Abstract
The use of covalent chemistry to track biomolecules in their native environment—a focus of bioorthogonal chemistry—has received considerable interests recently among chemical biologists and organic chemists alike. To facilitate wider adoption of bioorthogonal chemistry in biomedical research, a central effort in the last few years has been focused on the optimization of a few known bioorthogonal reactions, particularly with respective to reaction kinetics improvement, novel genetic encoding systems, and fluorogenic reactions for bioimaging. During these optimizations, three strategies have emerged, including the use of ring strain for substrate activation in the cycloaddition reactions, the discovery of new ligands and privileged substrates for accelerated metal-catalysed reactions, and the design of substrates with pre-fluorophore structures for rapid “turn-on” fluorescence after selective bioorthogonal reactions. In addition, new bioorthogonal reactions based on either modified or completely unprecedented reactant pairs have been reported. Finally, increasing attention has been directed toward the development of mutually exclusive bioorthogonal reactions and their applications in multiple labeling of a biomolecule in cell culture. In this feature article, we wish to present the recent progress in bioorthogonal reactions through the selected examples that highlight the above-mentioned strategies. Considering increasing sophistication in bioorthogonal chemistry development, we strive to project several exciting opportunities where bioorthogonal chemistry can make a unique contribution to biology in near future.
Introduction
The development of ectopically expressed green fluorescent protein (GFP) and its many variants has dramatically increased our understanding of protein dynamics and function in living systems.1–3 However, this genetic tagging approach is not well suited for the study of other biomolecules such as nucleic acids, glycans, lipids and small-molecule metabolites, as well as protein posttranslational modifications in living systems. To overcome this limitation, a reactivity-based bioorthogonal chemistry approach has been successfully developed recently.4 In this two-step approach, first, a small chemical reporter such as aldehyde, azide, alkyne or alkene is introduced into the biomolecule(s) of interest site-selectively via an appropriate biosynthetic or biochemical pathway; then, a bioorthogonal chemical reaction is performed in situ to allow selective ligation of biophysical probes carrying the cognate reactive groups with the pre-tagged biomolecule(s) of interest. New insights into a wide range of biological processes, e.g., glycome imaging,5 protein lipidation and lipid trafficking,6 and activity-based protein profiling,7 to name a few, have been gained using the bioorthogonal chemistry-based approaches.
While the use of a unique reactive group as “chemical handle” to direct selective reaction in complex biological environment was reported long ago,8, 9 the coinage of “bioorthogonal” term by Bertozzi in 2003 and formalization of “bioorthogonal chemistry” shortly afterwards10, 11 unleashed a flurry of activities by many research groups in searching for new bioorthogonal reactions and their biological applications. Since our last feature article published in this Journal,12 substantial progress has been made with the existing bioorthogonal reactions, including: (i) the optimization of reaction kinetics through either substrate activation or the use of metal catalysts; (ii) the development of novel genetic encoding systems to harvest the power of bioorthogonal reactions for biological applications; and (iii) the design of fluorogenic substrates to allow facile monitoring and interrogation of living systems. Besides, new bioorthogonal reactions involving novel reactant pairs have also been reported. Considering that a special issue on bioorthogonal chemistry was published else where recently,13 we decided to focus on the major advance in the field since 2010. We will use the selected examples to highlight the chemical strategies for reaction optimization and new reaction development. We will conclude by striving to project some exciting new opportunities in near future for the next phase of bioorthogonal chemistry development.
Copper-catalysed azide–alkyne cycloaddition
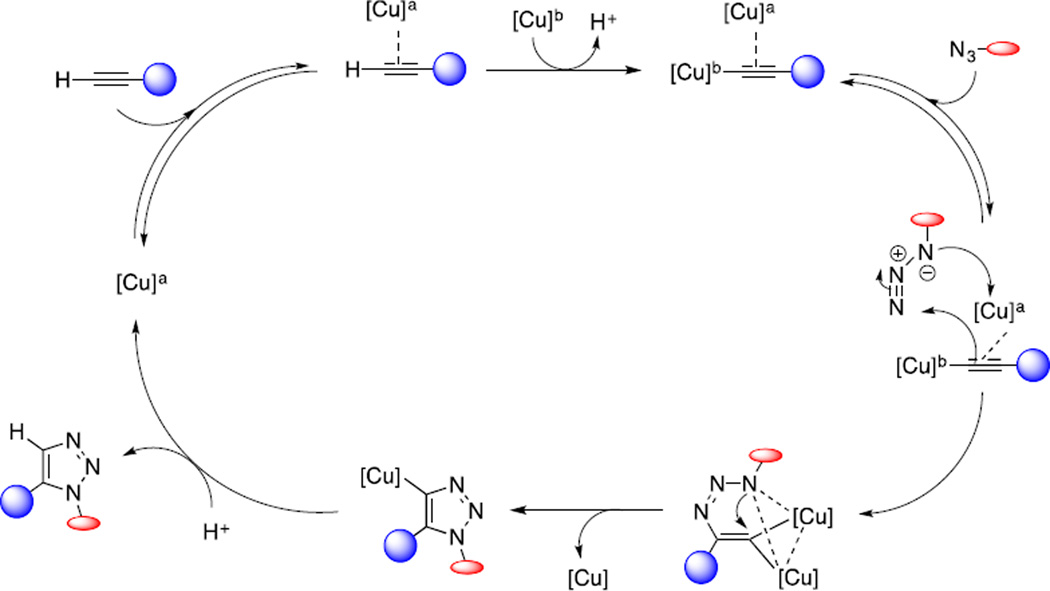
Transition metals are used by enzymes to catalyse chemical reactions inside living cells. The most popular metal catalysed reaction that took the center stage of bioorthogonal chemistry in the last decade is copper-catalysed azide-alkyne cycloaddition (CuAAC), also known widely as “click chemistry”. The 1,3-dipolar cycloaddition reaction between azide and alkyne was initially reported by Huisgen in 1963.14 In 2002,Sharpless and Meldal independently discovered dramatic rate acceleration when Cu(I) salt was used.15, 16 Based on DFT calculations, Sharpless and co-workers proposed a stepwise mechanism involving a mononuclear Cu-acetylide intermediate.17 Later, kinetic studies suggested the presence of two copper centers interacting with one or two alkynes and one azide, bringing out questions about the precise nature of dinuclear system.18 A recent report provided direct evidence about the presence of dinuclear copper complex,19 a major step toward finally unraveling CuAAC mechanism (Scheme 1). This proposed mechanism is consistent with two experimental observations: first, using heat-flow calorimetry to follow the cycloaddition reaction, the pre-formed mononuclear copper acetylide was unreactive before addition of an exogenous copper salt; and second, in crossover studies with the isotopically enriched exogenous copper catalyst, direct-injection time-of-flight mass spectrometry (TOF-MS) indicated that the two copper centers were equivalent and the formation of carbon-nitrogen bonds proceeded in a stepwise manner.
Scheme 1.
Proposed mechanism for the copper-catalysed azide-alkyne cycloaddition with two copper atoms.19
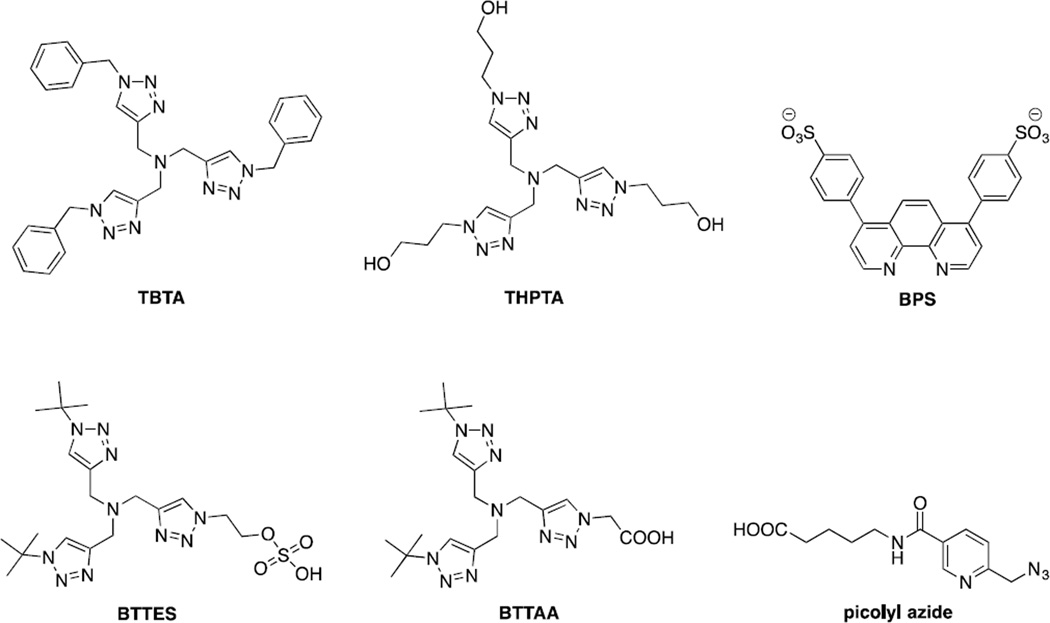
The CuAAC can be further accelerated by the use of ligands with stabilizing effect such as tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA)20 and tris(3-hydroxypropyltriazolylmethyl)amine (THPTA)21 (Figure 1). The more water-soluble THPTA ligand was successfully employed for the CuAAC-mediated functionalization of cowpea mosaic virus which was also achieved previously using sulfonated bathophenanthroline (BPS) ligand22 (Figure 1). Recently, Wu and co-workers reported two new biocompatible ligands for CuAAC: BTTES23 and BTTAA24 (Figure 1). These new ligands allowed CuAAC to be employed in noninvasive imaging of fucosylated glycans during early embryogenesis of zebrafish. Among the various ligands, BTTAA exhibited faster reaction rate than BTTES, TBTA and THPTA.24 Because of known cytotoxicity of Cu(I) salts, Pezacki and co-workers examined the toxicity of various copper complexes on four commonly used human cell lines by measuring their effect on mitochondrial activity based on the metabolism of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT), using inductively coupled plasma mass spectrometry to study their cellular uptake, and using coherent anti-Stokes Raman scattering (CARS) microscopy to study their effects on lipid metabolism.25 They found that the ligand environment around copper center influences all three parameters. Interestingly, the copper complex with the simplest ligand, L-histidine, showed no apparent toxicity when CuAAC was used to label the alkyne-containing glycans on the surface of human hepatoma cells.
Figure 1.
Structure of ligands for biocompatible copper-catalysed azide-alkyne cycloaddition.
