Abstract
Natural killer-like (NK) T, regulatory T (TR), and memory type T cells display surface phenotypes reminiscent of activated T cells. Previously, we reported that the generation of TR cells and, to a lesser extent, of memory type T cells, depends on IκB kinase 2. Here, we show that T cell-specific ablation of IκB kinase 2, in addition, completely precludes NKT cell development. T cell antigen receptor (TCR)-induced signals to activate NF-κB are essential for mature T cell activation, leading us to hypothesize that this pathway could play an important role in the generation of the antigen-driven T cell subsets comprising TR, memory type T, and NKT cells. TCR-mediated NF-κB activation critically depends on Bcl10 and PKCθ. By using mice deficient for these proteins, we demonstrate that the generation of TR and, to a lesser extent, of memory type T cells, depends on Bcl10 and PKCθ, and therefore, most likely on NF-κB activation initiated by TCR engagement. NKT cells, on the other hand, require PKCθ for thymic development, whereas absence of Bcl10 leads primarily to the reduction of peripheral NKT cell numbers.
Double-positive thymocytes, whose T cell antigen receptor (TCR) recognize major histocompatibility complex molecules, are positively selected to develop into mature T cells, whereas strong binding of agonistic ligand results in thymocyte apoptosis (negative selection) (1). Regulatory T (TR) cells and natural killer-like T (NKT) cells are specialized T cell subsets that play important roles in maintaining immune homeostasis. The development of TR and NKT cells, phenotypically resembling activated T cells, has been postulated to depend on recognition of (self)-antigen/MHC complexes with higher affinity/avidity than that leading to development of conventional T cells (2, 3). How induction of a regulatory phenotype is distinguished from elimination of thymocytes by negative selections is not clear, but might depend on the cell that expresses the TCR ligand.
TR cells are characterized by the expression of the IL-2 receptor α-chain (IL-2Rα; CD25) and the transcription factor Foxp3 (2). They down-regulate immune responses and inhibit development of autoimmune diseases (4). Evidence that TR cells are selected by self-antigen stems from experiments with T cells expressing a high-affinity transgenic TCR for influenza hemagglutinin, that develop into functional TR cells in mice expressing hemagglutinin (5, 6), whereas T cell bearing a low-affinity transgenic TCR for hemagglutinin do not (5). Selection through antigen expressed by thymic epithelium was shown to be the most effective way to induce CD4+CD25+ TR cells (7). NKT cells are characterized by expression of TCR and certain NK cell receptors. After antigen-receptor stimulation, they secrete large quantities of cytokines, most notably IL-4 and IFN-γ. The most extensively characterized NKT cells recognize the marine sponge-derived glycolipid α-galactosyl ceramide (α-GalCer) and have a highly skewed T cell repertoire, with a conserved Vα14-Jα18 rearrangement in mice. These cells have been termed Vα14i NKT cells (3). In mice, Vα14i NKT cells are derived from double-positive thymocytes (8) and are selected by self-lipids presented on the nonclassical MHC class I-like molecule CD1d, which is present on double- and single-positive thymocytes (9–11). NKT cell differentiation depends on RelB and on NF-κB-inducing kinase expressed in nonhematopoietic cells (12, 13). Expression of a mutant form of IκBα in T cells leads to strongly reduced thymic NKT cell numbers, although the peripheral NKT cell compartment is only modestly reduced (13).
After exposure to antigen, naïve peripheral CD4 T cells differentiate under the influence of cytokines into effector populations, producing distinct cytokine patterns. Whereas the majority of the effector populations undergo apoptosis, a small fraction develops into long-lived memory T cells without apparent further need for antigen, MHC interaction, or cytokines (14, 15).
Engagement of the TCR by antigen leads to a multitude of intracellular signaling events, ultimately causing activation of transcription factors, among them the NF-κB transcription factor family. NF-κB or Rel proteins are crucial for the regulation of genes important for immune reactions, cellular response to stress, and protection against apoptosis. In mammals, five Rel/NF-κB proteins are known: p65/RelA, c-Rel, RelB, NF-κB1/p50, and NF-κB2/p52. These proteins are usually kept inactive through interaction with inhibitor of κB (IκB) proteins, including IκBα, β, and ε, as well as p105 and p100, the precursors of p50 and p52, respectively (16, 17). After activation, the IκB kinase (IKK) complex, consisting of two kinases IKK1/α and IKK2/β and the regulatory subunit NEMO/IKK-γ, phosphorylates IκB proteins at two conserved serine residues, thereby inducing their polyubiquitination and proteosomal degradation. This pathway, termed the “classical” NF-κB activation pathway, is induced in response to numerous stimuli, such as proinflammatory cytokines and antigen-receptor ligation.
In conventional T cells, stimulation of the antigen receptor leads to IKK-mediated NF-κB activation through PKCθ (18), Bcl10 (19), Carma1/CARD11 (20–24), and MALT1 (25, 26). Ablation of any of these proteins leads to a specific block of NF-κB activation in response to TCR crosslinking, and therefore deficient activation of T cells.
Recently, we reported that ablation of IKK2 specifically in T cells through deletion of Ikk2FL alleles by Cre expressed under control of the CD4cre transgene (27) in CD4cre/Ikk2FL mice leads to the generation of mature conventional Ikk2–/– T cells, but impaired CD4+CD25+ TR and memory type T cell development (28). Here, we show that T cell-specific deletion of IKK2 prevents the generation of NKT cells altogether. We hypothesized that antigen (and therefore TCR)-mediated signaling through IKK2 to NF-κB plays a crucial role in the homeostasis of T cell subsets whose differentiation/expansion depend on antigen, such as memory type T, TR, and NKT cells. To test this notion, we analyze the homeostasis of these T cell subsets in mice deficient for PKCθ and Bcl10.
