Abstract
Arsenic has played a key medicinal role against a variety of ailments for several millennia, but during the past century its prominence has been displaced by modern therapeutics. Recently, attention has been drawn to arsenic by its dramatic clinical efficacy against acute promyelocytic leukemia. Although toxic reactive oxygen species (ROS) induced in cancer cells exposed to arsenic could mediate cancer cell death, how arsenic induces ROS remains undefined. Through the use of gene expression profiling, interference RNA, and genetically engineered cells, we report here that NADPH oxidase, an enzyme complex required for the normal antibacterial function of white blood cells, is the main target of arsenic-induced ROS production. Because NADPH oxidase enzyme activity can also be stimulated by phorbol myristate acetate, a synergism between arsenic and the clinically used phorbol myristate acetate analog, bryostatin 1, through enhanced ROS production can be expected. We show that this synergism exists, and that the use of very low doses of both arsenic and bryostatin 1 can effectively kill leukemic cells. Our findings pinpoint the arsenic target of ROS production and provide a conceptual basis for an anticancer regimen.
Although arsenic has played a significant therapeutic role in various diseases for >2,000 years (1, 2), it was not used clinically for decades, until recently when clinical trials world-wide confirmed its dramatic therapeutic effects in acute promyelocytic leukemia (APL) (3, 4). APL is a subtype of acute myelocytic leukemia with most cases carrying the characteristic chromosomal translocation t(15, 17) that results in the PML-RARα fusion protein (5). Although APL is highly responsive to arsenic, the presence of PML-RARα fusion protein is neither absolutely necessary nor sufficient for sensitivity to arsenic (3, 6, 7). The mechanism by which arsenic is effective against APL remains elusive, despite studies suggesting that arsenic can promote degradation of the oncogenic PML-RARα fusion protein (8, 9). Paradoxically, arsenic is also an established human carcinogen that can induce reactive oxygen species (ROS), leading to DNA damage or cell death (10–13).
Some previous mechanistic studies (14, 15) were limited to exposure of cells other than myeloid cells, or to arsenite rather than arsenic trioxide for brief periods, and hence do not reflect the clinical setting for cytotoxic effects of arsenic on APL cells. To explore the molecular mechanisms of arsenic's therapeutic effects in the treatment of APL patients with daily continuous infusion of arsenic trioxide, we treated a human APL cell line, NB4, for >1 week with arsenic trioxide at a dose lower than the plasma trough level achieved in APL patients. We reported previously that arsenic at this dose was able to down-regulate human telomerase hTERT transcription (16). In this report, we determined changes in gene expression profiles by using oligonucleotide microarrays, and we found that NADPH oxidase components were dramatically up-regulated within days in myeloid cells treated with low-dose arsenic. NADPH oxidase, which is an enzyme complex consisting of multiple membrane-associated and cytosolic subunits, can be stimulated by phorbol myristate acetate (PMA) through protein kinase C-mediated phosphorylation of the p47PHOX subunit (17, 18). This complex is responsible for the production of superoxide anion (respiratory burst) of professional phagocytes encountering microbial pathogens, and its importance in host immunity is underscored by the immunocompromised congenital disease, chronic granulomatous disease (CGD), which results from mutations in one of the subunits of NADPH oxidase (19, 20). Our biochemical and molecular biological studies reported here have uncovered a major role of this enzyme complex in arsenic-induced ROS production and cytotoxicity. We have also exploited the synergistic induction of NADPH oxidase activity and ROS production by arsenic and PMA to provide proof-of-concept that this synergy may be clinically applicable.
Methods
Cell Lines. NB4, U937, PLB-985, X-CGD, and HL60 cells were cultured in RPMI medium 1640 supplemented with 10% FBS. ML1 was maintained in RPMI medium 1640 with 7.5% FBS and 3.4 g of Hepes/500 ml, pH 7.4.
Microarray Analysis. NB4 cells were grown to a density of 105/ml and were treated with 0.75 μM arsenic trioxide for 10 days. mRNA was isolated with the Qiagen RNeasy minikit and was subjected to Affymetrix oligonucleotide microarray analysis by using an HG_U133A chip. Five replicates, including two control and three arsenic-treated NB4 samples, were studied. With the expectation that only a small fraction of genes is differentially expressed between samples under different treatments, the brightness of chips for the samples was adjusted to comparable level by normalizing the CEL file of signal values and the probe pair (perfect match and mismatch) level data of the Affymetrix expression chips, with the method of “invariant set normalization” (21). The normalized CEL data were then used to estimate the perfect match/mismatch-model-based expression index (with SE) for the probe sets (22), leading to the further computation of the fold changes and their 90% confidence intervals (21). The lower bound of a 90% confidence interval, a conservative estimate of the fold change, was then used to identify differentially expressed genes. The computing was performed with dchip 1.2.