In 2009, Zhu and co-workers reported chelation-assisted CuAAC in which the azide substrates containing auxiliary ligands such as 2-picolylazide (Figure 1) and 2-azidomethyl-quinoline were privileged substrates for the cycloaddition reactions.26 The use of oligotriazole ligands such as TBTA was found to further accelerate the reaction.27 Based on kinetic and structural data, they proposed a mechanism for chelation-assisted, Cu(OAc)2-accelarated cycloaddition involving the formation of critical Cu(I)/Cu(II) dinuclear core.28 The two copper centers serve discrete roles, one being Lewis acid and the other being active Cu(I) catalyst. The fast chelation of the copper by the azide substrates proceeds prior to the formation of Cu-acetylide with deprotonation of the terminal alkyne as the rate-limiting step. With this insight, the Ting group applied picolyl azide as a privileged substrate in CuAAC for protein labeling in live mammalian cells.29 To target picolyl azide to a protein of interest, they employed the probe incorporation mediated by enzyme (PRIME) method developed in their lab.30 Lipoic acid ligase from E. coli (LplA) was engineered to recognize the picolyl azide-containing substrate for its efficient incorporation into LplA acceptor peptide (LAP)-conjugated cyan fluorescent protein (CFP) expressed on the surface of human embryonic kidney (HEK) cells. CuAAC was then performed with the Alexa Fluor-647-conjugated alkyne. The optimum fluorescent labeling was achieved when BTTAA and 40 µM CuSO4 were used.
Strain promoted azide–alkyne cycloaddition
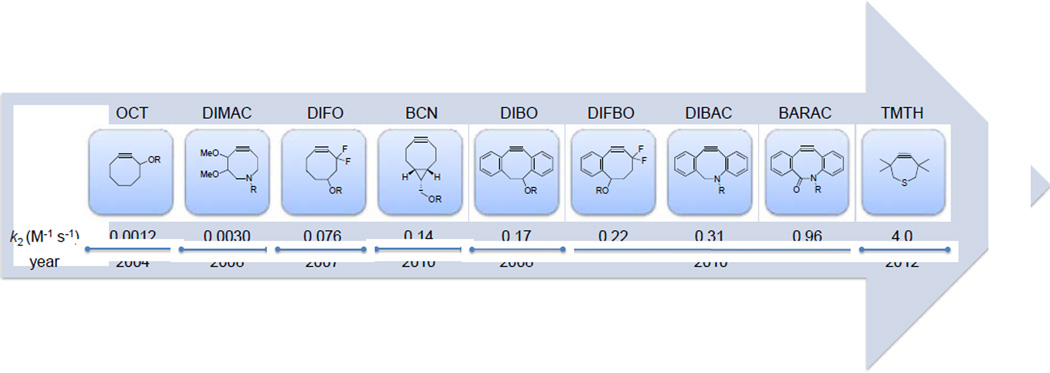
To avoid copper toxicity,25 several groups have sought to accelerate the rate of azide–alkyne cycloaddition through alkyne substrate activation, leading to successful development of the strain-promoted azide–alkyne cycloaddition (SPAAC), also known as Cu-free click chemistry. The first example of SPAAC was reported by Bertozzi and co-workers in 2004 when they used cyclooctyne (OCT) as an activated reaction partner for azide with the second-order rate constant k2 of 0.0012 M−1s−1.31 Further optimization led to the introduction of difluorocyclooctyne (DIFO, Figure 2) that showed 63-fold rate enhancement in the cycloaddition with benzyl azide.32 Another early work on cyclooctyne derivatives was reported by Boons and co-workers in which the reactivity of cyclooctyne was increased by fusing two benzene rings to cyclooctyne to generate dibenzocyclooctyne (DIBO).33 The rate constant of the DIBO-mediated cycloaddition reaction with benzyl azide was almost three orders of magnitude faster than simple cyclooctyne.
Figure 2.
Evolution of strained alkyne reagents for strain-promoted azide-alkyne cycloaddition showing improved kinetics.
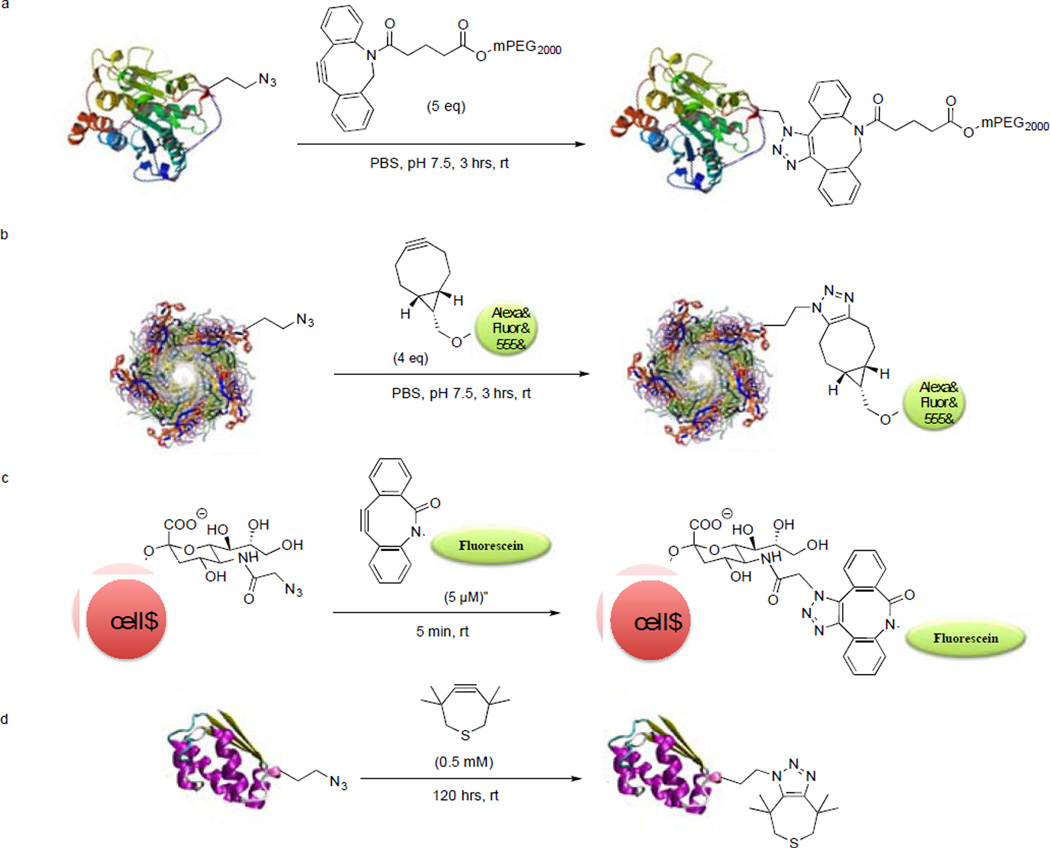
In the following years, numerous cyclooctyne derivatives displaying various improvements were reported. The van Delft group reported two cyclooctyne analogs, aza-dibenzocyclooctyne (DIBAC)34 and bicyclo[6.1.0]nonyne (BCN)35 that are synthetically more accessible (Figure 2). DIBAC represents a hybrid structure of DIBO and aza-dimethoxycyclooctyne (DIMAC);36 by combining favorable kinetics of DIBO and hydrophilicity of DIMAC, it showed faster kinetics than DIBO, DIFO and DIMAC (Figure 2). To demonstrate the efficiency of DIBAC in SPAAC in vitro, azidohomoalanine-encoded Candida antarctica lipase B (AHA-CalB)37 was treated with 5 equiv PEG2000-linked DIBAC for 3 hours, and the PEGylated product was obtained in essentially quantitative yield (Scheme 2a). Moreover, horseradish peroxidase (HRP) was modified via the diazo transfer38 to bear azide functionality, and the resulting HRP-N3 was PEGylated under the same conditions as with AHA-CalB with similar results.
Scheme 2.
Bioorthogonal labeling via strain-promoted azide-alkyne cycloaddition.
One of the factors hindering broader biological applications of SPAAC is the limited commercial availability of the cyclooctyne reagents and the typically lengthy synthetic routes for their preparation. This prompted the van Delft group to develop a synthetically readily accessible alkyne BCN.35 BCN was prepared via cyclopropanation of the commercially available 1,5-cyclooctadiene in 4 steps with an overall yield of 61%. The symmetric structure of BCN also simplifies NMR analysis of the cycloadducts. The design of BCN was inspired by the known effect of fusing a cyclopropane ring to enhance reactivity first reported in 1981.39 Indeed, the fusion of cyclopropane ring led to 70-fold enhancement in reactivity (k2 = 0.14 M−1s−1 with benzyl azide vs. 0.0012 M−1s−1 for OCT). To test the suitability of BCN derivatives for protein labeling, the azidohomoalanine-encoded virus capsid proteins were treated with BCN-Alexa Fluor 555 in phosphate buffer, pH 7.5, for 3 hours (Scheme 2b). The protein conjugate was detected by in-gel fluorescence analysis and the identity of the conjugate was confirmed by mass spectrometry. In addition, living human melanoma MV3 cells known to produce abundant surface glycans were metabolically labeled with N-azidoacetyl-D-mannosamine. The azido-labeled cells were treated with 60µM BCN-biotin for 1 hour at 20°C followed by staining with streptavidin-Alexa Fluor 488. Both confocal microscopy and flow cytometry analysis revealed a higher efficiency of azido-glycan labeling by BCN than by DIBO. Furthermore, three-dimensional (3D) imaging of MV3 cells was achieved by seeding cells into 3D collagen lattices resulting in spontaneous and vigorous invasion. Consistent with the known function of sialic acid in cell adhesion and migration, the fluorescently labeled glycans were distributed at the leading edge of filopodia, focal clusters at actin-rich contact sites to collagen fibers, and heavy deposits into the tissue matrix from the trailing edge, which were observed at submicron resolution.
In the same year, Bertozzi and co-workers reported two new cyclooctyne derivatives, biarylazacyclooctynone (BARAC)40 and difluorobenzocyclooctyne (DIFBO).41 Aside from improvement in reaction kinetics, these two compounds were readily prepared. The development of BARAC was inspired by the reported rate enhancement brought about by the dibenzo system in DIBO and the addition of an sp2-like center in DIBAC. The reaction of BARAC with benzyl azide is 12 times faster (k2= 0.96 M−1s−1) than DIFO and 800 times faster than OCT. The utility of BARAC in live cell imaging was demonstrated by modifying the azido-encoded glycans in Jurkat cells (Scheme 2c). The cells were treated with various concentrations of BARAC-biotin for 1 hour followed by FITC-avidin. Flow cytometry analysis showed specific labeling by BARAC-biotin at concentrations as low as 50 nM. On the other hand, computational studies predicted the rate-enhancing effect of fluorine substitution and aryl ring fusion with cyclooctyne.42–45 Therefore, combining two effects in the same structure was expected to lead to even faster cycloaddition than DIFO and DIBO. Unexpectedly, the triple bond in DIFBO became so reactive that DIFBO underwent spontaneous trimerization in solution.41 They ingenious strategy solved this instability problem by encapsulating DIFBO in β-cyclodextrin to form the inclusion complex. The cyclodextrin-stabilized DIFBO was then used successfully in SPAAC with a rate constant of 0.22 M−1s−1, almost three times greater than that of DIFO and DIBO.
