Abstract
Macrophages (Mϕ) play a central role as effector cells in immunity to intracellular pathogens such as Mycobacterium. Paradoxically, they also provide a habitat for intracellular bacterial survival. This paradoxical role of Mϕ remains poorly understood. Here we report that this dual role may emanate from the functional plasticity of Mϕ: Whereas Mϕ-1 polarized in the presence of granulocyte–Mϕ colony-stimulating factor promoted type 1 immunity, Mϕ-2 polarized with Mϕ colony-stimulating factor subverted type 1 immunity and thus may promote immune escape and chronic infection. Importantly, Mϕ-1 secreted high levels of IL-23 (p40/p19) but no IL-12 (p40/p35) after (myco)bacterial activation. In contrast, activated Mϕ-2 produced neither IL-23 nor IL-12 but predominantly secreted IL-10. Mϕ-1 required IFN-γ as a secondary signal to induce IL-12p35 gene transcription and IL-12 secretion. Activated dendritic cells produced both IL-12 and IL-23, but unlike Mϕ-1 they slightly reduced their IL-23 secretion after addition of IFN-γ. Binding, uptake, and outgrowth of a mycobacterial reporter strain was supported by both Mϕ subsets, but more efficiently by Mϕ-2 than Mϕ-1. Whereas Mϕ-1 efficiently stimulated type 1 helper cells, Mϕ-2 only poorly supported type 1 helper function. Accordingly, activated Mϕ-2 but not Mϕ-1 down-modulated their antigen-presenting and costimulatory molecules (HLA-DR, CD86, and CD40). These findings indicate that (i)Mϕ-1 and Mϕ-2 play opposing roles in cellular immunity and (ii) IL-23 rather than IL-12 is the primary type 1 cytokine produced by activated proinflammatory Mϕ-1. Mϕ heterogeneity thus may be an important determinant of immunity and disease outcome in intracellular bacterial infection.
Mycobacteria can infect human macrophages (Mϕ) and cause serious chronic infectious diseases such as tuberculosis and leprosy. Mϕ play a crucial role in human host defense by secreting cytokines and chemokines, presenting antigen to T lymphocytes and clearing infectious agents. Type 1 cell-mediated immunity is required for granuloma formation and effective host defense against intracellular pathogens (1), but mycobacteria are able to escape immunity and persist in a nonreplicating state inside Mϕ for many years (2). The molecular and cellular mechanisms that underlie the development of effective immunity versus latent infection (or immune escape) and the induction of immunopathology after Mycobacterium tuberculosis infection, however, remain poorly understood.
Mononuclear phagocytes including Mϕ are activated through ligation of pattern-recognition receptors such as Toll-like receptors (TLRs) by microbial ligands (3, 4), which is generally considered to potentiate the production of the type 1 cytokine IL-12. IL-12 is a heterodimer of p40 and p35 that drives polarization of naive T cells toward type 1 helper (Th1) cells and induces the release of IFN-γ from T and natural killer cells (5). IFN-γ, in turn, activates Mϕ and enhances cytokine secretion, antigen presentation and, supposedly, the bactericidal activity of Mϕ (6). The IL-12/IFN-γ axis is critical indeed for the establishment of effective host defense against intracellular pathogens: We and others have reported that human genetic deficiencies in this type 1 cytokine signaling cascade (affecting IL-12p40, IL-12R, IFN-γR, or STAT-1) lead to increased susceptibility to otherwise weakly pathogenic mycobacteria and salmonellae (reviewed in ref. 1).
Dendritic cells (DCs) are highly potent phagocytes that prime naive T cells and control the development of Th1 cells (7, 8). The roleofMϕ in Th1-mediated immunity, however, is less clear. It has been reported that Mϕ fail to release IL-12 (p40/p35) heterodimer after mycobacterial stimulation (9, 10). Recently, IL-23 was identified as an IL-12-like heterodimer, consisting of IL-12p40 and a novel p19 chain. IL-23, similar to IL-12, induces IFN-γ secretion from T cells (11) and may be involved in type 1 immune defense against mycobacteria (1, 12).
Beside the classical route of Mϕ activation, it has become evident in recent years that alternative activation modes of Mϕ can be distinguished (for recent reviews see refs. 13 and 14). Whereas classical activation of Mϕ by microbial compounds yields a phenotype that is hallmarked by the production of proinflammatory cytokines, alternative activation can lead to an antiinflammatory phenotype, hallmarked by IL-10 as the signature cytokine. Non-classical antiinflammatory Mϕ may also evolve by natural neuroendocrine control mechanisms and play a role in homeostatic processes such as dampening inflammation, scavenging debris, angiogenesis, and wound healing (but also tumor outgrowth) (14–16). The characterization of functional human Mϕ profiles thus far is incomplete, and the role of Mϕ in immunity to mycobacteria remains elusive.
Here we show that highly pure subsets of Mϕ with polarized proand antiinflammatory phenotypes can be obtained by differentiating human blood monocytes in the presence of the lineage-determining cytokines granulocyte–Mϕ colony-stimulating factor (GM-CSF) and Mϕ CSF (M-CSF), respectively. Pro- and antiinflammatory Mϕ, designated Mϕ-1 and Mϕ-2, respectively, were characterized for their potential to produce IL-12 and IL-23 to support intracellular mycobacterial growth and present antigens to Th1 lymphocytes. Both Mϕ subsets supported the outgrowth of a mycobacterial reporter strain in the absence of any other immune components, but our findings also indicate that Mϕ-1 promote whereas Mϕ-2 subvert type 1 immunity in the face of (myco)bacterial infection. These results have important implications for our understanding of Mϕ biology in cell-mediated immunity to intracellular infections.