Real-Time PCR. Detection of hTERT was described (16). Expression of other genes was determined by reverse transcription followed by SYBR green real-time PCR. All primer sequences for the genes tested are available on request. cDNA was generated by first heating a 15-μl mixture containing 15 μg of total RNA and 1 μg of random primers (Promega) to 70°C for 5 min. After immediate chilling on ice, 5 μl of 5× reaction buffer, 5 μl of dNTP (2.5 mM each), 40 units of RNase inhibitor, and 200 units of Moloney murine leukemia virus reverse transcriptase (Promega) were added, and the mixture was incubated at 37°C for 1 h. Ten nanograms of cDNA was subjected to SYBR green quantitative real-time PCR. Every tube of 20 μl contained 500 nM each of primer, 200 μM dNTP, and PCR buffer with 1.75 mM MgCl2/0.5 μl of 15,000-fold diluted SYBR (Molecular Probes)/0.5 units of PlatinumTaq (Invitrogen). All primers were designed to cross introns and span <400 base pairs of the mRNA. All PCRs were performed at 95°C for 5 min, followed by 40 cycles of 95°C for 30 sec, 60°C for 30 sec, and 72°C for 30 sec. The signals were detected with the ABI 7700 sequence detection system. All of the signals were normalized by the expression levels of large acidic ribosomal protein (RPLP0).
Immunohistochemical Staining. The cells were prepared by cytospin and were fixed with methanol and then acetone for 2 min each. The staining procedures were performed with the Vectastain kit (Vector Laboratories). After blocking, the cells were stained with 5 μg/ml antieosinophil peroxidase antibody (Research Diagnostics, Flanders, NJ), followed by washes and anti-mouse secondary antibody. The reaction was stopped by washing with water. The slides were counterstained with hematoxylin/eosin.
Detection of ROS by Flow Cytometry. NB4 cells treated with 0.75 μM arsenic for 10 days were washed with PBS and were resuspended in complete medium with original concentration of arsenic, followed by incubation with 0.5 μM dihydrorhodamine 123 (Sigma) for 30 min at 37°C. The ROS was determined by the fluorescent intensity by flow cytometry with excitation at 490 nm and emission at 520 nm.
ROS Detection by Luminol Chemiluminescence. To detect extracellular and intracellular ROS, 10 μM luminol was added to 1 × 106 cells in 2 ml of aerated complete PBS (PBS with 0.5 mM MgCl2/0.7 mM CaCl2/0.1% glucose) supplemented with 10 μg/ml horseradish peroxidase (HRP). The chemiluminescence was measured continuously in a Berthold LB9505 (Pforzheim, Germany) six-channel luminometer at 37°C for 30 min.
Superoxide Detection by Lucigenin-Derived Chemiluminescence. One million arsenic-treated or control cells were suspended in 2 ml of air-aerated complete PBS. Lucigenin (5 μM) was added to the cells, and the chemiluminescence was detected as described above.
PMA Stimulation. After 30 min of recording as described above, the chemiluminescence signals were than recorded for another 30 min after the addition of 50 nM of PMA into the reaction mixture.
Superoxide Detection by Cytochrome c Reduction. The procedures were performed by adding 1.5 mg/ml cytochrome c to 1 × 106 cells with or without 50 nM PMA or 300 units/ml superoxide dismutase. The mixture was shaken at 37°C for 1 h. The supernatant was measured by spectrophotometry at 550 nm. The amount of reduced cytochrome c was determined by converting the absorbance with extinction coefficient of 28 per mM. For inhibitor studies, 10 μM diphenyleneiodonium (DPI) was added to cells in complete PBS and was incubated for 5 min at 37°C before measurement of chemiluminescence or cytochrome c reduction.
RNA Interference. Three million NB4 cells growing in log phase were washed twice and were resuspended in 500 μl of electroporation buffer (21 mM Hepes/137 mM NaCl/5 mM KCl/0.7 mM Na2HPO4/6 mM glucose, pH 7.15) containing either 0.5 nmol of scrambled small interference RNA (siRNA) (5′-CACGCUCGGUCAAAAGGUUdTdT-3′) or p47PHOX siRNA (5′-GAGUACCGCGACAGACAUCdTdT-3′, Dharmacon, Lafayette, CO) in a 4-mm gap cuvette (BTX, Holliston, MA). The mixture was then electroporated with 1,500 μF and 200 volts by using Gene Pulser II (Bio-Rad). After 48 h, the cells were treated with 1.5 μM arsenic for another 48 h. The cells were harvested for ROS detection and immunoblotting.