Materials and Methods
Mice and Flow Cytometry. CD4cre (27), Ikk2FL (29), and Pkcθ–/– (18) mice were maintained on a C57BL/6 and Bcl10–/– mice were maintained on a C57BL/6–129 mixed genetic background. All mice were housed in a conventional animal facility according to Harvard guidelines. Fluorescence-activated cell sorter (FACS) analyses and purifications of cells by magnetic cell sorting and FACS were conducted as published (30). Liver mononuclear cells were isolated as described (31). Briefly, livers were homogenized to single-cell suspensions with a metal cell strainer. Mononuclear cells were then isolated from the interface of a Percoll gradient (70% and 40%). mAbs to CD4, CD8, CD25, CD49B (DX5), CD45Rb, CD62L, CD94 (NK-G2A/C/E), CD122, NK1.1, TCRβ, and CD19 were purchased from Pharmingen. Phycoerythrin-labeled recombinant mouse CD1d molecules were expressed in SC2 Drosophila melanogaster cells and purified essentially as previously described (32), except that the SC2 cell supernatant was run over a noncharged nitrilotriacetic acid column and subsequently eluted with 100 mM imidazole in PBS pH 7.4. Unloaded CD1d tetramers did not yield specific staining above background. Absolute cell numbers for each T cell population were calculated by multiplying the percentage of each T cell subset of all live cells obtained from the FACS analysis with the total number of live cells recovered from the respective organ, determined by cell counting.
αCD3 Ab and α-GalCer Injections and PS-1145 Administration. Mice were injected with 10 μg per mouse of αCD3 mAb (Pharmingen, 553057) i.p. or with 2 μg per mouse of α-GalCer i.p. PS-1145 was resuspended in methyl cellulose solution (0.5% methyl cellulose, Aldrich 188042; and 0.2% Tween-80, Sigma P-8074). The suspension was vortexed vigorously for 2 min and was sonicated at 37°C for 20 min. PS-1145 was administrated to mice through forced oral feeding at 50 mg/kg, 1.5 h before α-GalCer injections.
RT-PCR and Real-Time Quantitative PCR. RNA was prepared from purified cells by using TRIzol reagent (Invitrogen) supplemented with 2 μg of glycogen (Invitrogen). RT-PCR was then performed with the ThermoScript RT-PCR system (Invitrogen). PCR on cDNA was performed essentially as described (33). Real-time quantitative PCR was performed by using published primers for Foxp3 and Hprt (33) and SYBR green PCR core reagents (PE Biosystems) with the following program: 10 min at 95°C followed by 40 cycles of 30 s at 94°C, 30 s at 65°C, and 30 s at 72°C. Each reaction was performed with cDNA corresponding to 25,000 purified cells and each sample was measured at least twice in independent experiments. All samples were run out on a 2% agarose gel after quantification to confirm the specificity of the PCR.
NKT Cell in Vitro Activation. Splenocyte suspension was seeded into a 96-well plate at 5 × 105 cells per well, preincubated with PS-1145 in 0.1% DMSO at room temperature for 30 min, and stimulated with 200 ng/ml α-GalCer at 37°C for 3 days. A half-volume of supernatant was removed to measure cytokines, and the remaining cells were pulsed for the last 6–16 h with 1 μCi per well (Ci = 37 GBq) of thymidine. Cytokines were measured by using R & D Systems ELISA kits or by Pierce's Searchlight service.
Results
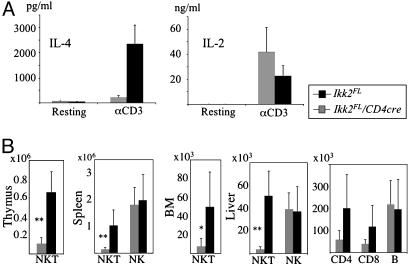
T Cell-Specific Ablation of IKK2 Leads to a Complete Absence of NKT Cells. To test the responses of Ikk2–/– T cell in vivo, we injected CD4-Cre/Ikk2FL and control mice with αCD3 Abs and measured serum cytokine levels after 2 h. There were no significant differences in IL-2, IFN-γ, and tumor necrosis factor levels between sera from CD4-Cre/Ikk2FL and control mice (Fig. 1A and data not shown). However, αCD3 administration failed to augment IL-4 serum levels in CD4-Cre/Ikk2FL mice, whereas it induced a robust increase of IL-4 levels in the serum of WT mice (Fig. 1 A). This result suggested that NKT development or function was affected by lack of IKK2 in T lineage cells. Indeed double-positive thymocytes, which give rise to NKT cells, have deleted their Ikk2 alleles in CD4-Cre/Ikk2FL mice (28). FACS analysis showed a strong reduction of TCRβ+NK1.1+ NKT cells in thymus, spleen, bone marrow (BM), and liver of CD4-Cre/Ikk2FL, compared with control mice. Calculation of absolute NKT cell numbers confirmed the deficiency of TCRβ+NK1.1+ NKT cells in CD4-Cre/Ikk2FL mice (Fig. 1B). Liver CD4 and CD8 T cell numbers were also reduced in CD4-Cre/Ikk2FL, compared with control mice (Fig. 1B), most likely reflecting the fact that most of the intrahepatic T cells display an activated phenotype (34), and are therefore more dependent on IKK2-mediated signals than on naïve T cells.
Fig. 1.
Reduction of NKT cells in CD4-Cre/Ikk2FL mice. (A) Histograms showing serum levels of IL-4 and IL-2 2 h after i.p. injection of αCD3 Abs in CD4-Cre/Ikk2FL (gray bars, n = 5) and Ikk2FL control mice (black bars, n = 5). Error bars indicate SEM. P ≤ 0.001; Student's t test. (B) Bar charts of absolute TCRβ+NK1.1+ cell numbers in thymus, spleen, BM, and liver of CD4-Cre/Ikk2FL and control mice. Absolute NK cell numbers are shown for spleen and liver, and CD4 T, CD8 T, and B cell numbers are shown for the liver. Gray bars represent CD4-Cre/Ikk2FL mice (n ≥ 9), and black bars represent control mice (Ikk2FL; n ≥ 9). Error bars indicate SD. *, P ≤ 0.05; **, P ≤ 0.001; Student's t test.