Materials and Methods
Cells and Microbial Reagents. Monocytes were isolated to high purity by magnetic cell sorting using anti-CD14-coated beads (per manufacturer recommendations, Miltenyi Biotec, Auburn, CA) and subsequently cultured for 6 days in medium (RPMI medium 1640, GIBCO/Invitrogen) with 10% FCS (HyClone) and either 50 units/ml recombinant human GM-CSF (Novartis Pharma, Arnhem, The Netherlands) to generate Mϕ-1 or 50 ng/ml recombinant human M-CSF (R & D Systems) to generate Mϕ-2. As a control, DCs [monocyte-derived DCs (mo.DCs)] were generated with 1,000 units/ml GM-CSF and 500 units/ml IL-4 as described (17).
Lipopolysaccharide (LPS) from Escherichia coli (serotype 055:B5, Sigma–Aldrich) was used to stimulate Mϕ and DCs at 10 ng/ml (unless indicated otherwise). Mycobacterial lysate was obtained by ultrasonication of heat-inactivated M. tuberculosis H37Rv, lyophilized, and resuspended in PBS as described (18). The lysate was quantified on the basis of bacterial dry weight; cells were stimulated with 10 μg/ml unless indicated otherwise.
Cell Cytometry. To analyze cell surface marker expression, aliquots of 105 Mϕ were stained for 30 min at 4°C by using appropriate isotype controls and phycoerythrin-conjugated anti-CD14, anti-CD1a, anti-CD83, anti-HLA-DR, anti-CD80, anti-CD86, and anti-CD40 (BD Biosciences/Pharmingen). Samples were analyzed on a FACSCalibur using cellquest software (BD Biosciences).
Cytokine and Chemokine Measurements. IFN-γ secretion was quantified by ELISA (U-CyTech, Utrecht, The Netherlands) with a sensitivity of 20 pg/ml. Specific ELISAs for IL-12p40 and IL-10 were purchased from BioSource International (Camarillo, CA; sensitivity: 20 pg/ml), and IL-12p40/p35 heterodimer was measured by using the cytometric bead assay (BD Biosciences/Pharmingen; sensitivity: 40 pg/ml). IL-23 was measured by ELISA using anti-IL-12p40 monoclonal antibody BP40 (Diaclone) for coating and rat-anti-hp19 monoclonal antibody 12F12 for detection (with a sensitivity of 60 pg/ml).
Polarized Mϕ were harvested by using trypsin-EDTA in Hanks' balanced salt solution without Ca/Mg (GIBCO/Invitrogen). DCs and Mϕ were washed, counted, and seeded in triplicate at 105 cells per 200 μl in 96-well flat-bottom culture plates (Corning Life Sciences) in the presence or absence of stimulating agents as indicated. Supernatants were collected after 24 h (unless indicated otherwise).
mRNA levels of IL-12p40, IL-12p35, and IL-23p19 were measured by real-time quantitative PCR 8 h after activation. RNA was extracted by using an RNeasy kit (Qiagen, Valencia, CA) according to manufacturer protocol and reverse-transcribed with oligo(dT)14–18 (Life Technologies) and random hexamer primers (Promega) by using standard protocols. cDNA was analyzed by PCR with a Perkin–Elmer ABI Prism 7700 sequence-detection system. Gene expression was quantified by correcting for the relative expression to 18S rRNA levels (19).
Mycobacterial Binding, Uptake, and Survival Assays. Mycobacterial binding and uptake were measured by incubating Mϕ at 0 and 37°C with chemically killed (streptomycin/amikacin), GFP-tagged Mycobacterium bovis bacillus Calmette–Guérin for 20 min. Mϕ were washed and analyzed by flow cytometry. Intracellular mycobacterial survival was measured by using luciferase-transfected M. bovis bacillus Calmette–Guérin as a reporter strain (20). Transfected M. bovis bacillus Calmette–Guérin was a generous gift from D. Young (Imperial College, London, U.K.). Briefly, polarized Mϕ were harvested, counted, and seeded at 105 cells per 100 μl per well in 96-well plates in culture medium. After overnight incubation of the Mϕ, 100 μl of log-phase M. bovis bacillus Calmette–Guérin were added in tissue-culture medium at the concentration indicated. After 2–4 h of infection, Mϕ were washed twice with warm medium and incubated in a final volume of 200 μl. After 6 days, cells were washed, and luciferase activity was measured after adding Triton X-100 for 10 min [25 μl of a 10% (vol/vol) solution] by the conversion of n-decyl aldehyde [25 μl of a 1% (wt/vol) solution in ethanol]. Luminescence was measured in a Victor2 multilabel plate reader (Perkin–Elmer) equipped with an automatic injector. Host cell viability was determined with a fluorescent calcein viability stain (Molecular Probes). To this end, Mϕ were washed and incubated for 30 min at room temperature in the dark with 50 μl of 1 mM calcein in anhydrous DMSO (Sigma–Aldrich). Subsequently, the cells were washed twice with PBS, resuspended in 100 μl of 1% Triton X-100, and measured in the multilabel plate reader.