Determination of Viability. NB4 cells (105 per ml) without or with arsenic (0.75 μM) treatment for 10 days were then exposed to PMA (0.2 nM) or bryostatin 1 (0.75 or 1 nM) with or without concomitant presence of 10 mM N-acetylcysteine (NAC), and the cell viability was followed up for another 6 days by using the Trypan blue exclusion method. At the fourth day of viability assay, control or arsenic-treated cells without or with NAC coincubation were evaluated by luminol plus HRP chemiluminescence. Bryostatin 1 (1 nM) was added in the middle of the assay to detect the induction of ROS production.
Results
Arsenic Effects on Gene Expression Profiles of NB4 Cells. To explore the molecular mechanisms of arsenic's therapeutic effects in the treatment of APL patients with daily continuous infusion of arsenic trioxide, we treated a human APL cell line, NB4, for 10 days with 0.75 μM arsenic trioxide, a dose slightly lower than the plasma trough levels achieved in APL patients (23). We reported previously that arsenic at this dose was able to down-regulate human telomerase hTERT transcription (16). Multiple replicate experiments were analyzed by microarray hybridizations, including three microarrays for arsenic-treated and two microarrays for control NB4 cells. The effect of arsenic treatment verified by real-time PCR showed the >99% down-regulation of hTERT expression, which is too low for reliable microarray analysis (data not shown). With arsenic exposure, NB4 cells continued to proliferate although at a slower rate compared with control (data not shown). Gene expression index was estimated for the samples with the perfect match/mismatch multiplicative statistical (22). The high correlation of gene expression index between samples ranges from 0.970 to 0.991, suggesting only a small fraction of genes is differentially expressed between the samples with and without arsenic exposure (data not shown). To identify the up- and down-regulated genes, a 90% confidence interval was computed for the fold change of the averaged expression index of each gene between the samples with and without arsenic treatment. The lower bound of a 90% confidence interval, a conservative estimate of the fold change, served as the practical way of identifying differentially expressed genes (21). Of 22,000 genes on the array, 15 and 9 were up- and down-regulated, respectively, by arsenic with a lower bound fold change ≥5 (Fig. 1). Real-time PCR confirmed the microarray results for all 14 genes randomly selected for verification (data not shown).
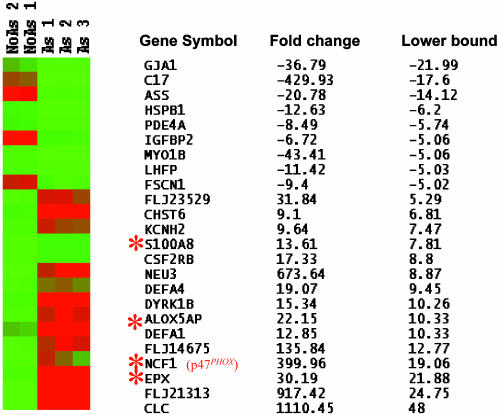
Fig. 1.
Microarray analysis of NB4 cells before and after arsenic treatment. Expression profiles of the 24 genes (with the lower bound fold change of the 90% confidence intervals ≥5) were shown across the five samples in Eisen's heat map. No As, untreated NB4 cells; As, arsenic-treated NB4 cells. Red and green colors represent high and low expression levels, respectively. The corresponding gene symbols, fold changes, and the lower bound fold change of the 90% confidence intervals are also listed. –, down-regulated gene expression. Those genes related to ROS production were marked with an asterisk. The genes are ordered from the most down-regulated genes to the highest up-regulated genes, based on the lower bound fold change.
Up-Regulation of NADPH Oxidase Components by Arsenic. Among those 14 selected were genes involved in oxidant production such as NADPH oxidase components p47PHOX (NCF1) (17), NADPH oxidase assembly scaffolding protein (S100A8) (24), arachidonate lipoxygenase activating protein (ALOX5AP) (25), and oxidation stress-related protein (eosinophil peroxidase, EPX) (refs. 26 and 27 and Fig. 1, asterisk). These findings strongly suggest that ROS production and NADPH oxidase activity are induced by arsenic. Further inspection of the raw microarray data revealed up-regulation (>2-fold) of another scaffolding protein, S100A9 (24), and all of the other NADPH oxidase components, including p67PHOX, CYBA (p22 subunit of cytochrome b558), CYBB (gp91PHOX), p40PHOX, and rac2, which is a small G protein important in activating the NADPH oxidase complex (data not shown). To confirm the increased levels of these ROS-related proteins, we performed immunohistochemical staining for eosinophil peroxidase (Fig. 2A) and immunoblotting for p47PHOX and p67PHOX (Fig. 2B). We also used real-time PCR and confirmed that mRNAs for all of the NADPH oxidase components were significantly up-regulated in NB4 cells after arsenic treatment (Fig. 2C). In these experiments, NB4 cellular morphology or expression of CD11b and CD15 were unaltered (data not shown), suggesting that differentiation does not play a major role, although induction of myeloid differentiation by arsenic (28) could trigger NADPH oxidase expression. These data indicated that arsenic potently induces components of NADPH oxidase.
