (Invitrogen), followed by Q-PCR reaction using Sybr green master mix (Qiagen) and specific primers. For Q-PCR using single cells, Single-cell cDNA preparation was performed as described in detail previously (Kurimoto et al., 2007). The putative number of transcripts were estimated by referring in vitro-transcribed control transcripts (Kurimoto et al., 2007). cDNAs was used for each 20ul Q-PCR reaction with SyberGreen Master Mix (Qiagen) and 1μM of each primer and was amplified under following condition: 95°C for 10min, and then 40 cycles of 95°C for 15sec and 60°C for 1min. Primer sequences used in this study are available on request.
CHIP analysis
FACS-sorted 106 ESCs or EpiSCs and 3×107 3T3 cells, as carrier cells, were fixed with 1% Formaldehyde, lysed and sonicated to obtain 200 to 500 bp DNA fragment conjugated with nucleosomes. The sonicated lysates were immunoprecipitated with rabbit polyclonal anti-acetyl H3K9 (Abcam), mouse monoclonal anti-trimethyl H3K4 (Abcam) or rabbit polyclonal anti-tri-methyl H3K27 (Abcam) antibodies that were in advance reacted with secondary antibodies conjugated with magnetic beads (Dynal). After incubation with each antibody for 1 day, immunoprecipitants were recovered and washed by using magnetic stand (Dynal). Then DNA fragments contained in the immunoprecipitants were purified by incubation with proteinaseK. The DNA fragments were subjected to Q-PCR. Whole cell lysates before incubation with antibodies were used as input. DNA fragments from whole lysates and immunoprecipitants were subject to pre-PCR at 15 cycles and 20 cycles, respectively. After purified the pre-PCR products by PCR purification kit, 1μl of the product was used for Q-PCR reaction 1μM of nested primers. Negative control samples using only secondary antibodies conjugated with magnetic beads showed similar levels of very low background or no detectable background. Primer sequences used in this study are available on request.
Bisulfite sequence analysis
Bisulphite sequence analysis of the genomic DNA isolated from FACS-sorted ESCs was carried out by EpiTect Bisulfite Kit (Qiagen). The primers sequences and PCR conditions for amplification of LINE1 and IAP sequences are previously described (Lane et al., 2003). The PCR products were cloned using pGEM-T Easy Vector System I (Promega) and were sequenced by Cogenics.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to T. Nakano for anti-PGC7, and P. Beverly for mouse anti-SSEA1. We thank C. Lee for technical support, C. Mummery for continuing support, and A. McLaren, B. Payer, P. Hajkova and S. Jeffries for useful comments on the manuscript. This work was supported by the Netherlands Organization for Scientific Research (NWO, TALENT 809.67.024) to S.C.S.L., and the Japan Society for Promotion of Science to K.H., The Technology Programme (DTI Project No: TP/4/BIO/6/I/22020) of CellCentric Ltd and The Technology Strategy Board sponsored by Department for Innovation,Universities and Skills (DIUS), and the Wellcome Trust (062801).
REFERENCES
- Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003;4:616–623. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chambers I, Smith A. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene. 2004;23:7150–7160. doi: 10.1038/sj.onc.1207930. [DOI] [PubMed] [Google Scholar]
- Chapman G, Remiszewski JL, Webb GC, Schulz TC, Bottema CD, Rathjen PD. The mouse homeobox gene, Gbx2: genomic organization and expression in pluripotent cells in vitro and in vivo. Genomics. 1997;46:223–233. doi: 10.1006/geno.1997.4969. [DOI] [PubMed] [Google Scholar]
- Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AT, Rodriguez RT, Bodnar MS, Abeyta MJ, Cedars MI, Turek PJ, Firpo MT, Reijo Pera RA. Human STELLAR, NANOG, and GDF3 genes are expressed in pluripotent cells and map to chromosome 12p13, a hotspot for teratocarcinoma. Stem Cells. 2004;22:169–179. doi: 10.1634/stemcells.22-2-169. [DOI] [PubMed] [Google Scholar]
- Cui L, Johkura K, Yue F, Ogiwara N, Okouchi Y, Asanuma K, Sasaki K. Spatial distribution and initial changes of SSEA-1 and other cell adhesion-related molecules on mouse embryonic stem cells before and during differentiation. J Histochem Cytochem. 2004;52:1447–1457. doi: 10.1369/jhc.3A6241.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa Lopes SM, Roelen BA, Monteiro RM, Emmens R, Lin HY, Li E, Lawson KA, Mummery CL. BMP signaling mediated by ALK2 in the visceral endoderm is necessary for the generation of primordial germ cells in the mouse embryo. Genes Dev. 2004;18:1838–1849. doi: 10.1101/gad.294004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa T, Ikeda M, Inoue F, Ohkoshi K, Hamano T, Tokunaga T. Gene expression profiling of mouse embryonic stem cell subpopulations. Biol Reprod. 2006;75:555–561. doi: 10.1095/biolreprod.105.049502. [DOI] [PubMed] [Google Scholar]