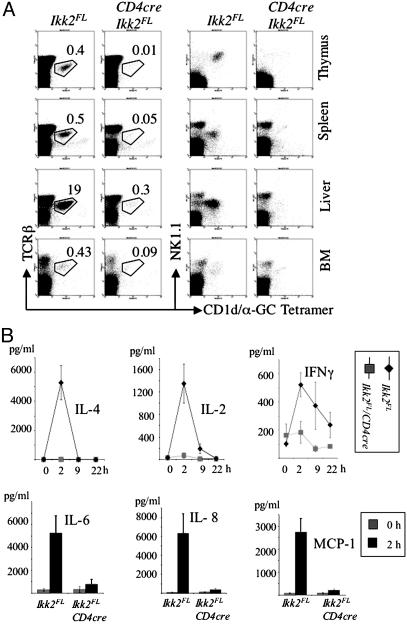
To identify specifically CD1d-dependent NKT cells, we stained cell suspensions from thymus, spleen, liver, and BM with CD1d/α-GalCer tetramers (ref. 32 and Fig. 2A). No specific staining above background was seen in single-cell suspensions from CD4-Cre/Ikk2FL mice with the CD1d/α-GalCer tetramers in combination with TCRβ, NK1.1 and CD4 (Fig. 2 A and data not shown). Levels of CD1d were normal on thymocytes from CD4-Cre/Ikk2FL mice (data not shown).
Fig. 2.
Analysis of presence and function of α-GalCer-reactive NKT cells in CD4-Cre/Ikk2FL mice. (A) FACS analysis of NKT cells in thymus, spleen, liver, and BM of CD4-Cre/Ikk2FL and control mice by using CD1d/α-GalCer tetramers. Numbers indicate mean percentages of gated cell population of live lymphocytes calculated from two mice, respectively. (B) Histograms and bar charts of cytokine serum levels after injection of α-GalCer. Cytokine levels were determined in the serum collected at various time points from CD4-Cre/Ikk2FL (n = 4) and control mice (n = 5). Error bars represent SEM.
To test specifically for NKT cell function in vivo, we measured serum cytokine levels at various time points after α-GalCer injection. Whereas α-GalCer administration led to a transient increase in IL-4, IL-2, IFN-γ, IL-6, IL-8, and MCP-1 serum levels in WT mice, it did not elicit any detectable release of cytokines into the serum of CD4-Cre/Ikk2FL mice at 2–22 h after injection (Fig. 2B). These findings demonstrate a systemic absence of NKT cells in CD4-Cre/Ikk2FL mice.
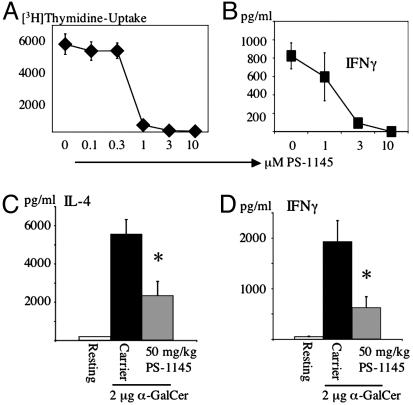
The IKK2 Inhibitor PS-1145 Interferes with NKT Cell Function. To test for a functional role of IKK2 in NKT cell activation, we made use of the specific IKK2 inhibitor, PS-1145 (35, 36). WT splenocytes were treated with α-GalCer in the presence of various concentrations of PS-1145. PS-1145 reduced proliferation and IFN-γ production by α-GalCer-treated splenocyte cultures in a dose-dependent manner (Fig. 3 A and B). To assay for NKT cell function in vivo, we administered PS-1145 at various doses perorally 1.5 h before injection of α-GalCer. PS-1145 inhibited α-GalCer induced IL-4 and IFN-γ release in vivo (Fig. 3 C and D), supporting an essential role for IKK2 in NKT cell activation, in addition to its requirement for NKT cell development.
Fig. 3.
Inhibition of NKT cell function by PS-1145. (A and B) Splenocyte cultures were activated for 3 days with α-GalCer (200 ng/ml) in the presence of various amounts of the IKK2-inhibitor PS-1145. After 3 days, proliferation was monitored by [3H]thymidine incorporation (A), and IFN-γ levels were determined by ELISA (B). (C and D) Mice were pretreated with PS-1145 or carrier perorally 1.5 h before injection of 2 μgof α-GalCer. Three hours after injection of α-GalCer, serum was removed to determine IL-4 and IFN-γ levels by ELISA. Each bar represents the mean calculated from five mice, error bars indicate the SEM. *, P ≤ 0.05; Student's t test.
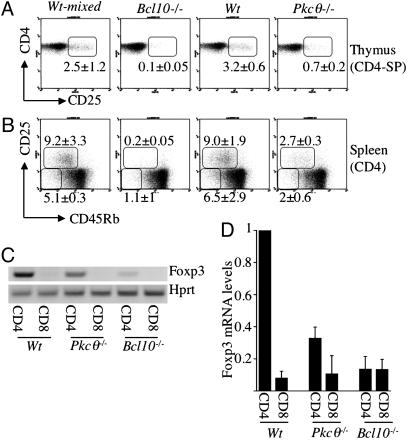
Antigen Receptor-Mediated Activation of NF-κB Is Required for the Generation of CD4+CD25+ TR Cells. In mice deficient for IKK2, specifically in T cells, the numbers of memory type T and TR cells are strongly reduced, in contrast with those of naïve CD4 T cells (28). To assess whether this result is due to NF-κB activation in response to TCR-mediated T cell activation, we made use of PKCθ (18)- and Bcl10-deficient mice (19), in which NF-κB activation is impaired specifically in response to cellular activation through the antigen receptor, but not in response to proinflammatory cytokines such as tumor necrosis factor. FACS analysis yielded strongly reduced CD4+CD25+CD45Rblow T cell numbers in the spleen and LN of Pkcθ–/– mice and absence of these cells in Bcl10–/– mice (Fig. 4B, Table 1, and data not shown). A similar picture is observed in the thymus (Fig. 4A and Table 1), indicating that TCR-mediated activation of NF-κB is crucial for TR development.