Fig. 2.
Up-regulation of NADPH oxidase and eosinophil peroxidase expression by arsenic. (A) Absent immunohistochemical staining in control cells (Left) and intense staining of eosinophil peroxidase in arsenic-treated NB4 cells (Right). The staining in arsenic-treated cells depended on the primary antibody (data not shown). As, arsenic. (B) Immunoblotting of p47PHOX and p67PHOX shows dramatic increases in the protein levels in NB4 cells after arsenic treatment. α-Tubulin was used as a loading control. (C) Arsenic increased mRNA of all of the NADPH oxidase subunits in NB4 cells as measured by SYBR green quantitative real-time PCR.
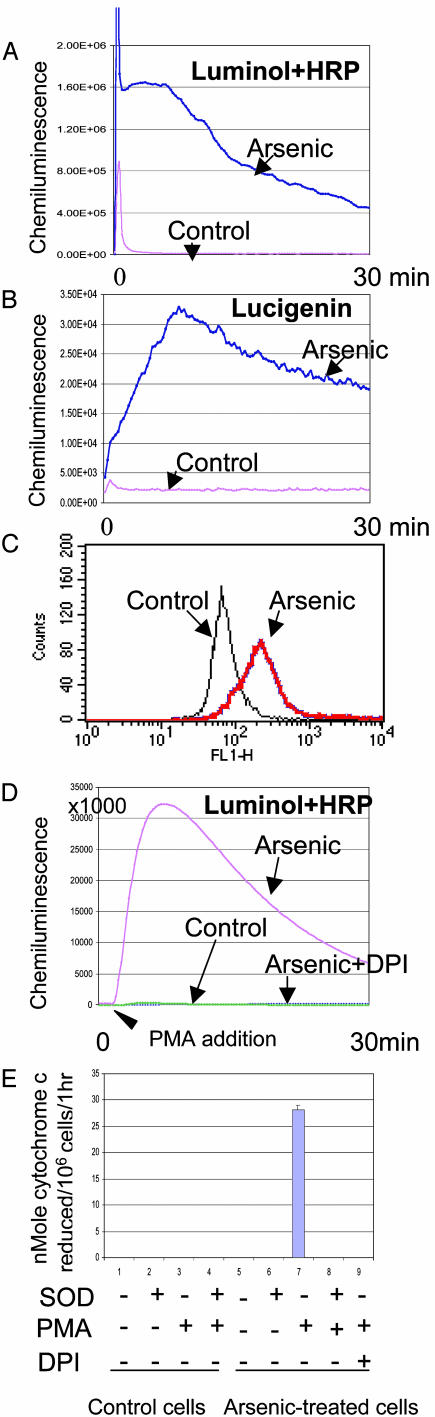
Induction of ROS by Arsenic in Myeloid Cells. Corresponding to the activation of NADPH oxidase, ROS was greatly enhanced by arsenic treatment as verified by several approaches for detection of ROS such as chemiluminescence of luminol with or without HRP, lucigenin, and flow cytometry by using dihydrorhodamine 123 as a probe (Fig. 3 A–C). The enhancement of chemiluminescence of either luminol or lucigenin was dramatically amplified by the addition of 50 nM PMA (Fig. 3D and data not shown) that is known to stimulate NADPH oxidase activity (17). These data indicate that arsenic dramatically induces the latent form of NADPH oxidase. We also verified the production of superoxide through its ability to reduce cytochrome c (Fig. 3E). The reduction of cytochrome c in arsenic-treated cells challenged with PMA was completely rescued by addition of superoxide dismutase, confirming the ROS produced in arsenic-treated NB4 cells was superoxide anion (Fig. 3E). The absence of signals of cytochrome c reduction in arsenic-treated cells without challenge with PMA might reflect the lower sensitivity of this assay compared with chemiluminescence. We also tested arsenic induction of ROS and NADPH oxidase activation in other myeloid cell lines, including ML1 (monocytic), PLB-985 (monoblastic leukemia), HL60 (promyelocytic leukemia), and U937 (monocytic leukemia). We found that all but U937 produced higher amount of ROS as measured by chemiluminescence after arsenic treatment, with the corresponding elevation of mRNAs of both p47 PHOX and p67PHOX (data not shown).
Fig. 3.