The most recent development on strained alkyne was reported by Bertozzi and co-workers wherein they replaced two methylene groups with a sulfur atom to generate 3,3,6,6-tetramethyl-thiacycloheptyne (TMTH).46 They initially introduced a sulfur atom to replace one methylene group in cyclooctyne, which led to a decreased reactivity despite improved stability. To increase reactivity, they envisioned ring contraction to a 7-membered ring, known previously to react with phenyl azide and other 1,3-dipoles more favorably.47 Thus, thiacycloheptyne TMTH was identified that gives the fastest rate to date in SPAAC; TMTH reacted with benzyl azide in CD3CN cleanly with the second-order rate constant of 4.0 ± 0.4 M−1s−1. The fast and clean reaction was attributed to the smaller ring size and the propargylic methyl groups that shield the alkyne from the side reactions. The use of TMTH as a bioorthogonal labeling reagent was demonstrated through cell surface modification of azido-functionalized glycans as well as selective labeling of barstar protein encoding azidohomoalanine (Scheme 2d). Notably, an attempt to further enhance TMTH reactivity by fusing with the aryl rings resulted in unstable compounds, indicating a delicate balance between the reactivity and chemical stability.
There is also a growing interest in developing fluorogenic probes for situations that require sensitive detection with minimum background fluorescence and that washing is not feasible, e.g., in live animals. To this end, a coumarin-conjugated, fluorogenic BARAC derivative, coumBARAC, was reported by the Bertozzi group in 2011, which showed 10-fold increase in fluorescence quantum yield after the cycloaddition with benzyl azide.48 In 2012, the same group developed the fluorogenic azidofluorescein for CuAAC49, and most recently, Kele and co-workers introduced a new family of fluorogenic probes based on a conjugated benzothiazole and azidostyrene motif for both CuAAC and SPAAC.50
Tetrazine ligation
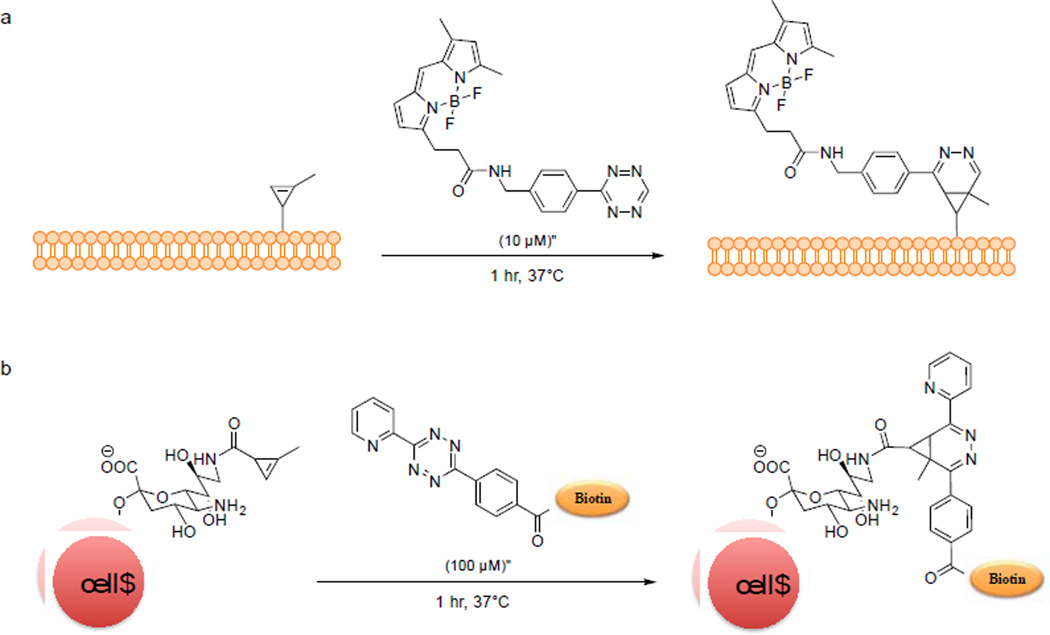
The tetrazine ligation was reported independently by the groups of Fox51 and Hilderbrand52 in 2008 based on the pioneering work of Sauer.53–55 The reaction proceeds extremely fast, with the second-order rate constant approaching 2000 M−1s−1. In addition to robust kinetics, the fluorophore-conjugated tetrazine reagents also exhibit “turn-on” fluorescence effect. Tetrazine quenches the fluorescence of a covalently linked fluorophore; upon cycloaddition with an alkene, the fluorescence is recovered due to the loss of the tetrazine moiety. The Weissleder group first demonstrated this effect when they used tetrazine-BODIPY reagent to imaging trans-cyclooctene-labeled taxol inside mammalian cells via tetrazine ligation.56 Because both norbornene and trans-cyclooctene are large in size, to harness the power of tetrazine ligation, the groups of Devaraj and Prescher independently explored the use of the smallest strained alkene, cyclopropene, as an activated reaction partner.57, 58 Due to its small size and absence in most eukaryotes, cyclopropene is an attractive bioorthogonal reporter. However, simple cyclopropene is susceptible to both nucleophilic attack and polymerization at room temperature. To overcome these limitations, Devaraj and co-workers placed a methyl group on the double bond, and the resulting methylcyclopropene carboxy amide showed improved stability along with a reduced reaction rate towards tetrazine benzyl alcohol (k2 = 0.137 ± 0.004M−1s−1).59 By switching carboxyamide moiety to carbamate moiety, they increased the rate by almost two orders of magnitude (k2 = 13 ± 2M−1s−1 in 8:1 H2O/DMSO). They then employed the methylcyclopropene carbamate as a bioorthogonal reporter to image phospholipids in SKBR3 breast cancer cells (Scheme 3a). At about the same time, Prescher and co-workers demonstrated the use of cyclopropene as a chemical reporter for proteins and glycans.57 In addition, the use of cyclopropene in tandem with azide for protein dual labeling was demonstrated by incubating a mixture of the cyclopropene- and azide-modified bovine serum albumin (BSA) with both tetrazine-rhodamine and DIBO-fluorescein. In-gel fluorescence analysis showed the selective labeling of both reporters by their respective probes. Furthermore, sialic acid was modified with methylcyclopropene (9-Cp-NeuAc) and the resulting modified carbohydrate was used for metabolic labeling of surface glycans on Jurkat cells. The presence of cyclopropene on the cell surface was probed with tetrazine-biotin (Scheme 3b). Flow cytometric analysis showed concentration-dependent labeling.
Scheme 3.
Cell surface labeling via tetrazine ligation using cyclopropene as a bioorthogonal reporter.
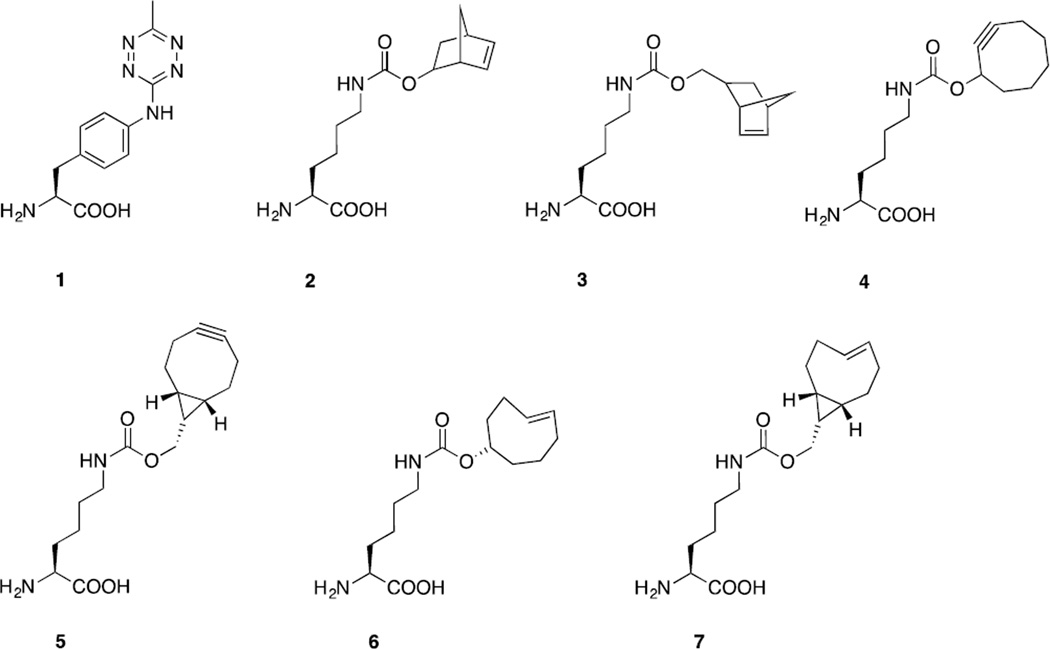
The challenge to apply this fast reaction to cellular studies was met through genetic encoding of unnatural amino acids (UAA) bearing either tetrazine or strained alkenes and alkyne such as norbornene, trans-cyclooctene (TCO), bicyclononyne (BCN), and strained trans-cyclooctene (s-TCO) (Figure 3). Fox, Mehl and co-workers developed a tetrazine derivative, 4-(6-methyl-s-tetrazine-3-yl)aminophenylalanine (1), with improved stability inside cells and excellent reactivity with strained alkenes.60 The addition of electron-donating methyl and amine groups to tetrazine reduced its susceptibility to nucleophilic attack. Genetic encoding of the tetrazine 1 was achieved via amber suppression using an evolved tyrosyl-tRNA synthetase/tRNACUApair from Methanococcus jannaschii. In vitro and in vivo studies were conducted using E. coli cells expressing tetrazine 1-modified GFP and ligated with the strained s-TCO. Rate constants were measured to be 880 ± 10 M−1s−1 in vitro and 330 ± 20 M−1s−1 in vivo.
The Chin group demonstrated the tetrazine ligation-mediated protein labeling on the mammalian cell surface by genetically encoding a norbornene-containing amino acid, Nε-5-norbornene-2-yloxycarbonyl-L-lysine (2). An engineered pyrrolysyl-tRNA synthetase/tRNACUA pair from Methanosacina mazei allowed efficient incorporation of norbornene 2 into EGFR at the cell surface. Treatment with tetrazine-TAMRA led to the specific labeling of the norbornene-encoded EGFR on cell surface.61 In another report, Chin and co-workers described genetic encoding of BCN (5), TCO (6), and s-TCO (7) using the same strategy.62 Both BCN and TCO UAAs were efficiently incorporated into proteins in E. coli and mammalian cells and their reactions with the diaryltetrazine gave the second-order rate constants of 1245 ± 45 M−1s−1 and 17248 ± 3132 M−1s−1, respectively. They also showed that rapid, fluorogenic reaction of BCN with the tetrazine afforded a single isomeric product, which can be advantageous to applications such as single-molecule spectroscopy, super-resolution microscopy and FRET experiments.