Fig. 4.
Regulatory and memory type T cells in Bcl10–/– in Pkcθ–/– mice. (A and B) FACS analysis of CD4+CD25+ TR cells in the thymus (A) and of TR and memory type T cells in the spleen (B) of Bcl10–/–, Pkcθ–/–, and appropriate control mice. Numbers indicate mean and SD of the percentages of CD4+CD25+ T cells of CD4-SP thymocytes (A) and of CD25+CD45Rbint T and CD25–CD45Rblow T cells of splenic CD4 T cells (B), determined from five mice, respectively. Pkcθ–/– mice were on a pure C57BL/6 genetic background, whereas the Bcl10–/– mice were on a C57BL/6–129 mixed genetic background; therefore, the data obtained from the respective age- and sex-matched control mice are shown separately. (C) RT-PCR showing levels of Foxp3- and Hprt-specific transcripts amplified from total mRNA purified from CD4 and CD8 T cells from Bcl10–/–, Pkcθ–/–, and control mice. One of at least three independent experiments is shown for each genotype. (D) Quantification of Foxp3 mRNA levels in indicated T cell subsets purified from WT (n = 6), Pkcθ–/– (n = 3), and Bcl10–/– (n = 3) mice by real-time PCR. Foxp3 mRNA expression is normalized to expression of Hprt mRNA: Foxp3Hprt-message. The mean Foxp3Hprt-message in CD4 T cells purified from WT mice was set to 1. Foxp3Hprt-message in WT CD8 T cells and CD4 and CD8 T cells purified from Pkcθ–/– and Bcl10–/– mice was calculated relative to Foxp3Hprt-message (WT CD4 T cells) = 1. Results are shown as the mean, and error bars indicate the SEM.
Table 1. Absolute numbers of T cell populations in Bcl10-/-, Pkcθ-/-, and control mice.
| Cell type | Organ | Bcl10-/- | WT mixed | Pkcθ-/- | WT BL/6 |
|---|---|---|---|---|---|
| CD4+CD25+ | Thy (× 106) | 0.02 ± 0.03 | 0.21 ± 0.09 | 0.04 ± 0.02 | 0.43 ± 0.26 |
| CD4+CD25+CD45Rbint | Spl (× 106) | 0.05 ± 0.03 | 1.35 ± 0.29 | 0.32 ± 0.07 | 1.4 ± 0.53 |
| CD4+CD44+ | Spl (× 106) | 0.57 ± 0.29 | 2.31 ± 0.66 | 0.98 ± 0.22 | 1.91 ± 0.99 |
| CD4+CD44+CD62Llow | Spl (× 106) | 0.52 ± 0.31 | 2.6 ± 0.57 | — | — |
| CD4+CD25-CD45Rblow | Spl (× 106) | 0.2 ± 0.06 | 0.77 ± 0.25 | 0.21 ± 0.06 | 0.87 ± 0.37 |
| NKT | Thy (× 106) | 0.11 ± 0.06 | 0.34 ± 0.15 | 0.11 ± 0.04 | 0.46 ± 0.07 |
| NKT | Spl (× 106) | 0.06 ± 0.03 | 0.85 ± 0.05 | 0.44 ± 0.13 | 0.47 ± 0.1 |
| NKT | Liver (× 103) | 0.21 ± 0.4 | 4.5 ± 0.15 | 2.7 ± 1.59 | 2.7 ± 0.58 |
| NKT | BM (× 103) | 0.18 ± 0.12 | 2.26 ± 0.14 | 3.6 ± 1.36 | 3.6 ± 1.23 |
| NK | Spl (× 106) | 0.69 ± 0.36 | 2.03 ± 0.17 | — | — |
| NK | Liver (× 103) | 0.53 ± 0.13 | 2.95 ± 1.07 | — | — |
Cell types were distinguished by FACS according to the indicated cell surface markers. NKT cells comprise CD 1d/α-GalCer tetramer-positive T cells, and NK cells are NK1.1+TCRβ- cells. Staining with tetramers identifies a subset, although the major one, of Nk1.1+ T cells. This fact, together with the lower background of staining with αTCRβ/tetramer compared with αTCRβ/αNK1.1, explains the lower absolute NKT cell numbers detected in control mice shown in this table, compared with NKT cell numbers in control mice shown in Fig. 2B. Data are shown as mean ± SD calculated from 4-5 mice.
Foxp3 expression is considered the best marker for TR cells to date (2). Because expression of CD25 is at least partially regulated by NF-κB, it became important to determine whether the absence of CD4+CD25+ T cells reflected merely NF-κB-dependent CD25 regulation or truly reduced numbers of bona fide TR cells. RT-PCR and real-time PCR analyses of Foxp3 mRNA levels in CD4 T cells purified from Pkcθ–/– and Bcl10–/– mice revealed a reduction of Foxp3 expression proportional to the reduction of CD25 expression in these mice (Fig. 4 C and D). Bcl10-deficient CD4 T cells contain levels of Foxp3 mRNA close to background levels, similar to CD8 T cells, whereas Pkcθ–/– CD4 T cells express one-third of Foxp3 mRNA, compared with WT CD4 T cells. (Fig. 4D).