Induction of ROS formation from NADPH oxidase by arsenic. Chemiluminescence measured with luminol plus HRP (A), lucigenin (B), and flow cytometry (C) by using dihydrorhodamine 123 showed significant ROS induction in NB4 cells treated with arsenic. (D and E) Luminol plus HRP (D) and cytochrome c reduction (E) showed that induction of ROS was further dramatically enhanced by the addition of 50 nM PMA. Incubation with DPI completely blunted the baseline, arsenic-induced, or PMA-stimulated chemiluminescence and cytochrome c reduction. SOD, superoxide dismutase.
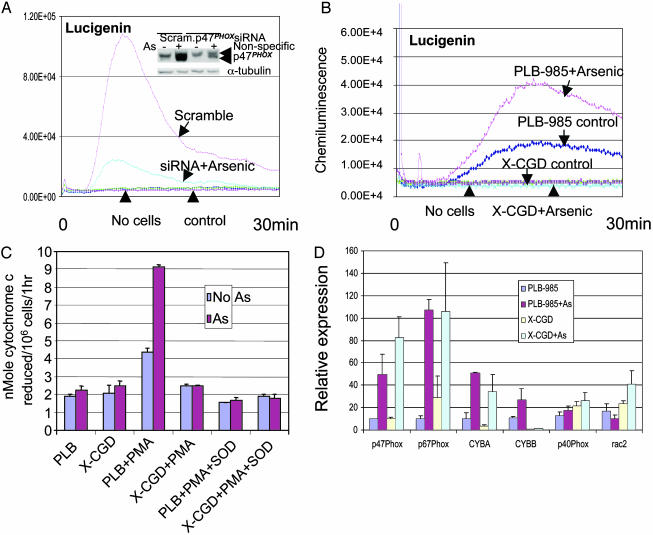
NADPH Oxidase Is the Main Source of Arsenic-Induced ROS Production. Although arsenic induced both NADPH oxidase and ROS production, whether the ROS came from NADPH oxidase remains to be determined. The dramatic PMA stimulation of ROS production by arsenic-treated NB4 cells strongly implicates NADPH oxidase as the source, because PMA is known to activate latent NADPH oxidase (17). To address this issue further, we first used DPI, a flavoprotein inhibitor of NADPH oxidase. We found that arsenic-treated cells did not exhibit any ROS production after the addition of DPI, even after PMA stimulation (Fig. 3 D and E). Although DPI is regarded as an NADPH oxidase inhibitor, it is not specific (29). Hence, we inhibited P47PHOX expression by siRNA in NB4 cells. ROS production as measured by chemiluminescence of lucigenin or luminol plus HRP after arsenic treatment and PMA stimulation was dramatically reduced after suppression of p47PHOX induction by arsenic (Fig. 4A and data not shown). We also used the myeloid cell line PLB-985 and its gp91PHOX-knockout derivative, X-CGD (30), as another genetic approach to test our hypothesis that NADPH oxidase is the main source of arsenic-induced ROS production. We found that X-CGD exhibited no baseline, arsenic-induced, or PMA-stimulated ROS as measured by luminol plus HRP (data not shown) and lucigenin chemiluminescence (Fig. 4B and data not shown). In contrast, the parental cells PLB-985 responded to arsenic with enhanced lucigenin signals (Fig. 4B) and luminol plus HRP (data not shown). PMA stimulation further augmented the chemiluminescence as measured by using luminol plus HRP and lucigenin in the parental cells (data not shown). Cytochrome c reduction also showed increased superoxide production in PLB-985 after arsenic treatment and PMA stimulation, but not in X-CGD (Fig. 4C). Most of the NADPH oxidase components were also up-regulated by arsenic in PLB-985 and X-CGD (Fig. 4D), but due to the lack of the functional gp91PHOX component in X-CGD cells (30), there was no functional NADPH oxidase enzyme complex. These results indicate that NADPH oxidase is the main source of arsenic-induced ROS production.
Fig. 4.
NADPH oxidase is the main source of arsenic-induced ROS. (A) p47PHOX protein was induced by arsenic in cells transfected with scrambled (Scram.) siRNA, but this induction is diminished in cells transfected with p47PHOX siRNA (Inset). The lucigenin chemiluminescence induced by arsenic in p47PHOX siRNA-transfected cells was significantly diminished compared with those transfected with scrambled siRNA. As, arsenic. (B) Lucigenin chemiluminescence was absent in X-CGD cells before and after arsenic treatment. In contrast, the baseline chemiluminescence of the parental cells, PLB-985, was enhanced by arsenic treatment. (C) Cytochrome c reduction assay showed PMA-enhanced superoxide production in arsenic-treated parental cells PLB-985 but not in X-CGD cells. (D) mRNAs of most NADPH oxidase subunits in PLB-985 or X-CGD cells were increased by arsenic, as determined by real-time PCR.