- Furusawa T, Ohkoshi K, Honda C, Takahashi S, Tokunaga T. Embryonic stem cells expressing both platelet endothelial cell adhesion molecule-1 and stage-specific embryonic antigen-1 differentiate predominantly into epiblast cells in a chimeric embryo. Biol Reprod. 2004;70:1452–1457. doi: 10.1095/biolreprod.103.024190. [DOI] [PubMed] [Google Scholar]
- Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, Cedar H, Razin A. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992;6:705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Shi Y, Xiang B, Qu B, Su W, Zhu M, Zhang M, Bao G, Wang F, Zhang X, et al. A nuclear function of beta-arrestin1 in GPCR signaling: regulation of histone acetylation and gene transcription. Cell. 2005;123:833–847. doi: 10.1016/j.cell.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Kemp C, Willems E, Abdo S, Lambiv L, Leyns L. Expression of all Wnt genes and their secreted antagonists during mouse blastocyst and postimplantation development. Dev Dyn. 2005;233:1064–1075. doi: 10.1002/dvdy.20408. [DOI] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Ono Y, Uno KD, Yamada RG, Ueda HR, Saitou M. An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic Acids Res. 2006;34:e42. doi: 10.1093/nar/gkl050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Saitou M. Global single-cell cDNA amplification to provide a template for representative high-density oligonucleotide microarray analysis. Nat Protoc. 2007;2:739–752. doi: 10.1038/nprot.2007.79. [DOI] [PubMed] [Google Scholar]
- Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- Lee JH, Hart SR, Skalnik DG. Histone deacetylase activity is required for embryonic stem cell differentiation. Genesis. 2004;38:32–38. doi: 10.1002/gene.10250. [DOI] [PubMed] [Google Scholar]
- Lei H, Oh SP, Okano M, Juttermann R, Goss KA, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- McCool KW, Xu X, Singer DB, Murdoch FE, Fritsch MK. The role of histone acetylation in regulating early gene expression patterns during early embryonic stem cell differentiation. J Biol Chem. 2007;282:6696–6706. doi: 10.1074/jbc.M609519200. [DOI] [PubMed] [Google Scholar]
- Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohinata Y, Payer B, O’Carroll D, Ancelin K, Ono Y, Sano M, Barton SC, Obukhanych T, Nussenzweig M, Tarakhovsky A, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Payer B, Chuva de Sousa Lopes SM, Barton SC, Lee C, Saitou M, Surani MA. Generation of stella-GFP transgenic mice: A novel tool to study germ cell development. Genesis. 2006;44:75–83. doi: 10.1002/gene.20187. [DOI] [PubMed] [Google Scholar]
- Pelton TA, Sharma S, Schulz TC, Rathjen J, Rathjen PD. Transient pluripotent cell populations during primitive ectoderm formation: correlation of in vivo and in vitro pluripotent cell development. J Cell Sci. 2002;115:329–339. doi: 10.1242/jcs.115.2.329. [DOI] [PubMed] [Google Scholar]
- Pesce M, Gross MK, Scholer HR. In line with our ancestors: Oct-4 and the mammalian germ. Bioessays. 1998;20:722–732. doi: 10.1002/(SICI)1521-1878(199809)20:9<722::AID-BIES5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Rathjen J, Lake JA, Bettess MD, Washington JM, Chapman G, Rathjen PD. Formation of a primitive ectoderm like cell population, EPL cells, from ES cells in response to biologically derived factors. J Cell Sci. 1999;112(Pt 5):601–612. doi: 10.1242/jcs.112.5.601. [DOI] [PubMed] [Google Scholar]
- Robson P, Stein P, Zhou B, Schultz RM, Baldwin HS. Inner cell mass-specific expression of a cell adhesion molecule (PECAM-1/CD31) in the mouse blastocyst. Dev Biol. 2001;234:317–329. doi: 10.1006/dbio.2001.0274. [DOI] [PubMed] [Google Scholar]
- Saitou M, Barton SC, Surani MA. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- Sato M, Kimura T, Kurokawa K, Fujita Y, Abe K, Masuhara M, Yasunaga T, Ryo A, Yamamoto M, Nakano T. Identification of PGC7, a new gene expressed specifically in preimplantation embryos and germ cells. Mech Dev. 2002;113:91–94. doi: 10.1016/s0925-4773(02)00002-3. [DOI] [PubMed] [Google Scholar]
- Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- van Grunsven LA, Verstappen G, Huylebroeck D, Verschueren K. Smads and chromatin modulation. Cytokine Growth Factor Rev. 2005;16:495–512. doi: 10.1016/j.cytogfr.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Zwaka TP, Thomson JA. A germ cell origin of embryonic stem cells? Development. 2005;132:227–233. doi: 10.1242/dev.01586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.