Antigen Receptor-Mediated Activation of NF-κB Is Involved in the Generation and/or Maintenance of CD4 Memory Type T Cells. Bcl10–/– mice contain 4- to 5-fold fewer CD4+CD44highCD62Llow memory type T cells than do WT mice (Table 1). However, because also nearly all CD4+ NKT cells are CD44highCD62Llow (3) and over one-third of the CD4+CD25+ T cells are CD44high (data not shown), this reduction could also reflect loss of NKT and TR cells. NKT cells are mostly CD45Rbint (37), therefore CD4+CD25–CD45Rblow T cells should represent a memory type T cell population largely devoid of regulatory and NKT cells. These CD4+CD25–CD45Rblow T cells are also CD44high (data not shown). Bcl10–/– and Pkcθ–/– mice have roughly four times less of these memory type T cell numbers than do WT mice in their spleens and LNs (Fig. 4B, Table 1, and data not shown). NF-κB activation upon TCR engagement is therefore critical for efficient generation and/or maintenance of CD4 memory type T cells. In contrast, we found no statistically significant difference in CD8+CD44+ memory type T cell numbers between Bcl10–/–, Pkcθ–/–, and control mice (data not shown).
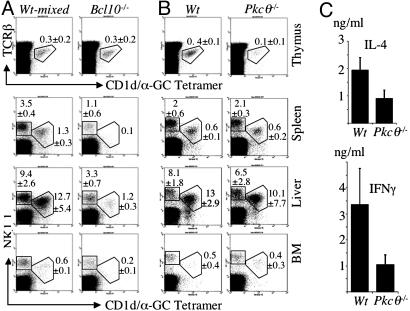
Bcl10 and PKCθ Are Involved in NKT Cell Homeostasis. Normal proportions of peripheral CD3+CD122+ NKT cells were found in Card11–/– (20) and Malt1–/– (26) mice, suggesting that TCR-induced activation of NF-κB has no major role in NKT cell homeostasis. In agreement with this notion, we find that in the thymus of Bcl10–/–mice, the proportion of NKT cells is normal (Fig. 5A), but because the thymi of adult Bcl10–/– mice are consistently much smaller than those of WT mice, there is a reduction in absolute NKT cell numbers in Bcl10–/– thymi (Table 1). However, Bcl10–/– mice have a >10-fold reduction in the numbers of NKT cells in spleen, liver, and BM (Fig. 5A and Table 1), indicating that Bcl10 plays a role in NKT cell persistence. We also observed a threefold reduction in NK cell numbers in spleen and liver of Bcl10–/– mice (Fig. 5A and Table 1), as seen in Card11–/– mice (20). We confirmed these findings by using Abs against TCRβ, CD122, NK-G2A/C/E, and the mouse Pan NK cell marker DX5 (data not shown).
Fig. 5.
Analysis of NK and NKT cells in Bcl10–/– and Pkcθ–/– mice. (A and B) FACS analysis of NKT and NK cells in thymus, spleen, liver, and BM of Bcl10–/– and control mice (A) and Pkcθ–/– and control mice (B) using CD1d/α-GalCer tetramers. Numbers indicate mean percentages and SD (n = 4–5 mice for each genotype) of NK or NKT cells of live cells in the lymphocyte gate. (C) Bar charts of IL-4 and IFN-γ serum levels after injection of α-GalCer. IL-4 and IFN-γ levels were determined in the serum collected 3 h after injection from Pkcθ–/– (n = 5) and control mice (n = 5). Error bars represent the SEM.
Pkcθ–/– mice, on the other hand, have normal NKT cell numbers in the periphery, but strongly reduced NKT cell numbers in the thymus (Fig. 5B and Table 1). The involvement of both proteins, but especially of PKCθ, in other signal transduction pathways than TCR-mediated activation of NF-κB could explain this discrepancy between the roles of Bcl10 and PKCθ in NKT cell homeostasis. The reduction in thymic NKT cells could be linked to the role of PKCθ in AP-1 activation (18). Measurement of cytokine levels in the serum after injection of α-GalCer showed ≈50% reduction in IL-4 and IFN-γ release by Pkcθ–/– NKT cells, compared with WT NKT cells (Fig. 5C). Because thymic NKT cells do not seem to be activated efficiently in response to α-GalCer injections in vivo (39), this reduction could reflect a functional defect in Pkcθ–/– NKT cells.
Discussion
We previously reported (28) that the survival of naïve T cells depends on a mode of IKK-mediated activation of NF-κB, which is independent of the TCR and in which IKK1 and IKK2 play redundant roles. T cell-specific ablation of IKK2 allows for the generation of mature naïve conventional T cells, but interferes with regulatory and memory type T cell development. NKT cells display surface markers characteristic for activated or memory type cells, similar to TR cells. Taking the importance of NF-κB transcription factors for T cell activation into account, it seemed feasible that also NKT cell development depends on IKK2. Indeed, while this manuscript was under preparation, a cell-intrinsic role for NF-κB in thymic NKT cell development was demonstrated by using mice that express mutant IκBα in T cells (13). However, residual NKT cells are present in the thymus and nearly normal numbers of NKT cells are detected in the periphery of these mice (13), suggesting that the requirement for NF-κB in NKT cell development is not absolute. The authors also propose that NF-κB could specifically play a role in vα14i NKT cells, but not in total NK1.1+ T cell development (13). Our data obtained by T cell-specific ablation of IKK2, on the other hand, demonstrate that activation of NF-κB mediated by IKK2 is strictly required for the generation of α-GalCer-reactive NKT cells, which are completely absent in CD4-Cre/Ikk2FL mice. We show that IKK2 activity is important for the development/homeostasis of all NKT cell subgroups, because we observe a global lack of NK1.1+ T cells and absence of IL-4 release after αCD3 administration in CD4-Cre/Ikk2FL mice. In addition, IKK2 seems to be involved in NKT cell activation, as administration of the inhibitor PS-1145 reduced α-GalCer-induced cytokine release in vivo.