A similar report was made by the Lemke group detailing the genetic encoding of a BCN UAA 5 and its application to in vitro and in vivo protein studies.63 FRET-based assays for measuring reaction rates were carried out using the BCN- or OCT-encoded GFP as FRET donor and TAMRA-tetrazine as FRET acceptor. Compared with OCT, tetrazine ligation with BCN proceeded 80 times faster with a k2 value of 29000 ± 7500 M−1s−1. Using an engineered pyrrolysyl-tRNA synthetase/tRNACUA pair from Methanosacina mazei, they demonstrated the selective labeling of the BCN-encoded nuclear localization signal-maltose-binding protein-GFP fusion protein in HeLa Kyoto cells with Cy5-tetrazine in 10 minutes. The fast labeling of BCN via tetrazine ligation can be useful for monitoring the newly synthesized proteins or imaging proteins with short half-lives. The utility of BCN-modified proteins can be extended to SPAAC labeling since azido compounds are more accessible than tetrazine probes.63 A separate study by the same group succeeded in the genetic incorporation of norbornene amino acids 2 and 3, TCO amino acid 6, and cyclooctyne amino acid 4 (Figure 3). These UAAs allowed the labeling of proteins inside mammalian cells by means of tetrazine ligation, which was also shown to be orthogonal to SPAAC.64
Aside from genetic encoding, another means of introducing a chemical reporter into biomolecules is through the enzyme-catalyzed biochemical processes. The PRIME method developed by Ting and co-workers enabled targeting of TCO into proteins both inside and on the surface of cells. Their two-step process is comprised of (i) the ligation of a TCO derivative into a protein of interest by the engineered LplA enzyme, and (ii) the reaction of TCO with a fluorophore-conjugated tetrazine via the tetrazine ligation.65 Due to fluorescence quenching by tetrazine prior to ligation, the fluorescent labeling on the cell surface was shown to be highly sensitive and fluorogenic. In addition, both actin and vimentin at the cytoskeleton were specifically labeled inside cells.
Photoinduced tetrazole-alkene cycloaddition
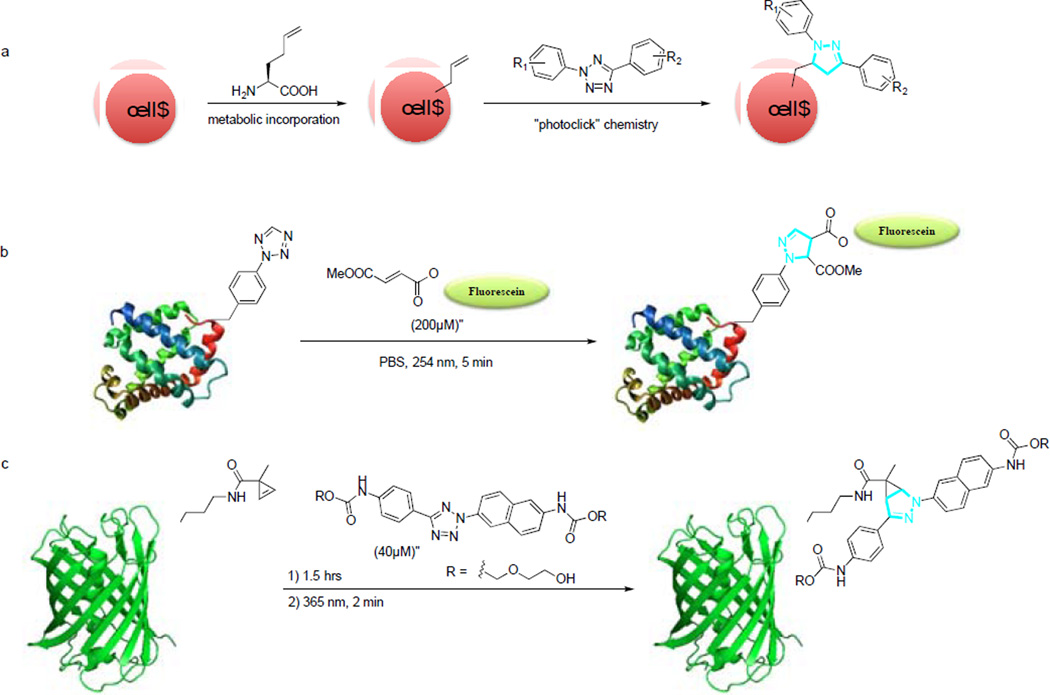
Photoinduced cycloaddition reactions provided a means of spatial and temporal control over chemical and biological processes. The level of control provided by photo induction is made possible by combining the speed and specificity of a click reaction and the versatility of a photochemical process.66 Inspired by the work of Huisgen and co-workers on the photoactivated cycloaddition reaction between 2,5-diphenyltetrazole and methyl crotonate,67 we harnessed the reactivity of an in situ photo-generated nitrile imine dipole from cycloreversion of a diaryltetrazolefor efficient cycloaddition reactions with the alkenes in aqueous buffer,68 for peptide side chain cross-linking,69 and for selective functionalization70 in a bioconjugation reaction we called “photoclick chemistry” of alkene-containing proteins in vitro71 and in vivo.72–75
We first reported the use of cyclic nitrile imines as strained dipoles for 1,3-dipolar cycloadditions.76 This idea arose from our photocrystallographic study where we observed the formation of the bent nitrile imine geometry in solid state upon photo irradiation of a substituted diphenyltetrazole.77 The bent geometry was reinforced by inserting a short bridge between the ortho positions of the two flanking phenyl rings to form a macrocyclic tetrazole. Reactions of the macrocyclic tetrazoles with both acyclic (4-penten-1-ol) and cyclic (norbornene) alkene gave higher yields than their acyclic tetrazole counterparts. These photoactivatable tetrazole reagents were then employed in the fluorescent labeling of norbornene-modified lysozyme.
Using a simple alkene tag, homoallyglycine (HAG), we demonstrated the capacity of photoclick chemistry in imaging the newly synthesized proteins in mammalian cells.74 This was achieved via a two-step process involving the metabolic incorporation of HAG into HeLa cells followed by photocontrolled chemical functionalization with a diaryl tetrazole (Scheme 4a). After a brief exposure to femtosecond 700 nm laser for 5 seconds, cellular fluorescence was recorded over 1 minute using a confocal microscope. Only those cells that were directly illuminated showed greater than 2-fold rapid increase in fluorescence, indicating a spatial and temporal control over the chemical modification.
Scheme 4.
Photoinduced protein labeling via photolick chemistry.
To better apply photoclick chemistry to protein imaging in cell culture, we first designed and synthesized a series photoreactive tetrazole amino acids.78 Among them, p-(2-tetrazole) phenylalanine (pTpa) was genetically incorporated into proteins site-selectively in E. coli using an engineered tyrosyl-tRNA synthetase/tRNACUA pair. The pTpa-encoded myoglobin (pTpa-Myo) was found to react selectively with the FITC-modified fumarate, affording a fluorescent product after 5-minute 302 nm photoirradiation in PBS (Scheme 4b).75 In parallel, a cyclopropene-modified lysine (CpK) was successfully incorporated into proteins both in bacteria and in mammalian cells.79 To demonstrate the use of CpK as a bioorthogonal reporter, HEK293 cells expressing CpK-encoded EGFP were treated with 40 µM tetrazole for 1.5 hours followed by 2 minutes of 365-nm photoirradiation before confocal microscopy (Scheme 4c). Only cells expressing CpK-encoded EGFP showed cyan fluorescence that correspond to the formation of pyrazoline adduct.
Since the photoclick chemistry in Scheme 4 is intrinsically fluorogenic, it can also be used to image the cellular structures via an intramolecular reaction.80 To this end, we appended the alkene-containing tetrazoles to position 7 of paclitaxel and obtained the photoactivatable microtubule probes that can be turned on in as little as 1 minute. A high fluorescence turn-on ratio of 112-fold in CH3CN/PBS (1:1) was observed. Using a long-wavelength photoactivatable taxoid-tetrazole, we demonstrated the spatially controlled imaging of microtubules in live CHO cells.
To tune photoactivation wavelength further away from 365-nm UV light,81, 82 which still causes significant phototoxicity, we recently succeeded in designing 405-nm laser light activatable tetrazoles.83 In a model reaction with mono-methyl fumarate amide, one of these tetrazoles also gave a high rate constant of about 1300 M−1s−1.
Strain-promoted alkyne-nitrone cycloaddition
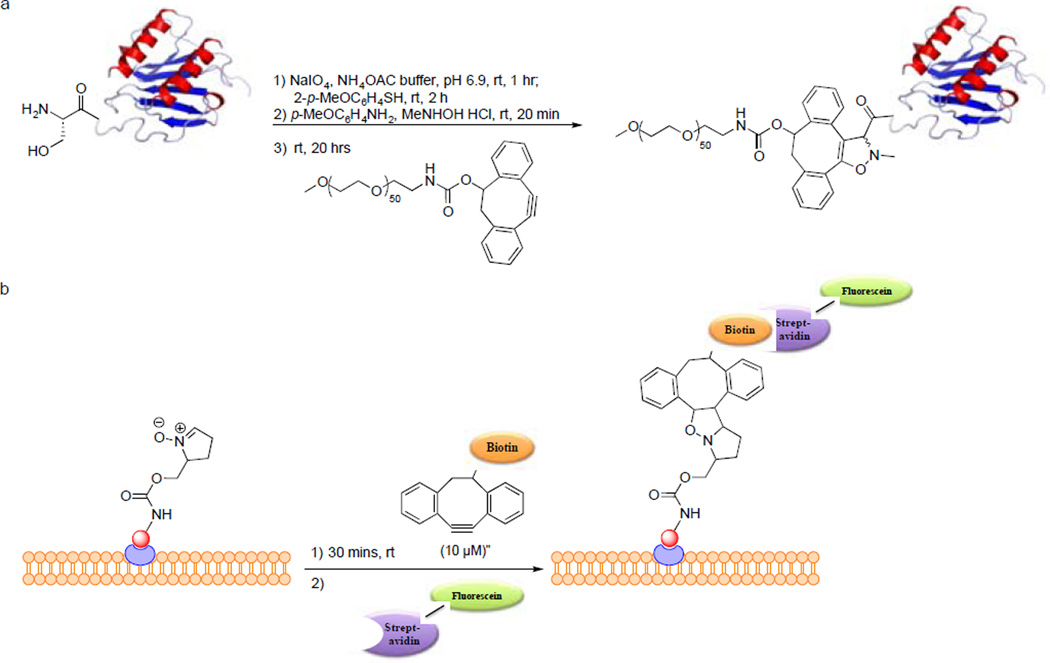
In 2010, van Delft and co-workers reported the strain-promoted alkyne-nitrone cycloaddition (SPANC) between dibenzocyclooctynol and nitrone to generate the N-alkylated isoxazoline.84 Reaction proceeded 32 times faster than the SPAAC reaction with DIBO. They demonstrated the utility of SPANC in protein modification. The chemokine interleukin-8 (IL-8), which has an N-terminal serine residue and a relatively low molecular weight, was successfully PEGylated in a one-pot three-step procedure in which IL-8 in ammonium acetate buffer, pH 6.9, was sequentially treated with 1.1 equiv of NaIO4 for 1 hour, 6.6 equiv of p-MeOC6H4SH for 2 hours, and then 10 equiv each of N-methylhydroxylamine and p-anisidine and 25 equiv of DIBO for 24 hours. Mass spectrometry analysis revealed a single product with mass corresponding to the isoxazoline-PEG conjugate of IL-8 (Scheme 5a). Unfortunately, attempts to label the nitrone-modified sugars in Jurkat cells were unsuccessful, which the authors attributed to either incompatibility of the nitrone modification with the biosynthetic pathway or potential nitrone hydrolysis in the acidic compartments of the cell. To broaden the application of SPANC for protein modification, they recently reported the dual functionalization of GFP. The serine residue at the N-terminal of GFP was modified using the same one-pot three-step procedure together with N-propargyl hydroxylamine to produce a propargyl function and an in situ-generated nitrone that reacts readily with BCN-biotin conjugate. Then CuAAC reaction was performed on the propargyl moiety with an azido-fluorescein conjugate. This demonstrates the potential of combining SPANC with CuAAC for dual labeling of proteins.85
Scheme 5.