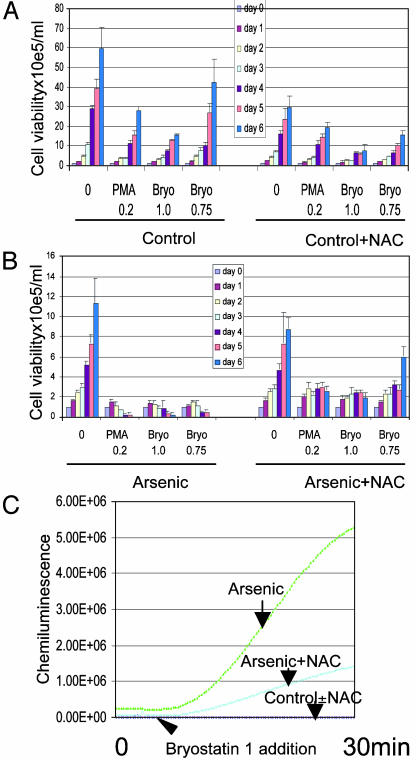
Synergism of Cytotoxicity Between Arsenic and PMA or Bryostatin 1. Because oxidants can exert cytotoxicity (12, 31–33), and PMA dramatically enhances oxidant production in cells pretreated with arsenic, we tested whether arsenic and PMA are synergistically cytotoxic. First, we identified a dose of PMA (0.2 nM) that did not significantly reduce cell viability (Fig. 5A), but could enhance ROS production in arsenic-treated NB4 cells (data not shown). At this dose, PMA potently synergized with arsenic in cytotoxicity (Fig. 5B). We extended our finding to a clinically used PMA analog, bryostatin 1, a macrocyclic lactone isolated from a marine bryozoan Bugula neritina (34). Its antileukemic effect was suggested by in vitro studies (35, 36) and preliminary clinical trials (37, 38). We identified doses of bryostatin 1 (0.5–1 nM) that still maintained cellular proliferation (Fig. 5A) and greatly enhanced ROS production (data not shown). Bryostatin 1 also synergized with arsenic in killing the cells (Fig. 5B). The synergism between arsenic and either PMA or bryostatin 1 diminished after the addition of the radical scavenger NAC (Fig. 5B), with corresponding blunting of the chemiluminescence induced by arsenic with or without 1 nM bryostatin 1 (Fig. 5C). We conclude that arsenic and either PMA or bryostatin 1 synergize to enhance ROS production and tumoricidal activity.
Fig. 5.
Synergistic cytotoxicity between arsenic and either PMA or bryostatin 1 (Bryo). (A) PMA or byrostatin 1 at the doses used did not prevent cell proliferation. Without arsenic, the cells continued proliferation in the presence of PMA or bryostatin 1 in absence (Left) or presence (Right) of NAC. NAC had mild toxicity to NB4 cells. (B Left) Synergistic toxicity is evident in cells pretreated with arsenic and then exposed to PMA or byrostatin 1, as compared with cells pretreated with arsenic only. (Right) NAC blocks the synergistic toxicity. (C) NAC decreased arsenic-induced ROS signals. After the addition of 1 nM bryostatin, the arsenic-treated cells showed significant induction of chemiluminescence of luminol plus HRP than those cells coincubated with NAC.
Discussion
Although arsenic has played a significant role in human medicinal history, the mechanisms underlying arsenic's antileukemic activity remain enigmatic. Its ability to induce ROS production has been reported but the source of the ROS remained unknown. Here, we provide evidence that NADPH oxidase induced by arsenic is central to the mechanism of arsenic-mediated ROS production. Not only were NADPH oxidase components induced by arsenic concordant with ROS production in different leukemic cells, leukemic cells with p47PHOX levels diminished by RNA interference were minimally responsive to arsenic. Moreover, cells depleted of the gp91PHOX subunit of NADPH oxidase by homologous recombination were totally unresponsive to arsenic as compared with the wild-type parental cells in ROS production.
The role of NADPH oxidase in apoptosis is implicated by the findings that zinc, vanadium, and brain-derived neurotropic factor could all induce NADPH oxidase activity and ROS production, leading to the death of nonmyeloid cells (12, 31–33). In our studies, which began with gene expression analysis, we observed that arsenic significantly induces components of the NADPH oxidase complex in the APL cell line NB4, as well as other leukemic cell lines. The protection of cells by the oxidant scavenger NAC against arsenic plus either PMA or bryostatin 1 further supports the role of ROS in the cytotoxicity of leukemic cells.