The fact that TR and NKT cells are thought to be selected by higher-affinity TCR to cognate ligand/MHC complex interactions than conventional T cells (2, 3), together with their dependence on IKK2, prompted us to hypothesize that generation and/or expansion of these T cell subsets could depend on TCR-mediated activation of NF-κB. We tested this hypothesis by analyzing T cell subset in Pkcθ–/– and Bcl10–/– mice. One should keep in mind, however, that Pkcθ–/– T cells have impairments in activation of AP-1, in addition to defects in NF-κB activation in response to TCR-crosslinking and that a role for PKCθ in NF-AT activation has been suggested (40), but is controversial (18). It has also been reported that residual NF-κB activity can be elicited by αCD3/αCD28 treatment in Pkcθ–/– T cells (40), demonstrating that signaling from the TCR to NF-κB is not completely blocked in the absence of PKCθ. Furthermore, absence of PKCθ does not affect αCD3-induced NF-κB activation in total thymocytes (18), but it remains unclear at which stage of T cell development PKCθ becomes essential for TCR-mediated NF-κB activation. Therefore, the defects observed in Pkcθ–/– mice could be due to defective AP-1 and potentially NF-AT activation in addition or instead of reduced NF-κB activation in response to TCR triggering. Deletion of Bcl10 provides a better model because it leaves AP-1 activation unaffected and elicits a more complete block of TCR-induced NF-κB activation than deletion of PKCθ (19).
Our results showing absence and very strong reduction of TR cells in thymus and periphery of Bcl10–/– and Pkcθ–/– mice, respectively, indicate that TCR-induced activation of NF-κB constitutes an essential signal for TR cell development and homeostasis. Our previous findings (28) suggest that IKK2 is indispensable in this pathway.
IL-2 is an essential factor for survival and expansion of TR cells (4). Il-2–/– mice were initially reported to lack CD4+CD25+ TR cells in thymus and periphery (41). Bcl10–/– and Pkcθ–/– T cells produce less IL-2 when activated in vitro (18, 19). Therefore, the argument could be made that the lack of TR cells in Bcl10–/– and Pkcθ–/– mice should be attributed to absence of IL-2 rather than to defective selection signals due to defective TCR-induced activation of NF-κB in TR cell precursors. However, two recent reports (42, 43) suggest that CD4+CD25+ TR cells are generated in the thymus of Il-2–/– mice. Malek et al. (42) find a 2- to 3-fold reduction of CD4+CD25+ TR cells in the thymi of Il-2–/– and Il-2rβ–/– mice, compared with heterozygous controls (42, whereas Antov et al. (43) show the presence of normal numbers of thymic CD4+CD25+ T cells in Il-2–/– mice (43). Therefore, it is likely that CD4+CD25+ TR cells are generated in the thymus in the absence of IL-2, and that IL-2 is essential for the expansion and survival of a stable functional CD4+CD25+ TR cell population in the periphery. Because TR cells are completely absent in the thymus of Bcl10–/– mice, it appears improbable that insufficient production of IL-2 by Bcl10-deficient cells is responsible for the lack of TR cells in these mice. In addition, Almeida et al. (44) report that adoptive transfer of Il-2–/– BM into Rag-2–/– hosts leads to generation of normal numbers of CD4+CD25+ TR cells, suggesting that resident non-T cells of the Rag-2–/– recipient provide a source of endogenous IL-2 sufficient for the generation of these T cells. Therefore, it seems unlikely that lack of IL-2 is the cause for the defect in TR cell generation in Bcl10–/– and Pkcθ–/– mice.
NF-κB was reported to play a critical role in the development of CD8+-memory-phenotype T cells (45), and Ikk2–/– CD8+CD44+ T cell are strongly counterselected in CD4-Cre/Ikk2FL mice (28). However, we did not find a statistically significant reduction of CD8+CD44+ T cell numbers in Bcl10–/– and Pkcθ–/– mice (data not shown). This result suggests that the NF-κB dependence of CD8+CD44+ T cells is based on a different mechanism than activation through the TCR.
On the contrary, Bcl10–/– and Pkcθ–/– mice contain significantly less memory type CD4 T cells, compared with WT mice. TCR-deficient CD4 memory type T cells persist normally, but expand less efficiently than TCR-proficient CD4 memory type T cells (46). Therefore, the reduction of CD4 memory type T cell numbers in Bcl10–/– and Pkcθ–/– mice is likely due to defective generation and/or expansion of such cells resulting from defective TCR-mediated activation of NF-κB.
The analysis of NKT cell populations led to unexpected results. Absence of Bcl10 allows development of NKT cells in the thymus but leads to dramatically reduced peripheral NKT cell numbers, whereas lack of PKCθ interferes with thymic NKT cell development, but normal numbers of NKT cells can accumulate in the periphery. However, it is clear that lack of Bcl10 or PKCθ does not lead to absence of NKT cell generation, as is observed after T cell-specific ablation of IKK2. This finding could indicate that absence of NKT cells caused by lack of IKK2 is due to defective NF-κB activation induced by receptors other than the TCR. Alternatively, it is possible that in NKT cells, the signaling pathway from the TCR to NF-κB activation employs different mediators than in conventional T cells, and that Bcl10 and PKCθ play roles in NKT cell generation and/or maintenance distinct from TCR-mediated NF-κB activation.
The defect in thymic T cells observed in Pkcθ–/– mice could be linked to the block of AP-1 activation in the absence of PKCθ, because transgenic expression of the AP-1 inhibitor basic leucine zipper transcription factor in T cells impairs NKT cell development (38). However, PKCθ does not play a major role in NKT cell persistence. Normal percentages of CD3+CD122+ NKT cells were seen in the spleen of Card11–/– (20) and in the liver of Malt1–/– (26) mice. It is therefore puzzling that peripheral NKT cell numbers are strongly reduced in Bcl10–/– mice. This result seems to indicate that Bcl10 has a role in peripheral NKT cell persistence through a mechanism distinct from TCR-mediated NF-κB activation. Indeed Bcl10-deficient marginal zone, but not follicular B cells, have defective responses to lipopolysaccharide, suggesting that in some cell-types Bcl10 can participate in signaling pathways that are not initiated by the antigen receptor (47).