T Cell Activation Assays. Antigen presentation by Mϕ and DCs was determined by measuring proliferation and IFN-γ secretion of the HLA-DR2-restricted CD4+ T cell clone R2F10 that is specific for the 60-kDa heat-shock protein (amino acids 418–427) of Mycobacterium leprae (21) or the HLA-DR1-restricted CD4+ T cell clone HA1.7 that is specific for the hemagglutinin (amino acids 306–318) of influenza virus (22). Briefly, antigen-presenting cells were harvested and seeded in triplicates at 2.5 × 103 cells per well in 96-well flat-bottom culture plates in 100 μl in the presence or absence of LPS or mycobacterial sonicate for 24 h. Subsequently, 104 R2F10 cells and recombinant protein or synthetic peptide were added to a final concentration of 10 μg/ml and 100 ng/ml, respectively, in a final volume of 200 μl and incubated for another 72 h. Aliquots of 50 μl of supernatant were harvested and pooled per triplicate to measure IFN-γ secretion (see above), and 0.5 μCi of [3H]thymidine (1 Ci = 37 GBq) was added for another 18 h to measure T cell proliferation.
Results
Polarization of Monocytes into Type 1 and Type 2 Mϕ Subsets. Highly pure proinflammatory (IL-12p40+) Mϕ-1 were obtained after culturing CD14+ human blood monocytes for 6 days in the presence of recombinant human GM-CSF, and antiinflammatory (IL-10+) Mϕ-2 were obtained by using M-CSF. Both procedures yielded CD14+CD1a– cells (Fig. 1A) that were predominantly adherent and had an apparent Mϕ morphology as judged by microscopy (data not shown). For control purposes, from the same donors CD14–CD1a+CD83+ DCs (mo.DCs) were generated (Fig. 1A). Fig. 1B illustrates that Mϕ-1, similar to mo.DCs, secreted high levels of IL-12p40 after activation with either the TLR4-agonist LPS or a sonicate of heat-killed M. tuberculosis that stimulates via TLR2 (23). Although Mϕ-1 and Mϕ-2 showed similar levels of (IFN-γ-induced) TLR2 and TLR4 expression (data not shown), activated Mϕ-2 did not secrete IL-12p40 (Fig. 1B) but produced high levels of IL-10 in comparison with Mϕ-1 or mo.DCs (Fig. 1C). These signature cytokine profiles were found consistently in >10 independent experiments using cells from different donors. Moreover, Mϕ-1 but not Mϕ-2 produced (high levels of) IL-18, tumor necrosis factor-α, IL-6, and IL-1β (F.A.W.V., unpublished observations).
Fig. 1.
Differentiation of human blood monocytes into functionally distinct Mϕ subsets, Mϕ-1 and Mϕ-2. (A) In contrast to mo.DCs, Mϕ-1 and Mϕ-2 highly expressed CD14 but showed no or only weak expression of CD1a or CD83 (the latter after activation by LPS) as determined by flow cytometry. Cytokine secretion was measured up to 72 h after stimulation with M. tuberculosis sonicate (myc) (▪), LPS (•), or medium control (♦). (B) Whereas activated Mϕ-1, similar to DCs, secreted high levels of IL-12p40, Mϕ-2 failed to secrete IL-12p40. (C) IL-10, in contrast, was most predominant in Mϕ-2. Similar cytokine profiles were obtained with cells from at least 10 independent donors.
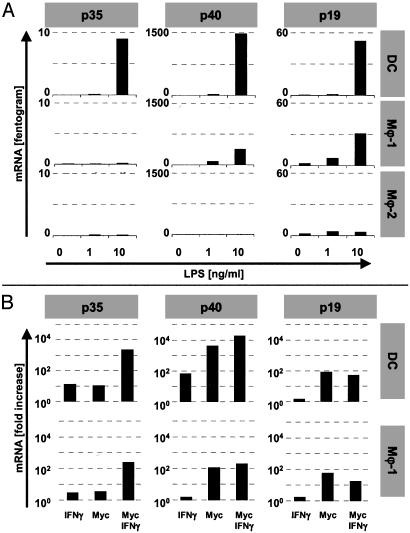
Regulation of IL-12 and IL-23 Production by Mϕ and DCs. Because IL-12p40 can pair with either p35 or p19 to form IL-12 (p40/p35) and IL-23 (p40/p19), respectively, we studied the capacity of Mϕ and DCs to produce these cytokines. Fig. 2 shows that Mϕ-1 and DCs required exogenous IFN-γ to secrete high levels of IL-12 in response to mycobacteria or LPS. Accordingly, IFN-γ was required to induce strong IL-12p35 gene transcription (Fig. 3B). In the absence of IFN-γ, only LPS-activated mo.DCs but not Mϕ-1 produced low levels of IL-12, concordant with the low level of LPS-induced p35 mRNA in these cells (Fig. 3A).
Fig. 2.
Type 1 cytokine secretion by monocyte-derived Mϕ and mo.DCs. The capacity of mo.DCs, Mϕ-1, and Mϕ-2 to secrete IL-12 (p40/p35) and IL-23 (p40/p19) proteins was determined 24 h after microbial stimulation in the absence (black bars) or presence (gray bars) of 500 units/ml IFN-γ. Mϕ-1 secreted IL-23 but failed to produce IL-12 after activation with mycobacterial sonicate (10 μg/ml) or LPS (10 ng/ml) unless IFN-γ was added. Also, DCs showed IFN-γ-enhanced IL-12 secretion, but unlike Mϕ-1 they yielded decreased rather than elevated IL-23 levels with IFN-γ. Mϕ-2 failed to produce IL-12 or IL-23. Depicted are average protein levels plus standard deviation (n = 3; n = 5 for IL-12 in DCs and Mϕ-1).