Protein modification via strain-promoted alkyne-nitrone cycloaddition.
In the same year, an independent report by Pezacki and co-workers described the use of aromatic acyclic and endocyclic nitrones as dipoles for rapid strain-promoted 1,3-dipolar cycloaddition with DIBO.86 The following year, they reported the use of cyclic nitrones for labeling human cancer cells via SPANC with DIBO.87 The reaction of cyclic nitrones with DIBO reaches a rate of 3.38 ± 0.31 M−1 s−1 that is 59 times greater than the corresponding SPAAC of DIBO. To demonstrate the ability of cyclic nitrones to modify proteins by SPANC, BSA was modified at the lysine residues with nitrones. Subsequent reaction with DIBO-Alexa Fluor 3488 conjugate at 0-60 minutes showed time-dependent labeling based on in-gel fluorescence scanning. They also extended the utility of SPANC with cyclic nitrones to cellular imaging. To this end, epidermal growth factor receptors (EGFR) that are over expressed on the surface of human breast cancer cell line MDA-MB-468 were labeled. To accomplish this, the epidermal growth factor (EGF) protein was coupled with a 5-membered cyclic nitrone then targeted to EGFR. Labeling was carried out using DIBO-biotin then followed by detection with streptavidin-FITC (Scheme 5b). A faster reaction of cyclic nitrones in SPANC was achieved with the use of BARAC with a rate constant of 47.3 M−1 s−1. This reaction is 47 times faster than SPAAC of BARAC and 14 times faster than the previously reported SPANC reaction.88
Palladium-mediated cross-coupling reactions
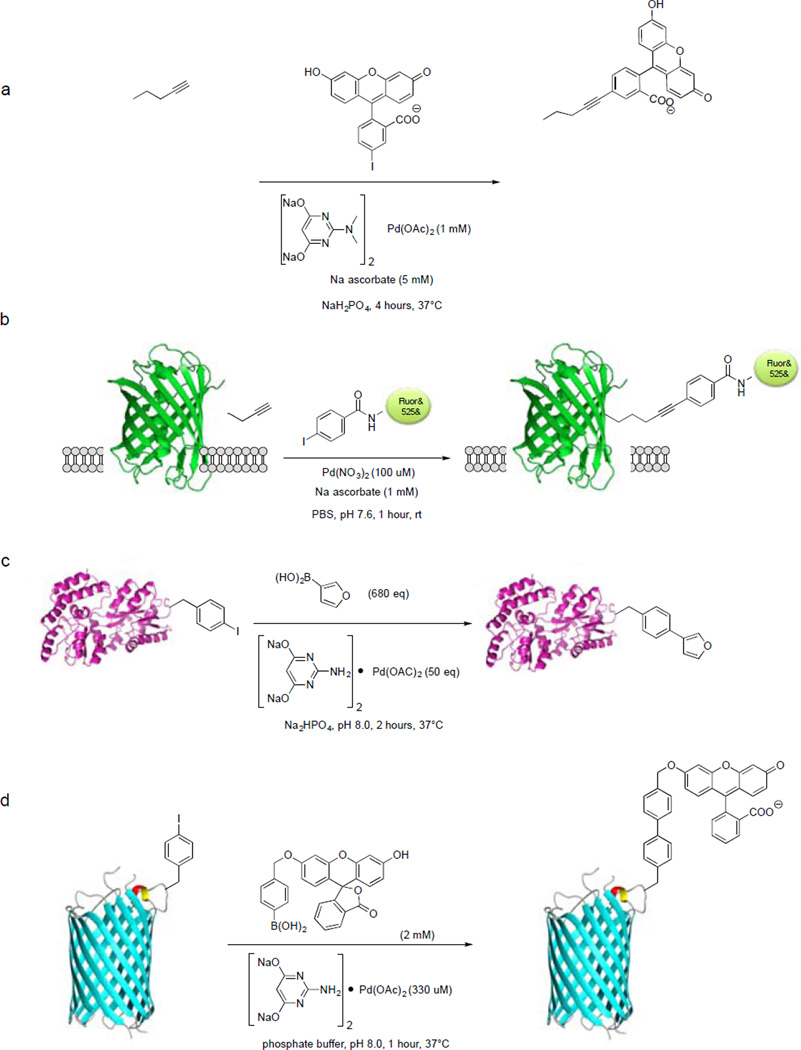
The interest in using palladium complexes as organometallic tools to investigate biological processes has been steadily growing in the past few years. In 2007, Yokoyama and co-workers first reported the use of Pd(OAc)2-triphenylphosphine-3,3’,3’’-trisulfonate complex to achieve Sonogashira cross-coupling between iodophenylalanine-encoded Ras protein and propargylic biotin in aqueous medium containing 12% DMSO and 0.7 mM CuOTf with 25% yield.89To improve reaction efficiency, we reported the discovery of a water-soluble palladium–2-amino-4,6-dihydroxypyrimidine (ADHP) complex that allowed copper-free Sonogashira cross-coupling reactions with alkyne-encoded proteins both in aqueous medium and inside E. coli cells.90 In brief, an alkyne amino acid homopropargyl-glycine (Hpg) was incorporated into the small ubiquitin protein as a methionine surrogate by expressing the protein in M15A, a methionine auxotroph, in the presence of Hpg. The Hpg-encoded ubiquitin (Ub-Hpg) was incubated with 50 equiv of fluorescein iodide and 50 equiv of palladium–ADHP complex (Scheme 6a) in phosphate buffer, and the cross-coupling reaction reached completion after 30 minutes based on LC-MS analysis. Furthermore, M15A cells overexpressing Ub-Hpg were treated with a solution of 1 mM Pd–ADHP complex, 100 µM fluorescein iodide, and 5 mM sodium ascorbate in sodium phosphate buffer for 4 hours, and fluorescent labeling of Ub-Hpg was detected by SDS-PAGE/in-gel fluorescence analysis.90 Recently, we showed that palladacycles can serve as convenient, storable reagents to effect selective functionalization of the Hpg-encoded proteins in biological buffer, though the Heck-type cross-coupling products were observed.91
Scheme 6.
Palladium-catalysed bioorthogonal cross-coupling reactions for protein modification.
More recently, Chen and co-workers reported a ligand-free Sonogashira cross-coupling reaction for fluorescent labeling of the intracellular proteins.92 In their study, they found Pd(NO3)2 was in itself sufficient to catalyse efficient cross-coupling between alkyne-encoded GFP (GFP-alkyne) and rhodamine-conjugated phenyl iodide. To assess cellular uptake of the palladium complex, E. coli cells were treated with 200 µM Pd(NO3)2 for 1 hour at room temperature and the intracellular palladium concentration was analyzed by inductively coupled plasma mass spectrometry (ICP-MS). A 50-fold increase in intracellular palladium concentration was observed compared to the untreated cells. This shows that bacteria are able to uptake the palladium species with no apparent toxicity, consistent with the previous studies on Pd-nanoparticles.93 To demonstrate that this new cross-coupling condition is suitable for protein labeling inside E. coli cells, GFP-alkyne was treated with 200 µMPd(NO3)2 and 200 µMrhodamine-conjugated phenyl iodide at room temperature for 1 hour (Scheme 6b). In-gel fluorescence and western blot analysis confirmed the specificity of the Pd(NO3)2 mediated intracellular Sonogashira cross-coupling inside bacterial cells. They further showed that this new reaction condition can be extended to intracellular protein labeling in gram-negative Shigella cells by labeling an alkyne-modified virulence protein, Type-III secretion (T3S) effector-OspF.