Taken together, the data presented in this paper support the hypothesis that activation of NF-κB through IKK2 initiated by TCR engagement with (self-) antigen is required for the development of TR cells and plays a role in the generation/expansion of memory type T cells. Our results also demonstrate that NKT cell development strictly depends on as-yet-unidentified signals mediated by IKK2.
Acknowledgments
We thank N. Bartenev for cell sorting; Luc Teyton for providing the CD1d and β2-microglobulin containing the SC2 D. melanogaster cell line; C. Cantu for technical advice; M. Kuroda for assistance on CD1d tetramer production; S. Akira, D. R. Littman, T. W. Mak, and C. B. Wilson for transgenic mice; M. Exley and K. Ngyuen for advice; and M. Exley, B. Greve, Z. Illes, K. Ngyuen, and Y. Sasaki for critical reading of the manuscript. This work was supported by National Institutes of Health Grant AI057947 (to K.R.). R.M. received a scholarship from the Boehringer Ingelheim Fonds.
Abbreviations: αGalCer, α-galactosyl ceramide; IκB, inhibitor of κB; IKK, IκB kinase; NKT, natural killer-like T cell; TCR, T cell receptor; TR, regulatory T cell; FACS, fluorescence-activated cell sorter; BM, bone marrow.
References
- 1.Von Boehmer, H., Aifantis, I., Gounari, F., Azogui, O., Haughn, L., Apostolou, I., Jaeckel, E., Grassi, F. & Klein, L. (2003) Immunol. Rev. 191, 62–78. [DOI] [PubMed] [Google Scholar]
- 2.Ramsdell, F. (2003) Immunity 19, 165–168. [DOI] [PubMed] [Google Scholar]
- 3.Kronenberg, M. & Gapin, L. (2002) Nat. Rev. Immunol. 2, 557–568. [DOI] [PubMed] [Google Scholar]
- 4.Shevach, E. M. (2002) Nat. Rev. Immunol. 2, 389–400. [DOI] [PubMed] [Google Scholar]
- 5.Jordan, M. S., Boesteanu, A., Reed, A. J., Petrone, A. L., Holenbeck, A. E., Lerman, M. A., Naji, A. & Caton, A. J. (2001) Nat. Immunol. 2, 301–306. [DOI] [PubMed] [Google Scholar]
- 6.Jordan, M. S., Riley, M. P., von Boehmer, H. & Caton, A. J. (2000) Eur. J. Immunol. 30, 136–144. [DOI] [PubMed] [Google Scholar]
- 7.Apostolou, I., Sarukhan, A., Klein, L. & von Boehmer, H. (2002) Nat. Immunol. 3, 756–763. [DOI] [PubMed] [Google Scholar]
- 8.Gapin, L., Matsuda, J. L., Surh, C. D. & Kronenberg, M. (2001) Nat. Immunol. 2, 971–978. [DOI] [PubMed] [Google Scholar]
- 9.Elewaut, D., Lawton, A. P., Nagarajan, N. A., Maverakis, E., Khurana, A., Honing, S., Benedict, C. A., Sercarz, E., Bakke, O., Kronenberg, M. & Prigozy, T. I. (2003) J. Exp. Med. 198, 1133–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiu, Y. H., Park, S. H., Benlagha, K., Forestier, C., Jayawardena-Wolf, J., Savage, P. B., Teyton, L. & Bendelac, A. (2002) Nat. Immunol. 3, 55–60. [DOI] [PubMed] [Google Scholar]
- 11.Vincent, M. S., Gumperz, J. E. & Brenner, M. B. (2003) Nat. Immunol. 4, 517–523. [DOI] [PubMed] [Google Scholar]
- 12.Elewaut, D., Shaikh, R. B., Hammond, K. J., De Winter, H., Leishman, A. J., Sidobre, S., Turovskaya, O., Prigozy, T. I., Ma, L., Banks, T. A., et al. (2003) J. Exp. Med. 197, 1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sivakumar, V., Hammond, K. J., Howells, N., Pfeffer, K. & Weih, F. (2003) J. Exp. Med. 197, 1613–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swain, S. L. (2003) Microbes Infect. 5, 213–219. [DOI] [PubMed] [Google Scholar]
- 15.Seder, R. A. & Ahmed, R. (2003) Nat. Immunol. 4, 835–842. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh, S. & Karin, M. (2002) Cell 109, Suppl., S81–S96. [DOI] [PubMed] [Google Scholar]
- 17.Li, Q. & Verma, I. M. (2002) Nat. Rev. Immunol. 2, 725–734. [DOI] [PubMed] [Google Scholar]
- 18.Sun, Z., Arendt, C. W., Ellmeier, W., Schaeffer, E. M., Sunshine, M. J., Gandhi, L., Annes, J., Petrzilka, D., Kupfer, A., Schwartzberg, P. L. & Littman, D. R. (2000) Nature 404, 402–407. [DOI] [PubMed] [Google Scholar]
- 19.Ruland, J., Duncan, G. S., Elia, A., del Barco Barrantes, I., Nguyen, L., Plyte, S., Millar, D. G., Bouchard, D., Wakeham, A., Ohashi, P. S. & Mak, T. W. (2001) Cell 104, 33–42. [DOI] [PubMed] [Google Scholar]
- 20.Hara, H., Wada, T., Bakal, C., Kozieradzki, I., Suzuki, S., Suzuki, N., Nghiem, M., Griffiths, E. K., Krawczyk, C., Bauer, B., et al. (2003) Immunity 18, 763–775. [DOI] [PubMed] [Google Scholar]