Fig. 3.
Type 1 cytokine mRNA levels in monocyte-derived Mϕ and mo.DCs. (A) Quantitative reverse transcription–PCR analyses 8 h after LPS stimulation of Mϕ-1 revealed that expression of IL-12-specific p35, but not p40 or p19 mRNA, required costimulation with IFN-γ (500 units/ml). DCs showed low but significant transcription of p35 in response to LPS and profound levels of p40 and p19 mRNA. Mϕ-2 failed to induce any of these cytokine transcripts. (B) Whereas stimulation with 10 μg/ml mycobacterial sonicate strongly induced the production of p40 and p19 mRNA, high-level transcription of p35 required costimulation with IFN-γ. (Note that the y axes are logarithmic in B!) Stimulation indexes were calculated from the average level of mRNA from two (DCs) to four (Mϕ-1) experiments. Activation-induced p19 transcription was inhibited by IFN-γ in both mo.DCs and Mϕ-1.
In contrast to IL-12, however, IL-23 was secreted by Mϕ-1 as well as mo.DCs in response to both LPS and mycobacteria without requiring exogenous IFN-γ (Fig. 2). These findings were supported by the induction of p40 and p19 mRNA in activated Mϕ-1 and mo.DCs (Fig. 3). The highest levels of IL-23 were reproducibly secreted from Mycobacterium-activated Mϕ-1. Together, mycobacterial activation of Mϕ-1 initially results in the induction of IL-23, whereas IL-12 production from these cells requires IFN-γ as an essential second signal. Remarkably, the addition of IFN-γ mildly but reproducibly suppressed the production of IL-23 by DCs, whereas it strongly enhanced IL-23 secretion by Mϕ-1 (Fig. 2). Although IFN-γ enhanced the transcription of p40 mRNA, activation-induced p19 transcription was slightly decreased after the addition of IFN-γ both in DCs and Mϕ-1 (Fig. 3B). This paradoxical finding of IFN-γ-mediated decrease of p19 gene transcription (at t = 8 h) and elevation of IL-23 secretion (at t = 24 h) in Mϕ-1 suggests an as-yet-unknown posttranscriptional mechanism that affects IL-23 production by Mϕ-1 differently than in DCs.
In agreement with the lack of IL-12p40 production, activated Mϕ-2 completely failed to secrete IL-12 and IL-23 and failed to induce p35, p40, or p19 mRNA (Figs. 2 and 3A).
Binding, Uptake, and Outgrowth of Mycobacteria Infecting Mϕ-1 and Mϕ-2. To compare binding and/or uptake of mycobacteria by Mϕ-1 versus Mϕ-2, cells were infected with M. bovis bacillus Calmette–Guérin-GFP and analyzed by flow cytometry. Within 20 min at 0°C, ≈4-fold more Mϕ-2 acquired mycobacteria as compared with Mϕ-1 (Fig. 4A). Similarly, metabolically active Mϕ-2 at 37°C showed at least 2-fold better binding and/or uptake of fluorescent M. bovis bacillus Calmette–Guérin (Fig. 4A).
Fig. 4.
Binding, uptake, and outgrowth of mycobacteria by Mϕ-1 and Mϕ-2. (A) Binding (at 0°C) and/or uptake (at 37°C) of an M. bovis bacillus Calmette–Guérin-GFP reporter strain was determined by flow cytometry. After 20 min, more Mϕ-2 than Mϕ-1 had acquired fluorescent mycobacteria (representative of three independent experiments). (B) Accordingly, over a wide range of multiplicities of infection (MOI) from 20 to 2.5 mycobacteria per host cell, Mϕ-2 displayed enhanced outgrowth of M. bovis bacillus Calmette–Guérin-lux at day 6 compared with Mϕ-1. Depicted are averaged luminescence signals as cps [+ standard deviation (n = 4)]. Similar results were found with cells from different donors in four independent experiments.
To study the capacity of mycobacteria to survive within Mϕ-1 versus Mϕ-2, we used a luciferase-transfected M. bovis bacillus Calmette–Guérin reporter strain (bacillus Calmette–Guérin-lux). As reported previously (20, 24), the luciferase activity of bacillus Calmette–Guérin-lux correlated well with bacterial viability as determined by classical colony-forming unit counting (data not shown). In accordance with the higher capacity to bind and/or phagocytose mycobacteria, we found reproducibly enhanced outgrowth of mycobacteria at day 6 after infection in Mϕ-2 over Mϕ-1 over a range of 20–2.5 infecting mycobacteria per host cell (Fig. 4B). The condition of the infected Mϕ-1 and Mϕ-2 as judged by light microscopy revealed no significant differences during the incubation period. This was corroborated further by calcein viability staining of the host cells [Mϕ-1: 31,977 (±3,449) cps; Mϕ-2: 28,936 (±3,656) cps (n = 3)]. Notably, the predominant cytokines in the supernatants of Mϕ-1 and Mϕ-2 at the end of the infection period remained IL-12p40 and IL-10, respectively (data not shown), suggesting that Mϕ-1 and Mϕ-2 represent stable subsets in the Mϕ spectrum.