The ability to perform Suzuki-Miyaura cross-coupling in proteins was first demonstrated by the group of Hamachi, wherein a chemically-synthesized WW domain of Pin1 protein was modified in aqueous solution under ligandless conditions using a soluble palladium catalyst (Na2PdCl4).94 A more general approach to performing Suzuki-Miyaura cross-coupling on proteins was presented by Schultz and co-workers in which a boronate-containing amino acid was genetically incorporated into a cysteine-free T4 lysozyme, and modified with BODIPY-phenyliodide.95 One disadvantage of the method presented by the group of Schultz is the use of high temperature (70°C) to achieve efficient cross-coupling. Subsequent studies conducted by Davis and co-workers met this challenge with the report on efficient Suzuki-Miyaura cross-coupling between the p-iodobenzene-modified subtilisin protein and various aryl and vinyl boronic acids in phosphate buffer.96 Using similar reaction conditions, Liu and co-workers was able to employ Suzuki-Miyaura cross-coupling on Z-domain protein bearing a genetically incorporated p-iodophenylalanine.97 Following this development, the Davis group showed that Suzuki-Miyaura cross-coupling can also be employed to modify a genetically encoded, iodophenylalanine-containing maltose-binding protein (MBP).98 A 50% yield was obtained when 50 equiv of Pd catalyst and 680 equiv of boronic acid were used at 37°C for 1 hour, and complete conversion was achieved after 2 hours of incubation (Scheme 6c). They also found that the treatment with 3-mercaptopropionic acid as a scavenger was critical as no ion peaks were detected by LC-MS in its absence. They later demonstrated the utility of Suzuki-Miyaura cross-coupling in labeling channel protein OmpC carrying the genetically encoded p-iodophenylalanine with boronic acid-fluorescein(BA-Fluo) on the surface of E. coli.99 Labeling was achieved with 600 µM BA-Fluo and 350 µM Pd catalyst for 1 hour (Scheme 6d). The same palladium complex was also employed to conjugate the boronic acid-modified carbohydrates onto the E. coli surface.100 Davis and co-workers reported recently that Suzuki-Miyaura cross-coupling also allowed the site-selective conjugation of polyethylene glycol (PEG) onto proteins using a water-soluble Pd salt without addition of ligands.101 Their strategy relies on the stabilizing effect of PEG observed previously with the Pd-nanoparticle-based catalysts.93, 102–105 Two model proteins were chosen for the demonstration of their system, 3-layer-α/β-Rossman-fold protein subtilisin from Bacillus lentus(SBL) and all-β-helix protein 275-276 from Nostoc punctiforme(Npβ), along with two different iodinated unnatural amino acids. The SBL protein was modified chemically with p-iodobenzylcysteine (Pic) at position 156 (SBL-156Pic) and Npβ was charged with p-iodophenylalanine (pIPhe) at position 69 via amber codon suppression (Npβ-69 pIPhe). The two modified proteins were allowed to react via Suzuki-Miyaura cross-coupling with 2 kDa monomethoxy PEG phenylboronic acid (mPEG2k-PBA) and K2PdCl4. SDS-PAGE analysis showed the successful PEGylation of SBL-156Pic and Npβ-69 pIPhe with 70% and 60% yield, respectively, under the ligand-free conditions. A very recent report by the Davis group described the application of Pd-catalyzed Suzuki-Miyaura cross-coupling to modifying DNA for the preparation of functional probes.106 This method uses mild conditions that are biologically relevant (37°C, pH 8.5) with a broad range of substrates and excellent yields. Their method utilizes an iododeoxyuridine (IdU) installed in the middle of an oligodeoxynucleotide (ODN). Using previously reported ligands, 2-aminopyrimidine-4,6-diol96 and its N,N’-dimethylated analogue that aid in Suzuki-Miyaura cross-coupling, in tandem with Tris buffer at pH 8.5, they were able to introduce a variety of functional groups into ODNs. To promote efficient entry of palladium into mammalian cells, Bradley and co-workers developed a nanoparticle-based system in which Pd0 is enclosed in polystyrene microspheres.107 The ability of the Pd0 microspheres to catalyse Suzuki-Miyaura cross-coupling inside HeLa cells was demonstrated through in situ synthesis of a fluorescent dye from alkylaminophenylboronate and a lipophilic mono-triflate conjugated to fluorescein. The microsphere delivery strategy makes it possible to anchor Pd0-based heterogeneous catalysts in a pseudo-organelle inside living cells for an extended period in order to perform intracellular palladium chemistry without posing toxicity risks. It remains to be seen whether this strategy works with protein modifications inside living cells.
Ruthenium-catalysed reactions
Aside from palladium, ruthenium as catalyst for olefin cross-metathesis on proteins has gained significant development in recent years. From the early example of functionalization of serine protease subtilisin Bacillus lentus(SBL) via olefin cross-metathesis (OCM) using allyl sulfides as privileged substrates,108 followed by the method development for installing S-allyl cysteine on protein surfaces reported by Davis and co-workers,109 and the genetic encoding of alkene-modified amino acids in yeast by Schultz and co-workers,110 OCM has emerged as a viable strategy for protein modification. The discovery of the reactivity enhancement brought by allyl sulfide and allylic hydroxy groups on OCM revealed the positive effect of allylic chalcogens in both synthesis and chemical biology applications of OCM.111, 112A recent example on the use of allyl sulfides as privileged substrates for OCM was reported by Vederas and co-workers in which cysteine was replaced by S-allyl cysteine in oxytocin leading to a stable analog via ring-closing metathesis.113 The latest development on the use of OCM was reported by the Davis group wherein Se-allyl-selenocysteine was found to be more reactive than S-ally sulfides, with a reaction rate reaching 0.3 M−1s−1. This strategy was applied to histone acetylation, mimicking an acetylated lysine, which is an important epigenetic marker.114
New bioorthogonal reactions
Most bioorthogonal reactions reported to date are based on either cycloaddition reactions or transition metal-catalysed reactions. The relatively small number of reactions in bioorthogonal chemistry toolkit warrants continuous exploration of new reactivity space that is compatible with biological systems, i.e., aqueous, oxygen-rich environment with abundant biological nucleophiles and electrophiles. In the following, we selected examples from the recent literature to highlight the strategies employed in their development and their potential drawbacks.
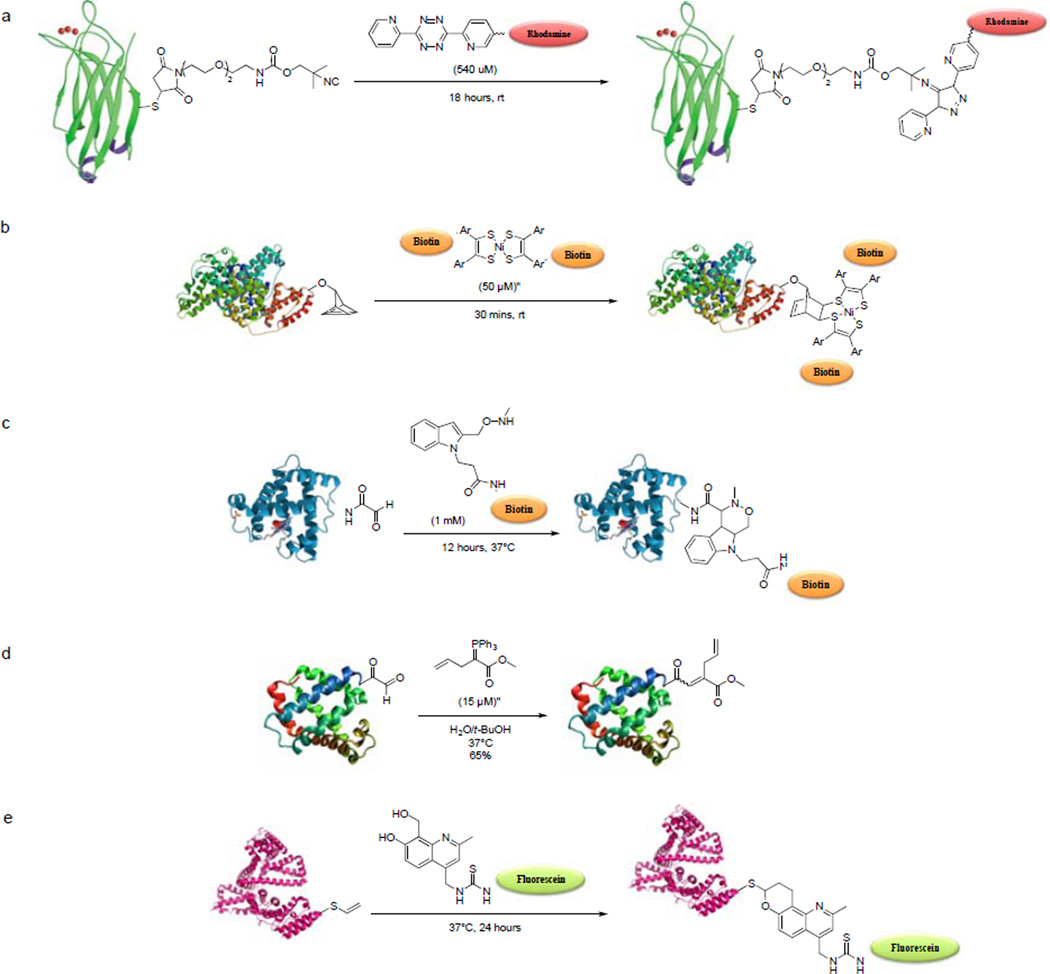
The first example of a [4+1] cycloaddition for biological systems was reported by the Leeper group in 2011, in which isonitriles were shown to participate the click-type reactions with tetrazines.115 This cycloaddition reaction was developed previously for the synthesis of aminopyrazoles from benzyl isonitriles and substituted tetrazines.116 The reaction proceeds as follows: tetrazine and isonitrile react via [4+1] cycloaddition to form norbornadienimine, which spontaneously undergoes [4+2] cycloreversion to form 4H-pyrazol-4-imine derivatives with subsequent release of N2, and tautomerize to aromatic pyrazoles, which is then hydrolyzed to aminopyrazoles. The rate of reaction was determined to be 0.052 ± 0.9 M−1s−1, comparable to SPAAC of the second-generation DIFO (k = 0.042 ± 0.2 M−1s−1). To test the biocompatibility of this reaction, a mutant C2A domain of synaptotagmin-I was modified to carry an isonitrile group at position 78 (C2Am-isonitrile). A solution of C2m-isonitrile (60 µM, pH 7.4) was incubated with 540 µM of tetrazine-rhodamine for 18 hours at room temperature (Scheme 7a). Mass spectrometric analysis revealed full conversion to the 4H-pyrazol-4-imine product with a small amount of the tert-amine hydrolysis product. Earlier this year, the isonitrile-tetrazine click chemistry was applied to metabolic glycan imaging.117 The labeling of cell surface glycans was achieved using isonitrile-modified sugars that are metabolically incorporated followed by a two-step labeling procedure with biotin-tetrazine and neutravidin-DyLight680. Leeper and co-workers also demonstrated that this chemistry is orthogonal to the azide-alkyne chemistry, which may allow simultaneous differential multi-sugar imaging.
Scheme 7.
Newly developed bioorthogonal reactions.