Phosphorylation of the p47PHOX subunit by PMA in protein kinase C-dependent mechanisms triggers the assembly and activation of a functional NADPH oxidase complex (17, 18). The abundant NADPH oxidase subunits induced by arsenic should facilitate the activation of NADPH oxidase by PMA or its clinically used analog, bryostatin 1. We exploited this molecular regulatory mechanism and used the synergism between arsenic and bryostatin 1 to kill leukemic cells. The concentrations of arsenic or bryostatin 1 were much lower than experimentally or clinically used, yet the synergistic tumoricidal effect is remarkable at these concentrations when the two are used together. Our biochemical and genetic studies reported here have uncovered a major role of NADPH oxidase in arsenic-induced ROS production and cytotoxicity, and also provided a conceptual basis for the development of clinical protocols for the treatment of leukemias, in particular APL, through the synergism between arsenic and bryostatin 1.
Acknowledgments
We thank M. Dinauer for PLB-985 and X-CGD cell lines and F. Racke for assistance with immunohistochemical staining and flow cytometry. This work was supported by National Institutes of Health Grants CA51497 (to C.V.D.) and ES03760 and ES03819 (to A.A.K. and M.A.T.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: APL, acute promyelocytic leukemia; ROS, reactive oxygen species; PMA, phorbol myristate acetate; DPI, diphenyleneiodonium; HRP, horseradish peroxidase; siRNA, small interference RNA; NAC, N-acetylcysteine.
References
- 1.Zhu, J., Chen, Z., Lallemand-Breitenbach, V. & de The, H. (2002) Nat. Rev. Cancer 2, 705–713. [DOI] [PubMed] [Google Scholar]
- 2.Miller, W. H., Jr., Schipper, H. M., Lee, J. S., Singer, J. & Waxman, S. (2002) Cancer Res. 62, 3893–3903. [PubMed] [Google Scholar]
- 3.Soignet, S. L., Maslak, P., Wang, Z. G., Jhanwar, S., Calleja, E., Dardashti, L. J., Corso, D., DeBlasio, A., Gabrilove, J., Scheinberg, D. A., et al. (1998) N. Engl. J. Med. 339, 1341–1348. [DOI] [PubMed] [Google Scholar]
- 4.Soignet, S. L., Frankel, S. R., Douer, D., Tallman, M. S., Kantarjian, H., Calleja, E., Stone, R. M., Kalaycio, M., Scheinberg, D. A., Steinherz, P., et al. (2001) J. Clin. Oncol. 19, 3852–3860. [DOI] [PubMed] [Google Scholar]
- 5.Warrell, R. P. J. (1993) N. Engl. J. Med. 329, 177–189. [DOI] [PubMed] [Google Scholar]
- 6.Zhu, X. H., Shen, Y. L., Jing, Y. K., Cai, X., Jia, P. M., Huang, Y., Tang, W., Shi, G. Y., Sun, Y. P., Dai, J., et al. (1999) J. Natl. Cancer Inst. 91, 772–778. [DOI] [PubMed] [Google Scholar]
- 7.Perkins, C., Kim, C. N., Fang, G. & Bhalla, K. N. (2000) Blood 95, 1014–1022. [PubMed] [Google Scholar]
- 8.Yoshida, H., Kitamura, K., Tanaka, K., Omura, S., Miyazaki, T., Hachiya, T., Ohno, R. & Naoe, T. (1996) Cancer Res. 56, 2945–2948. [PubMed] [Google Scholar]
- 9.Muller, S., Matunis, M. J. & Dejean, A. (1998) EMBO J. 17, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hei, T. K., Liu, S. X. & Waldren, C. (1998) Proc. Natl. Acad. Sci. USA 95, 8103–8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abernathy, C. O., Liu, Y. P., Longfellow, D., Aposhian, H. V., Beck, B., Fowler, B., Goyer, R., Menzer, R., Rossman, T., Thompson, C., et al. (1999) Environ. Health Perspect. 107, 593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lynn, S., Gurr, J. R., Lai, H. T. & Jan, K. Y. (2000) Circ. Res. 86, 514–519. [DOI] [PubMed] [Google Scholar]
- 13.Grad, J. M., Bahlis, N. J., Reis, I., Oshiro, M. M., Dalton, W. S. & Boise, L. H. (2001) Blood 98, 805–813. [DOI] [PubMed] [Google Scholar]
- 14.Rea, M. A., Gregg, J. P., Qin, Q., Phillips, M. A. & Rice, R. H. (2003) Carcinogenesis 24, 747–756. [DOI] [PubMed] [Google Scholar]