Mϕ-1 but Not Mϕ-2 Support Th1 Function After Mycobacterial Stimulation. To address the antigen-presenting capacity of Mϕ-1 and Mϕ-2, we used an HLA-DR2-restricted Th1 reporter clone (R2F10) that recognizes a M. leprae-specific HSP60 epitope that is lacking from M. tuberculosis. Fig. 5A shows that Mϕ-1 and mo.DCs both were effective in inducing proliferation of R2F10 when presenting M. leprae HSP60 protein or peptide antigen. Proliferation of R2F10 Th1 cells induced by Mϕ-1 was not significantly affected by preactivation of Mϕ-1 with M. tuberculosis sonicate before adding specific antigen (Fig. 5A). In contrast to Mϕ-1, however, Mϕ-2 stimulated R2F10 relatively poorly, and both protein- and peptide-mediated T cell activation were reduced further when Mϕ-2 had been preactivated by mycobacterial stimulation (Fig. 5A). Similar poor antigen presentation by Mϕ-2 was observed when LPS was used to preactivate the cells (Fig. 5B), indicating that the subversion of Th1 cell activation by Mϕ-2 was not unique to M. tuberculosis activation. Similar results were obtained with the HLA-DR1-restricted, influenza hemagglutinin-specific T cell clone HA1.7, as illustrated by the dose-dependent reduction in the proliferation of these cells induced by Mϕ-2 that had been preactivated with increasing doses of M. tuberculosis sonicate (Fig. 5C). Fig. 5D shows that (preactivated) Mϕ-1, albeit less efficiently than mo.DCs, supported antigen-specific IFN-γ secretion by R2F10 cells. In contrast, IFN-γ secretion was poorly supported by Mϕ-2, and preactivation further reduced this ability (Fig. 5D). In accordance with their poor antigen-presenting capacity, activated Mϕ-2 but not Mϕ-1 or mo.DCs down-regulated the expression of HLA-DR and CD86 (but not CD80; Fig. 6) as well as CD40 (data not shown).
Fig. 5.
Antigen-presenting capacity of Mϕ-1 versus Mϕ-2. To determine the antigen-presenting capacity of Mϕ-1 and Mϕ-2, cells were pretreated (black bars) or not (gray bars) for 24 h with a (myco)bacterial stimulus before incubation with antigen and antigen-specific Th1 cells. Mo.DCs were included for control purposes. (A) In contrast to Mϕ-1 and mo.DCs, Mϕ-2 relatively poorly supported proliferation of the M. leprae-specific Th1 clone R2F10 toward protein or peptide antigen, which was reduced further after activation of Mϕ-2 by M. tuberculosis sonicate (myc). (B) Similar results were obtained when antigen-presenting cells were pretreated with LPS. (C) The antigen-presenting capacity of Mϕ-2 toward the influenza hemagglutinin-specific T cell clone HA1.7 was reduced also by mycobacterial stimulation in a dose-dependent fashion. (D) Protein or peptide antigen-specific IFN-γ secretion of R2F10 Th1 cells was supported by Mϕ-1, albeit less efficiently than by mo.DCs. IFN-γ secretion by R2F10 cells responding to Mϕ-2 was substantially lower and reduced further by mycobacterial activation of Mϕ-2. IFN-γ production is depicted as secreted protein in the pooled supernatant of triplicate cultures. Proliferation is depicted as the average incorporation of [3H]thymidine in triplicate cultures (cpm; + standard deviation). Experiments were repeated at least twice using independent donors to generate Mϕ and DCs.
Fig. 6.
Expression of HLA and costimulatory molecules on Mϕ-1 and Mϕ-2. Mϕ were pretreated (gray bars) or not (black bars) with mycobacterial sonicate for 48 h before flow-cytometric analysis of HLA-DR, CD80, and CD86 expression. Mϕ-2 but not Mϕ-1 or mo.DCs down-regulated their cell surface levels of HLA-DR and the costimulatory molecule CD86 (but not CD80) after activation. Depicted are the mean fluorescent intensities (mfi; + standard deviation) of cells from three independent donors.
Thus, although Mϕ-1 (and mo.DCs) promote activation of Th1 cells, Mϕ-2 fail to promote Th1-mediated immunity efficiently after activation by microbial components.
Discussion
Mϕ are the major population of tissue-resident mononuclear phagocytes and the predominant targets for infection by intracellular pathogens including mycobacteria. Mϕ play a dual role in antimycobacterial host defense that currently is poorly understood: They contribute to cell-mediated immunity and bacterial elimination but also provide an essential niche for intracellular bacterial survival and escape from host defense mechanisms. Here we identify two distinct human Mϕ subsets, Mϕ-1 and Mϕ-2, that display largely opposite functions. Although both Mϕ populations can be infected and support the outgrowth of mycobacteria (in the absence of any other immune components), Mϕ-1 promote type 1 cellular immunity, whereas Mϕ-2 are poor antigen-presenting cells for supporting type 1 immunity. Mycobacterium-activated Mϕ-1 secrete IL-23 (p40/p19) but no IL-12 (p40/p35), whereas activated Mϕ-2 fail to produce IL-23 or IL-12 and predominantly secrete IL-10.