Bertozzi and co-workers reported two new bioorthogonal reactions in the past two years: the quadricyclane ligation between the highly strained hydrocarbon quadricyclane and the Ni bis(dithiolene) reagents,118 and the Pictet-Spengler ligation for chemical modification of aldehyde-functionalized proteins.119 The suitability of quadricylclane ligation for protein modification was demonstrated by incubating the quadricyclane-modified bovine serum albumin (QC-BSA) with varying concentrations of Ni bis(dithiolene) for various times (Scheme 7b); a time- and concentration-dependent protein labeling was detected. Since quadricyclane is relatively small, it should be amenable to genetic or metabolic encoding in various biomolecules. At this point, we have yet to see the application of this reaction to live cell labeling. The potential drawbacks of quadricyclane ligation includes susceptibility of the Ni bis(dithiolene) to reduction in biological media where stabilizing additives are not suitable. On the other hand, the Pictet-Spengler ligation involves the condensation of an aldehyde and an alkoxyamine, to generate an oxyiminium intermediate, which undergoes spontaneous intramolecular C-C bond formation with the indole nucleophile to form a stable oxycarboline product. To demonstrate that Pictet-Spengler ligation is suitable for protein modifcation, horse heart myoglobin with an N-terminal glyoxyl moiety was prepared via pyridoxal phosphate-mediated transamination reaction. The alkoxyamine-modified myoglobin was then treated with biotinylated aminoxy-indole (Scheme 7c), and the conjugated product was analyzed by SDS-PAGE and Western blotting. A concentration and time-dependent biotinylation of myoglobin was observed. Furthermore, a 6-residue peptide (LCTPSR) was appended to the formylglycine-modified MBP and a thrombin cleavage site was engineered at N-terminal to the aldehyde tag. After incubation with 1 mM aminooxy-indole at 37°C for 1 hour, a quantitative conversion to a single adduct was observed by ESI-MS. Trypsin digestion of the adduct released the 8-residue peptide at the C-terminus and thrombin cleavage confirmed that the modification occurred at the FGly residue.
The venerable Wittig reaction was introduced in 1954 by George Wittig,120 and has proven to be one of the most powerful methods for C-C double-bond construction. To investigate whether Wittig reaction is suitable for protein modification, the Ye group modified the N-terminal serine residue of chemokine interleukin-8 (IL-8) by oxidation with NaIO4 at room temperature to generate an aldehyde,121 and then treated the aldehyde-containing IL-8 with an ylide reagent in 4:1 H2O/t-BuOH. MALDI-TOF analysis indicated the formation of the desired Wittig product with complete conversion.122 In another example, the N-terminally aldehyde-modified myoglobin was allowed to react with 15 µM ylide reagent at 37°C for 30 minutes (Scheme 7d), and the Wittig product was formed at 65% yield based on ESI-MS analysis. MALDI-TOF/TOF analysis confirmed that the modification occurred at the N-terminus of myoglobin.
Earlier this year, a new bioorthogonal reaction based on the hetero-Diels-Alder (HDA) cycloaddition of o-quinolinone quinone methide (oQQM) and vinyl thioether (VT) was reported by the Lei group.123 The synthetic intermediates o-quinone methides (oQM) have been widely used in natural product synthesis.124 The generation of these intermediates typically requires hash conditions that are not compatible with biological systems. For example, the Popik group described the use of UV light to generate o-naphthoquinone methides, which limits their use in cellular systems because of potential phototoxicity. To overcome this, Lei and co-workers designed 8-(hydroxymethyl)-2-methylquinolin-7-ol as the oQQM precursor and 2-(vinylthio)ethanol as the dienophile. The HDA reaction between this new pair proceeded smoothly in water at 37°C, giving 92% yield after 24-hour incubation. The second-order rate constant was measured to be 0.0015 M−1s−1, comparable to those of Staudinger ligation (k2= 0.0025M−1s−1) and SPAAC of OCT (k2= 0.0012 M−1s−1). After verying that both reactants were stable against oxidation, they tested the reaction first for selective protein labeling in vitro by modifying the lysine residues in BSA with N-succinimidyl-2-(vinylthio)-ethyl carbonate to generate the vinylthio-modified BSA (VT-BSA). They then incubated VT-BSA with either a biotinylated oQQM or a fluorescein-conjugated oQQM (Scheme 7e). Western blot and in-gel fluorescence analyses showed selective labeling of VT-BSA with minimum background. Furthermore, oQQM–VT ligation was also successfully applied to fluorescent labeling of the VT-modified taxol inside live HeLa cells.
Mutually exclusive bioorthogonal reactions
The use of two mutually exclusive bioorthogonal reactions for dual labeling of biomolecules has been gaining popularity recently. Analogous to the development of fluorescent proteins with a full colour pallet, the ability to use mutually exclusive bioorthogonal reactions for rapid, simultaneous, dual labeling of biomolecules should offer a powerful tool to study of protein conformational changes and protein-protein interactions in living system, e.g., through FRET-based studies.
To achieve dual labeling, two chemical reporters need to be incorporated into the target biomolecule. To this end, Anthony, Davis, and co-workers reported a chemical tagging strategy that allow multiple modifications to protein scaffolds expressed in bacteria, mimicking the post-translational modification that is only seen in higher organisms.125 In this study, azide-bearing protein was modified with the alkyne-containing sugars via click chemistry, or vice versa, to produce glycoproteins in conjunction with modification of cysteine with glycomethanethiosulphonates. In a later study, Schultz, Deniz, and co-workers reported the genetic incorporation of a ketone amino acid into T4 lysozyme in addition to the presence of native cysteine to allow dual fluorescent labeling through the condensation reaction with alkoxyamine-Alexa 488 and the Michael addition with maleimide-Alexa 594, respectively.126 A more general approach is to incorporate two different UAAs. To this end, the Liu group reported the genetic incorporation of two UAAs in a single protein via suppression of two stop codons, amber and ochre.127 Later, they reported that p-azidophenylalanine and 2-amino-8-oxononanoic acid were incorporated into the glutamine-binding protein (QBP) and labeled with DIBAC-rhodamine and coumarin-alkoxyamine, respectively, via SPAAC and oxime formation reactions inside E. coli. cells.128
The Overkleeft group reported the first use of tandem bioorthogonal reactions, i.e., Staudinger ligation and Diels-Alder cycloaddition, in activity-based protein profiling in a complex biological environment.129 They developed the diene-containing probes that are directed to the enzyme active site, and maleimide-modified BODIPY(TMR)-tag as dienophile. They showed that this reaction is fully orthogonal to the Staudinger ligation, which can be used for simultaneous tracking of enzymatic activities. This strategy, although successful in monitoring the activity of endogenous proteases in cell lysates, suffers from nonspecific labeling due to the presence of free thiol groups that can participate in Michael addition with the diene probe. The same group reported a more elaborate use of mutually exclusive bioorthogonal reactions, wherein they demonstrated a triple bioorthogonal ligation strategy for activity-based protein profiling.130 The triple ligation strategy employs the tetrazine ligation, Staudinger ligation and CuAAC. In a single experiment, they were able to label multiple enzymatic activities in the 20S proteasome that contains three catalytic subunits. To achieve this, they used a newly developed two-step tetrazine ligation using a norbornene activity-based probe (ABP) as a ligation handle and tetrazine-conjugated fluorophore as a labeling probe, together with an azide ABP-phosphine probe pair for the Staudinger ligation and an alkyne-ABP-azide probe for CuAAC.
Another example of employing two bioorthogonal reactions for simultaneous labeling of cells was reported by Hilderbrand and co-workers wherein tetrazine-transcyclooctene and azide-cyclooctyne cycloaddition was shown to be mutually orthogonal and allow labeling of two targets in a complex biological environment.131 This was accomplished by co-culturing SKBR-3 and A431 cell lines then modifying with Cetuximab-AF488-DIBAC and Herceptin-AF568-TCO conjugates, respectively. Labeling was done simultaneously using AF67-azide and AF750-tetrazine that target DIBAC-modified SKBR-3 and TCO-modified A431 cells, respectively. Flow cytometry analysis and confocal imaging confirmed the specificity and orthogonality of both reactions showing the fluorescent labeling of two different cell types in a single experiment. A similar strategy was recently employed by Wittmann and co-workers wherein they demonstrated the double labeling of two differently modified monosaccharides, which were metabolically incorporated, by combining tetrazine ligation and SPAAC.132 In this study, they used a terminal alkene as reporter for tetrazine ligation and showed its efficient metabolic incorporation into cell surface glycans, which has not been reported before. They incorporated two different monosaccharides onto the cell surface that carry a terminal alkene and an azide, and then labeled them with tetrazine-biotin/AF647-streatavidin and AF488-DIBO, respectively. By combining tetrazine ligation and SPAAC, the imaging of two different glycan structures can be carried out in a single experiment.
Conclusions and Future Directions
Since its inception more than a decade ago, bioorthogonal chemistry has matured to the stage where it has been routinely employed in the study of biological processes in living systems. Many aspects of bioorthogonal reactions have been optimized over the last few years, including faster reaction rates, expanded sets of unnatural substrates for efficient introduction into proteins, glycans, nucleic acids, and lipids, new methods for simultaneous introduction of multiple bioorthogonal reporters, fluorogenic reactions for facile biological imaging, and robust photoactivatable reagents for imaging with improved spatial and temporal control with minimal phototoxicity. During the optimization of reaction kinetics, several successful strategies have emerged, including the use of ring strain to activate substrate and the design of new ligands or privileged substrates to accelerate the transition metal-catalysed reactions. In addition, a greater attention has been directed toward the physiochemical properties of the reagents and their interactions with the biological environments where bioconjugation reactions operate, including reagent solubility, stability, permeability and toxicity, as well as photophysical properties of the bioconjugates.
Looking ahead, while there is a continuing need to develop new bioorthogonal reactions, it would be particularly valuable if these reactions are mutually exclusive with respect to the existing ones. The emergence of genetic and metabolic encoding of multiple bioorthogonal reporters together with their cognate mutually exclusive bioorthogonal reactions promises to open new venues of biological investigations, e.g., the study of conformational dynamics of biomolecules and their interactions of other biomolecules, and the multiplexed interrogation of the subfamilies in a proteomic and glycomic study. Finally, with the metabolic133 and genetic encoding134, 135 of bioorthogonal groups in animals becoming possible, it is exciting to see what new biological insights will be gleaned in the near future with these unique chemical reactivity-based tools.
Figure 3.
Genetically encoded amino acids for tetrazine ligation.
Acknowledgements
Our work on bioorthogonal chemistry and its applications in living systems is supported by U. S. National Institutes of Health (GM 085092) and National Science Foundation (CHE-1305826).