- 21.Egawa, T., Albrecht, B., Favier, B., Sunshine, M. J., Mirchandani, K., O'Brien, W., Thome, M. & Littman, D. R. (2003) Curr. Biol. 13, 1252–1258. [DOI] [PubMed] [Google Scholar]
- 22.Newton, K. & Dixit, V. M. (2003) Curr. Biol. 13, 1247–1251. [DOI] [PubMed] [Google Scholar]
- 23.Jun, J. E., Wilson, L. E., Vinuesa, C. G., Lesage, S., Blery, M., Miosge, L. A., Cook, M. C., Kucharska, E. M., Hara, H., Penninger, J. M., et al. (2003) Immunity 18, 751–762. [DOI] [PubMed] [Google Scholar]
- 24.Wang, D., You, Y., Case, S. M., McAllister-Lucas, L. M., Wang, L., DiStefano, P. S., Nunez, G., Bertin, J. & Lin, X. (2002) Nat. Immunol. 3, 830–835. [DOI] [PubMed] [Google Scholar]
- 25.Ruefli-Brasse, A. A., French, D. M. & Dixit, V. M. (2003) Science 302, 1581–1584. [DOI] [PubMed] [Google Scholar]
- 26.Ruland, J., Duncan, G. S., Wakeham, A. & Mak, T. W. (2003) Immunity 19, 749–758. [DOI] [PubMed] [Google Scholar]
- 27.Lee, P. P., Fitzpatrick, D. R., Beard, C., Jessup, H. K., Lehar, S., Makar, K. W., Perez-Melgosa, M., Sweetser, M. T., Schlissel, M. S., Nguyen, S., et al. (2001) Immunity 15, 763–774. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt-Supprian, M., Courtois, G., Tian, J., Coyle, A. J., Israel, A., Rajewsky, K. & Pasparakis, M. (2003) Immunity 19, 377–389. [DOI] [PubMed] [Google Scholar]
- 29.Pasparakis, M., Courtois, G., Hafner, M., Schmidt-Supprian, M., Nenci, A., Toksoy, A., Krampert, M., Goebeler, M., Gillitzer, R., Israel, A., et al. (2002) Nature 417, 861–866. [DOI] [PubMed] [Google Scholar]
- 30.Pasparakis, M., Schmidt-Supprian, M. & Rajewsky, K. (2002) J. Exp. Med. 196, 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuda, J. L., Gapin, L., Sidobre, S., Kieper, W. C., Tan, J. T., Ceredig, R., Surh, C. D. & Kronenberg, M. (2002) Nat. Immunol. 3, 966–974. [DOI] [PubMed] [Google Scholar]
- 32.Benlagha, K., Weiss, A., Beavis, A., Teyton, L. & Bendelac, A. (2000) J. Exp. Med. 191, 1895–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hori, S., Nomura, T. & Sakaguchi, S. (2003) Science 299, 1057–1061.12522256 [Google Scholar]
- 34.Crispe, I. N. (2003) Nat. Rev. Immunol. 3, 51–62. [DOI] [PubMed] [Google Scholar]
- 35.Hideshima, T., Chauhan, D., Richardson, P., Mitsiades, C., Mitsiades, N., Hayashi, T., Munshi, N., Dang, L., Castro, A., Palombella, V., Adams, J. & Anderson, K. C. (2002) J. Biol. Chem. 277, 16639–16647. [DOI] [PubMed] [Google Scholar]
- 36.Muller, J. R. & Siebenlist, U. (2003) J. Biol. Chem. 278, 12006–12012. [DOI] [PubMed] [Google Scholar]
- 37.Hammond, K. J., Pelikan, S. B., Crowe, N. Y., Randle-Barrett, E., Nakayama, T., Taniguchi, M., Smyth, M. J., van Driel, I. R., Scollay, R., Baxter, A. G. & Godfrey, D. I. (1999) Eur. J. Immunol. 29, 3768–3781. [DOI] [PubMed] [Google Scholar]
- 38.Williams, K. L., Zullo, A. J., Kaplan, M. H., Brutkiewicz, R. R., Deppmann, C. D., Vinson, C. & Taparowsky, E. J. (2003) J. Immunol. 170, 2417–2426. [DOI] [PubMed] [Google Scholar]
- 39.Osman, Y., Kawamura, T., Naito, T., Takeda, K., Van Kaer, L., Okumura, K. & Abo, T. (2000) Eur. J. Immunol. 30, 1919–1928. [DOI] [PubMed] [Google Scholar]
- 40.Pfeifhofer, C., Kofler, K., Gruber, T., Tabrizi, N. G., Lutz, C., Maly, K., Leitges, M. & Baier, G. (2003) J. Exp. Med. 197, 1525–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Papiernik, M., de Moraes, M. L., Pontoux, C., Vasseur, F. & Penit, C. (1998) Int. Immunol. 10, 371–378. [DOI] [PubMed] [Google Scholar]
- 42.Malek, T. R., Yu, A., Vincek, V., Scibelli, P. & Kong, L. (2002) Immunity 17, 167–178. [DOI] [PubMed] [Google Scholar]
- 43.Antov, A., Yang, L., Vig, M., Baltimore, D. & Van Parijs, L. (2003) J. Immunol. 171, 3435–3441. [DOI] [PubMed] [Google Scholar]
- 44.Almeida, A. R., Legrand, N., Papiernik, M. & Freitas, A. A. (2002) J. Immunol. 169, 4850–4860. [DOI] [PubMed] [Google Scholar]
- 45.Hettmann, T., Opferman, J. T., Leiden, J. M. & Ashton-Rickardt, P. G. (2003) Immunol. Lett. 85, 297–300. [DOI] [PubMed] [Google Scholar]
- 46.Polic, B., Kunkel, D., Scheffold, A. & Rajewsky, K. (2001) Proc. Natl. Acad. Sci. USA 98, 8744–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xue, L., Morris, S. W., Orihuela, C., Tuomanen, E., Cui, X., Wen, R. & Wang, D. (2003) Nat. Immunol. 4, 857–865. [DOI] [PubMed] [Google Scholar]