It is well established that type 1 cell-mediated immunity is essential for optimal host defense against intracellular pathogens (1, 6, 25), but it is unresolved how (different functional) Mϕ (subsets) contribute to type 1 immunity in antimycobacterial host defense. In the present study, we generated proinflammatory Mϕ-1 using the lineage-determining cytokine GM-CSF, which is associated with inflammation. These Mϕ-1 efficiently supported the antigen-specific function of Th1 cells. Importantly, our findings indicate that these Mϕ-1 initially secrete IL-23 (p40/p19) but no IL-12 (p40/p35) after mycobacterial stimulation. Similar to that for monocytes and mo.DCs (26–28), enhancement of IL-12 production after microbial stimulation of Mϕ-1 required IFN-γ, which activates transcription of the IL-12p35 gene. This regulation of IL-12 production may reflect an important function of IFN-γ in enhancing type 1 cellular immunity against intracellular pathogens. Although activation of both DCs and Mϕ-1 in the presence of IFN-γ reduced the induction of IL-23-specific p19 mRNA, only DCs reduced IL-23 protein levels under these conditions. Mϕ-1, in contrast, enhanced IL-23 secretion after (myco)bacterial stimulation in the presence of IFN-γ. Additional studies are required to unravel the (posttranscriptional) mechanism that is responsible for this differential regulation of IL-23 secretion between mo.DCs and Mϕ-1. Although we confirm previous studies that were unable to detect IL-12 heterodimer (9, 10), our study now demonstrates that mycobacterial stimulation of human proinflammatory Mϕ triggers IL-23 secretion.
The finding that IL-23 rather than IL-12 is the initial type 1 cytokine released by activated proinflammatory Mϕ may point toward an important role for IL-23 in type 1 immunity and anti(myco)bacterial host defense. Because IL-23 has been shown to induce IFN-γ in memory T cells (11), Mϕ-derived IL-23 may play a significant role in immunological memory and/or the effector phase of T cell-dependent immunity toward mycobacteria. Murine knock-out models in which either IL-12 or both IL-12 and IL-23 signaling were abrogated indeed have suggested a significant role for IL-23 in host resistance to intracellular pathogens including mycobacteria (12, 29). Interestingly, IL-23 rather than IL-12 was reported to be critical for inflammation in mice (30–32). In support of its unique function, IL-23 rather than IL-12 stimulates mouse T cells to produce GM-CSF (33). Thus, by enhancing GM-CSF production, IL-23 may drive differentiation of newly recruited monocytes to a proinflammatory Mϕ phenotype.
Over the last few years it has become clear that Mϕ are highly heterogeneous, and nonclassical antiinflammatory subsets have been identified (13, 14). Our results show that human Mϕ polarized by M-CSF stably display such a nonclassical phenotype, secreting IL-10 but no IL-12(p40) in response to (myco)bacteria. The lack of type 1 cytokine secretion, the high production of IL-10, and the profound down-regulation of HLA, CD86, and CD40 by activated Mϕ-2 are likely to contribute to their poor Th1-activating capacity (34–37). Mϕ-2 expressed similar levels of TLR4 and TLR2 as Mϕ-1, suggesting that the functional differentiation between these two subsets emanates from differential signaling or gene-expression profiles rather than from divergent patterns of innate immune recognition. The enhanced binding and uptake of mycobacteria by Mϕ-2 compared with Mϕ-1 (already at 0°C) suggests differential expression of a cell surface receptor and also fits with the notion that nonclassical Mϕ display enhanced endocytosis (15). This may account for the elevated outgrowth of M. bovis bacillus Calmette–Guérin in Mϕ-2.
Gordon and coworkers (13, 38) have described alternatively activated Mϕ after treatment with IL-4 or IL-13, which in contrast to Mϕ-2 produce IL-10 without microbial stimulation. Also unlike alternatively activated Mϕ, activated Mϕ-2 failed to release Mϕ-derived chemokine (MDC/CCL22) or thymus- and activation-regulated chemokine (TARC/CCL17) (unpublished observations). However, activated Mϕ-2 readily produced other chemokines (e.g., MCP-1/CCL2, IP-10/CXCL10, and MIP-1β/CLL4; unpublished data), suggesting that Mϕ-2 can attract and regulate other immune cells such as monocytes and lymphocytes. The term “type2Mϕ” has been used to describe IL-12–IL-10+ Mϕ in the mouse, obtained by stimulation through TLR, CD40, or CD44 ligation in the presence of FcγR-ligating immune complexes (39). Additional analyses should reveal how these type 2 Mϕ relate to the human Mϕ-2 in this study.
A murine Leishmania infection model has indicated that IL-10 plays a central role in the maintenance of latent infection by intracellular pathogens (40). Moreover, IL-10-deficient mice display increased antimycobacterial immunity with concordant higher levels of tumor necrosis factor-α and lower bacterial burden (41). Therefore, IL-10 produced by activated Mϕ-2 may inhibit optimal host defense and promote latent infection and immune escape by mycobacteria. M-CSF used to generate Mϕ-2 is a ubiquitous serum protein, which may indicate that under normal homeostatic conditions Mϕ are switched to an antiinflammatory mode. M-CSF-treated Mϕ have been described to induce T cell hyporesponsiveness in an indoleamine 2,3-dehydrogenase-dependent fashion and have been implicated in the maintenance of peripheral tolerance (42–44). Furthermore, MCP-1, which is strongly secreted by Mϕ-2 (see above) and up-regulated by IL-10, stimulates type 2 helper polarization, and overexpression of MCP-1 in mice increased their susceptibility to infection by M. tuberculosis (45–47). Altogether, whereas Mϕ-1 promote cell-mediated immunity, Mϕ-2 seem to down-regulate type 1 cell-mediated immunity by various mechanisms and may promote chronic mycobacterial infection.