- 15.Zheng, X. H., Watts, G. S., Vaught, S. & Gandolfi, A. J. (2003) Toxicology 187, 39–48. [DOI] [PubMed] [Google Scholar]
- 16.Chou, W. C., Hawkins, A. L., Barrett, J. F., Griffin, C. A. & Dang, C. V. (2001) J. Clin. Invest. 108, 1541–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Babior, B. M. (1999) Blood 93, 1464–1476. [PubMed] [Google Scholar]
- 18.Groemping, Y., Lapouge, K., Smerdon, S. J. & Rittinger, K. (2003) Cell 113, 343–355. [DOI] [PubMed] [Google Scholar]
- 19.Royer-Pokora, B., Kunkel, L. M., Monaco, A. P., Goff, S. C., Newburger, P. E., Baehner, R. L., Cole, F. S., Curnutte, J. T. & Orkin, S. H. (1986) Nature 322, 32–38. [DOI] [PubMed] [Google Scholar]
- 20.Dinauer, M. C. & Orkin, S. H. (1992) Annu. Rev. Med. 43, 117–124. [DOI] [PubMed] [Google Scholar]
- 21.Li, C. & Hung Wong, W. (2001) Genome Biol. 2, RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, C. & Wong, W. H. (2001) Proc. Natl. Acad. Sci. USA 98, 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen, Z. X., Chen, G. Q., Ni, J. H., Li, X. S., Xiong, S. M., Qiu, Q. Y., Zhu, J., Tang, W., Sun, G. L., Yang, K. Q., et al. (1997) Blood 89, 3354–3360. [PubMed] [Google Scholar]
- 24.Doussiere, J., Bouzidi, F. & Vignais, P. V. (2002) Eur. J. Biochem. 269, 3246–3255. [DOI] [PubMed] [Google Scholar]
- 25.Bonizzi, G., Piette, J., Schoonbroodt, S., Greimers, R., Havard, L., Merville, M. P. & Bours, V. (1999) Mol. Cell. Biol. 19, 1950–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henderson, J. P., Byun, J., Williams, M. V., McCormick, M. L., Parks, W. C., Ridnour, L. A. & Heinecke, J. W. (2001) Proc. Natl. Acad. Sci. USA 98, 1631–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Dalen, C. J. & Kettle, A. J. (2001) Biochem. J. 358, 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guillemin, M. C., Raffoux, E., Vitoux, D., Kogan, S., Soilihi, H., Lallemand-Breitenbach, V., Zhu, J., Janin, A., Daniel, M. T., et al. (2002) J. Exp. Med. 196, 1373–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, Y. & Trush, M. A. (1998) Biochem. Biophys. Res. Commun. 253, 295–299. [DOI] [PubMed] [Google Scholar]
- 30.Zhen, L., King, A. A., Xiao, Y., Chanock, S. J., Orkin, S. H. & Dinauer, M. C. (1993) Proc. Natl. Acad. Sci. USA 90, 9832–9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, S. H., Won, S. J., Sohn, S., Kwon, H. J., Lee, J. Y., Park, J. H. & Gwag, B. J. (2002) J. Cell Biol. 159, 821–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noh, K. M. & Koh, J. Y. (2000) J. Neurosci. 20, RC111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, L., Medan, D., Mercer, R., Overmiller, D., Leornard, S., Castranova, V., Shi, X., Ding, M., Huang, C. & Rojanasakul, Y. (2003) J. Cell Physiol. 195, 99–107. [DOI] [PubMed] [Google Scholar]
- 34.Pettit, G. R., Herald, C. L., Doubek, D. L., Herald, D. L., Arnold, E. & Clardy, J. (1982) J. Am. Chem. Soc. 104, 6846–6848. [Google Scholar]
- 35.Jones, R. J., Sharkis, S. J., Miller, C. B., Rowinsky, E. K., Burke, P. J. & May, W. S. (1990) Blood 75, 1319–1323. [PubMed] [Google Scholar]
- 36.Wang, S., Wang, Z., Dent, P. & Grant, S. (2003) Blood 101, 3648–3657. [DOI] [PubMed] [Google Scholar]
- 37.Varterasian, M. L., Mohammad, R. M., Eilender, D. S., Hulburd, K., Rodriguez, D. H., Pemberton, P. A., Pluda, J. M., Dan, M. D., Pettit, G. R., Chen, B. D., et al. (1998) J. Clin. Oncol. 16, 56–62. [DOI] [PubMed] [Google Scholar]
- 38.Cragg, L. H., Andreeff, M., Feldman, E., Roberts, J., Murgo, A., Winning, M., Tombes, M. B., Roboz, G., Kramer, L. & Grant, S. (2002) Clin. Cancer Res. 8, 2123–2133. [PubMed] [Google Scholar]