Intracellular pathogens generally interact with and modulate their host cells, and M. tuberculosis has an intrinsic capacity to interfere with the classical IFN-γ- and TLR-mediated activation pathways of Mϕ (48, 49). Our findings imply that not only active interference of pathogens with Mϕ signaling pathways but also the plasticity in the human Mϕ compartment may critically affect host defense against intracellular infections. Beside providing a rationale for the paradoxical role of Mϕ in combating intracellular pathogens while also providing a niche that is sequestered from immunosurveillance, our study also provides a model for additional investigation of Mϕ plasticity and Mϕ-derived IL-23 during inflammation, protective immunity, and immunopathology in mycobacterial infections and immunological diseases in general.
Acknowledgments
We thank Drs. F. Koning and D. Roelen for critically reading the manuscript. Our work was supported by the Netherlands Leprosy Foundation, the Netherlands Organization for Scientific Research, the Commission of the European Community, and the Royal Netherlands Academy of Arts and Sciences.
Abbreviations: Mϕ, macrophage(s); TLR, Toll-like receptor; Th1, type 1 helper; DC, dendritic cell; M-CSF, Mϕ colony-stimulating factor; GM-CSF, granulocyte–Mϕ CSF; mo.DC, monocyte-derived DC; LPS, lipopolysaccharide.
References
- 1.Ottenhoff, T. H., Verreck, F. A., Lichtenauer-Kaligis, E. G., Hoeve, M. A., Sanal, O. & Van Dissel, J. T. (2002) Nat. Genet. 32, 97–105. [DOI] [PubMed] [Google Scholar]
- 2.Kaufmann, S. H. (2001) Nat. Rev. Immunol. 1, 20–30. [DOI] [PubMed] [Google Scholar]
- 3.Janeway, C. A., Jr., & Medzhitov, R. (2002) Annu. Rev. Immunol. 20, 197–216. [DOI] [PubMed] [Google Scholar]
- 4.Sieling, P. A. & Modlin, R. L. (2002) Curr. Opin. Microbiol. 5, 70–75. [DOI] [PubMed] [Google Scholar]
- 5.Trinchieri, G. (1995) Annu. Rev. Immunol. 13, 251–276. [DOI] [PubMed] [Google Scholar]
- 6.Raupach, B. & Kaufmann, S. H. (2001) Curr. Opin. Immunol. 13, 417–428. [DOI] [PubMed] [Google Scholar]
- 7.Kalinski, P., Hilkens, C. M., Wierenga, E. A. & Kapsenberg, M. L. (1999) Immunol. Today 20, 561–567. [DOI] [PubMed] [Google Scholar]
- 8.Reid, S. D., Penna, G. & Adorini, L. (2000) Curr. Opin. Immunol. 12, 114–121. [DOI] [PubMed] [Google Scholar]
- 9.Giacomini, E., Iona, E., Ferroni, L., Miettinen, M., Fattorini, L., Orefici, G., Julkunen, I. & Coccia, E. M. (2001) J. Immunol. 166, 7033–7041. [DOI] [PubMed] [Google Scholar]
- 10.Hickman, S. P., Chan, J. & Salgame, P. (2002) J. Immunol. 168, 4636–4642. [DOI] [PubMed] [Google Scholar]
- 11.Oppmann, B., Lesley, R., Blom, B., Timans, J. C., Xu, Y., Hunte, B., Vega, F., Yu, N., Wang, J., Singh, K., et al. (2000) Immunity 13, 715–725. [DOI] [PubMed] [Google Scholar]
- 12.Cooper, A. M., Kipnis, A., Turner, J., Magram, J., Ferrante, J. & Orme, I. M. (2002) J. Immunol. 168, 1322–1327. [DOI] [PubMed] [Google Scholar]
- 13.Gordon, S. (2003) Nat. Rev. Immunol. 3, 23–35. [DOI] [PubMed] [Google Scholar]
- 14.Mosser, D. M. (2003) J. Leukocyte Biol. 73, 209–212. [DOI] [PubMed] [Google Scholar]
- 15.Goerdt, S. & Orfanos, C. E. (1999) Immunity 10, 137–142. [DOI] [PubMed] [Google Scholar]
- 16.Mantovani, A., Sozzani, S., Locati, M., Allavena, P. & Sica, A. (2002) Trends Immunol. 23, 549–555. [DOI] [PubMed] [Google Scholar]
- 17.Sallusto, F. & Lanzavecchia, A. (1994) J. Exp. Med. 179, 1109–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verbon, A., Kuijper, S., Jansen, H. M., Speelman, P. & Kolk, A. H. (1990) J. Gen. Microbiol. 136, 955–964. [DOI] [PubMed] [Google Scholar]
- 19.Scheerens, H., Hessel, E., Waal-Malefyt, R., Leach, M. W. & Rennick, D. (2001) Eur. J. Immunol. 31, 1465–1474. [DOI] [PubMed] [Google Scholar]
- 20.Snewin, V. A., Gares, M. P., Gaora, P. O., Hasan, Z., Brown, I. N. & Young, D. B. (1999) Infect. Immun. 67, 4586–4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ottenhoff, T. H., Klatser, P. R., Ivanyi, J., Elferink, D. G., de Wit, M. Y. & de Vries, R. R. (1986) Nature 319, 66–68. [DOI] [PubMed] [Google Scholar]
- 22.Lamb, J. R., Eckels, D. D., Lake, P., Woody, J. N. & Green, N. (1982) Nature 300, 66–69. [DOI] [PubMed] [Google Scholar]
- 23.Means, T. K., Wang, S., Lien, E., Yoshimura, A., Golenbock, D. T. & Fenton, M. J. (1999) J. Immunol. 163, 3920–3927. [PubMed] [Google Scholar]
- 24.Boechat, N., Bouchonnet, F., Bonay, M., Grodet, A., Pelicic, V., Gicquel, B. & Hance, A. J. (2001) J. Immunol. 166, 6203–6211. [DOI] [PubMed] [Google Scholar]
- 25.Flynn, J. L. & Chan, J. (2001) Annu. Rev. Immunol. 19, 93–129. [DOI] [PubMed] [Google Scholar]
- 26.Snijders, A., Hilkens, C. M., van der Pouw Kraan, T. C., Engel, M., Aarden, L. A. & Kapsenberg, M. L. (1996) J. Immunol. 156, 1207–1212. [PubMed] [Google Scholar]
- 27.Hayes, M. P., Wang, J. & Norcross, M. A. (1995) Blood 86, 646–650. [PubMed] [Google Scholar]
- 28.Trinchieri, G. (2003) Nat. Rev. Immunol. 3, 133–146. [DOI] [PubMed] [Google Scholar]
- 29.Decken, K., Kohler, G., Palmer-Lehmann, K., Wunderlin, A., Mattner, F., Magram, J., Gately, M. K. & Alber, G. (1998) Infect. Immun. 66, 4994–5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiekowski, M. T., Leach, M. W., Evans, E. W., Sullivan, L., Chen, S. C., Vassileva, G., Bazan, J. F., Gorman, D. M., Kastelein, R. A., Narula, S., et al. (2001) J. Immunol. 166, 7563–7570. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, G. X., Gran, B., Yu, S., Li, J., Siglienti, I., Chen, X., Kamoun, M. & Rostami, A. (2003) J. Immunol. 170, 2153–2160. [DOI] [PubMed] [Google Scholar]
- 32.Cua, D. J., Sherlock, J., Chen, Y., Murphy, C. A., Joyce, B., Seymour, B., Lucian, L., To, W., Kwan, S., Churakova, T., et al. (2003) Nature 421, 744–748. [DOI] [PubMed] [Google Scholar]
- 33.Aggarwal, S., Ghilardi, N., Xie, M. H., De Sauvage, F. J. & Gurney, A. L. (2003) J. Biol. Chem. 278, 1910–1914. [DOI] [PubMed] [Google Scholar]
- 34.de Waal Malefyt, R., Haanen, J., Spits, H., Roncarolo, M. G., te Velde, A., Figdor, C., Johnson, K., Kastelein, R., Yssel, H. & de Vries, J. E. (1991) J. Exp. Med. 174, 915–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Waal Malefyt, R., Abrams, J., Bennett, B., Figdor, C. G. & de Vries, J. E. (1991) J. Exp. Med. 174, 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koppelman, B., Neefjes, J. J., de Vries, J. E. & de Waal Malefyt, R. (1997) Immunity 7, 861–871. [DOI] [PubMed] [Google Scholar]
- 37.Lim, W., Ma, W., Gee, K., Aucoin, S., Nandan, D., Diaz-Mitoma, F., Kozlowski, M. & Kumar, A. (2002) J. Immunol. 168, 1759–1769. [DOI] [PubMed] [Google Scholar]
- 38.Stein, M., Keshav, S., Harris, N. & Gordon, S. (1992) J. Exp. Med. 176, 287–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson, C. F. & Mosser, D. M. (2002) J. Leukocyte Biol. 72, 101–106. [PubMed] [Google Scholar]
- 40.Belkaid, Y., Hoffmann, K. F., Mendez, S., Kamhawi, S., Udey, M. C., Wynn, T. A. & Sacks, D. L. (2001) J. Exp. Med. 194, 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murray, P. J. & Young, R. A. (1999) Infect. Immun. 67, 3087–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munn, D. H., Shafizadeh, E., Attwood, J. T., Bondarev, I., Pashine, A. & Mellor, A. L. (1999) J. Exp. Med. 189, 1363–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frumento, G., Rotondo, R., Tonetti, M., Damonte, G., Benatti, U. & Ferrara, G. B. (2002) J. Exp. Med. 196, 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terness, P., Bauer, T. M., Rose, L., Dufter, C., Watzlik, A., Simon, H. & Opelz, G. (2002) J. Exp. Med. 196, 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu, L., Tseng, S., Horner, R. M., Tam, C., Loda, M. & Rollins, B. J. (2000) Nature 404, 407–411. [DOI] [PubMed] [Google Scholar]
- 46.Gu, L., Rutledge, B., Fiorillo, J., Ernst, C., Grewal, I., Flavell, R., Gladue, R. & Rollins, B. (1997) J. Leukocyte Biol. 62, 577–580. [DOI] [PubMed] [Google Scholar]
- 47.Rutledge, B. J., Rayburn, H., Rosenberg, R., North, R. J., Gladue, R. P., Corless, C. L. & Rollins, B. J. (1995) J. Immunol. 155, 4838–4843. [PubMed] [Google Scholar]
- 48.Ting, L. M., Kim, A. C., Cattamanchi, A. & Ernst, J. D. (1999) J. Immunol. 163, 3898–3906. [PubMed] [Google Scholar]
- 49.Nau, G. J., Richmond, J. F., Schlesinger, A., Jennings, E. G., Lander, E. S. & Young, R. A. (2002) Proc. Natl. Acad. Sci. USA 99, 1503–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]