References
- 1.Johnson FH, Saiga Y. J. Cell. Physiol. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- 2.Tsien RY. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 3.Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 4.Sletten EM, Bertozzi CR. Angew. Chem. Intl. Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laughlin ST, Bertozzi CR. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12–17. doi: 10.1073/pnas.0811481106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hang HC, Wilson JP, Charron G. Acc. Chem. Res. 2011;44:699–708. doi: 10.1021/ar200063v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cravatt BF, Wright AT, Kozarich JW. Annual Review of Biochemistry. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 8.Rideout D. Science. 1986;233:561–563. doi: 10.1126/science.3523757. [DOI] [PubMed] [Google Scholar]
- 9.Cornish VW, Hahn KM, Schultz PG. J. Am. Chem. Soc. 1996;118:8150–8151. [Google Scholar]
- 10.Prescher JA, Bertozzi CR. Nat. Chem. Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 11.van Swieten PF, Leeuwenburgh MA, Kessler BM, Overkleeft HS. Org. Biomol. Chem. 2005;3:20–27. doi: 10.1039/b412558d. [DOI] [PubMed] [Google Scholar]
- 12.Lim RKV, Lin Q. Chem. Comm. 2010;46:1589–1600. doi: 10.1039/b925931g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertozzi CR. Acc Chem Res. 2011;44:651–653. doi: 10.1021/ar200193f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huisgen R. Angew. Chem. Intl. Ed. 1963;2:565–598. [Google Scholar]
- 15.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew. Chem. Intl. Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 16.Tornøe CW, Christensen C, Meldal M. J. Org. Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 17.Himo F, Lovell T, Hilgraf R, Rostovtsev VV, Noodleman L, Sharpless KB, Fokin VV. J. Am. Chem. Soc. 2004;127:210–216. doi: 10.1021/ja0471525. [DOI] [PubMed] [Google Scholar]
- 18.Rodionov VO, Fokin VV, Finn MG. Angew. Chem. Intl. Ed. 2005;44:2210–2215. doi: 10.1002/anie.200461496. [DOI] [PubMed] [Google Scholar]
- 19.Worrell BT, Malik JA, Fokin VV. Science. 2013;340:457–460. doi: 10.1126/science.1229506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan TR, Hilgraf R, Sharpless KB, Fokin VV. Org. Lett. 2004;6:2853–2855. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- 21.Hong V, Presolski SI, Ma C, Finn MG. Angew. Chem. Intl. Ed. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta SS, Kuzelka J, Singh P, Lewis WG, Manchester M, Finn MG. Bioconjugate Chem. 2005;16:1572–1579. doi: 10.1021/bc050147l. [DOI] [PubMed] [Google Scholar]
- 23.Soriano del Amo D, Wang W, Jiang H, Besanceney C, Yan AC, Levy M, Liu Y, Marlow FL, Wu P. J. Am. Chem. Soc. 2010;132:16893–16899. doi: 10.1021/ja106553e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Besanceney-Webler C, Jiang H, Zheng T, Feng L, Soriano del Amo D, Wang W, Klivansky LM, Marlow FL, Liu Y, Wu P. Angew. Chem. Intl. Ed. 2011;50:8051–8056. doi: 10.1002/anie.201101817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kennedy DC, McKay CS, Legault MC, Danielson DC, Blake JA, Pegoraro AF, Stolow A, Mester Z, Pezacki JP. J. Am. Chem. Soc. 2011;133:17993–18001. doi: 10.1021/ja2083027. [DOI] [PubMed] [Google Scholar]
- 26.Willems LI, Li N, Florea BI, Ruben M, van der Marel GA, Overkleeft HS. Angewandte Chemie. 2012;51:4431–4434. doi: 10.1002/anie.201200923. [DOI] [PubMed] [Google Scholar]
- 27.Michaels HA, Zhu L. Chemistry, an Asian journal. 2011;6:2825–2834. doi: 10.1002/asia.201100426. [DOI] [PubMed] [Google Scholar]
- 28.Kuang GC, Guha PM, Brotherton WS, Simmons JT, Stankee LA, Nguyen BT, Clark RJ, Zhu L. J. Am. Chem. Soc. 2011;133:13984–14001. doi: 10.1021/ja203733q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uttamapinant C, Tangpeerachaikul A, Grecian S, Clarke S, Singh U, Slade P, Gee KR, Ting AY. Angew. Chem. Intl. Ed. 2012;51:5852–5856. doi: 10.1002/anie.201108181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uttamapinant C, White KA, Baruah H, Thompson S, Fernández-Suárez M, Puthenveetil S, Ting AY. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10914–10919. doi: 10.1073/pnas.0914067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agard NJ, Prescher JA, Bertozzi CR. J. Am. Chem. Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 32.Codelli JA, Baskin JM, Agard NJ, Bertozzi CR. J. Am. Chem. Soc. 2008;130:11486–11493. doi: 10.1021/ja803086r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ning X, Guo J, Wolfert MA, Boons GJ. Angew. Chem. Intl. Ed. 2008;47:2253–2255. doi: 10.1002/anie.200705456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Debets MF, van Berkel SS, Schoffelen S, Rutjes FP, van Hest JC, van Delft FL. Chem. Comm. 2010;46:97–99. doi: 10.1039/b917797c. [DOI] [PubMed] [Google Scholar]
- 35.Dommerholt J, Schmidt S, Temming R, Hendriks LJ, Rutjes FP, van Hest JC, Lefeber DJ, Friedl P, van Delft FL. Angew. Chem. Intl. Ed. 2010;49:9422–9425. doi: 10.1002/anie.201003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sletten EM, Bertozzi CR. Org. Lett. 2008;10:3097–3099. doi: 10.1021/ol801141k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schoffelen S, Lambermon MHL, Eldijk MBv, Hest JCMv. Bioconjugate Chem. 2008;19:1127–1131. doi: 10.1021/bc800019v. [DOI] [PubMed] [Google Scholar]
- 38.van Dongen SFM, Teeuwen RLM, Nallani M, van Berkel SS, Cornelissen JJLM, Nolte RJM, van Hest JCM. Bioconjugate Chem. 2008;20:20–23. doi: 10.1021/bc8004304. [DOI] [PubMed] [Google Scholar]
- 39.Meier H, Schuh-Popitz C, Peiersen H. Angew. Chem. Intl. Ed. Engl. 1981;20:270–271. [Google Scholar]
- 40.Jewett JC, Sletten EM, Bertozzi CR. J. Am. Chem. Soc. 2010;132:3688–3690. doi: 10.1021/ja100014q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sletten EM, Nakamura H, Jewett JC, Bertozzi CR. J. Am. Chem. Soc. 2010;132:11799–11805. doi: 10.1021/ja105005t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ess DH, Jones GO, Houk KN. Org. Lett. 2008;10:1633–1636. doi: 10.1021/ol8003657. [DOI] [PubMed] [Google Scholar]
- 43.Bach RD. J. Am. Chem. Soc. 2009;131:5233–5243. doi: 10.1021/ja8094137. [DOI] [PubMed] [Google Scholar]
- 44.Chenoweth K, Chenoweth D, Goddard WA., Iii Org. Biomol. Chem. 2009;7:5255–5258. doi: 10.1039/b911482c. [DOI] [PubMed] [Google Scholar]
- 45.Schoenebeck F, Ess DH, Jones GO, Houk KN. J. Am. Chem. Soc. 2009;131:8121–8133. doi: 10.1021/ja9003624. [DOI] [PubMed] [Google Scholar]
- 46.de Almeida G, Sletten EM, Nakamura H, Palaniappan KK, Bertozzi CR. Angew Chem Int Ed Engl. 2012;51:2443–2447. doi: 10.1002/anie.201106325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krebs A, Kimling H. Tetrahedron Lett. 1970;11:761–764. [Google Scholar]
- 48.Jewett JC, Bertozzi CR. Org. Lett. 2011;13:5937–5939. doi: 10.1021/ol2025026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shieh P, Hangauer MJ, Bertozzi CR. J. Am. Chem. Soc. 2012;134:17428–17431. doi: 10.1021/ja308203h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herner A, Nikic I, Kallay M, Lemke EA, Kele P. Org. Biomol. Chem. 2013;11:3297–3306. doi: 10.1039/c3ob40296g. [DOI] [PubMed] [Google Scholar]
- 51.Blackman ML, Royzen M, Fox JM. J. Am. Chem. Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Devaraj NK, Weissleder R, Hilderbrand SA. Bioconjugate Chem. 2008;19:2297–2299. doi: 10.1021/bc8004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sauer J, Heldmann DK, Hetzenegger J, Krauthan J, Sichert H, Schuster J. Eur. J. Org. Chem. 1998;1998:2885–2896. [Google Scholar]
- 54.Thalhammer F, Wallfahrer U, Sauer J. Tetrahedron Lett. 1990;31:6851–6854. [Google Scholar]
- 55.Balcar J, Chrisam G, Huber FX, Sauer J. Tetrahedron Lett. 1983;24:1481–1484. [Google Scholar]
- 56.Devaraj NK, Hilderbrand S, Upadhyay R, Mazitschek R, Weissleder R. Angew. Chem. Intl. Ed. 2010;49:2869–2872. doi: 10.1002/anie.200906120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patterson DM, Nazarova LA, Xie B, Kamber DN, Prescher JA. J. Am. Chem. Soc. 2012;134:18638–18643. doi: 10.1021/ja3060436. [DOI] [PubMed] [Google Scholar]
- 58.Yang J, Šečkutė J, Cole CM, Devaraj NK. Angew. Chem. Intl. Ed. 2012;124:7594–7597. [Google Scholar]
- 59.Closs GL, Closs LE, Boll WA. J. Am. Chem. Soc. 1963;85:3796–3800. [Google Scholar]
- 60.Seitchik JL, Peeler JC, Taylor MT, Blackman ML, Rhoads TW, Cooley RB, Refakis C, Fox JM, Mehl RA. J. Am. Chem. Soc. 2012;134:2898–2901. doi: 10.1021/ja2109745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lang K, Davis L, Torres-Kolbus J, Chou C, Deiters A, Chin JW. Nat. Chem. 2012;4:298–304. doi: 10.1038/nchem.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lang K, Davis L, Wallace S, Mahesh M, Cox DJ, Blackman ML, Fox JM, Chin JW. J. Am. Chem. Soc. 2012;134:10317–10320. doi: 10.1021/ja302832g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Borrmann A, Milles S, Plass T, Dommerholt J, Verkade JM, Wiessler M, Schultz C, van Hest JC, van Delft FL, Lemke EA. Chembiochem. 2012;13:2094–2099. doi: 10.1002/cbic.201200407. [DOI] [PubMed] [Google Scholar]
- 64.Plass T, Milles S, Koehler C, Szymański J, Mueller R, Wießler M, Schultz C, Lemke EA. Angew. Chem. Intl. Ed. 2012;51:4166–4170. doi: 10.1002/anie.201108231. [DOI] [PubMed] [Google Scholar]
- 65.Liu DS, Tangpeerachaikul A, Selvaraj R, Taylor MT, Fox JM, Ting AY. J. Am. Chem. Soc. 2011;134:792–795. doi: 10.1021/ja209325n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tasdelen MA, Yagci Y. Angew. Chem. Intl. Ed. 2013;52:5930–5938. doi: 10.1002/anie.201208741. [DOI] [PubMed] [Google Scholar]
- 67.Clovis JS, Eckell A, Huisgen R, Sustmann R. Chem. Ber. 1967;100:60–70. [Google Scholar]
- 68.Wang Y, Rivera Vera CI, Lin Q. Org. Lett. 2007;9:4155–4158. doi: 10.1021/ol7017328. [DOI] [PubMed] [Google Scholar]
- 69.Madden MM, Rivera Vera CI, Song W, Lin Q. Chem. Comm. 2009;0:5588–5590. doi: 10.1039/b912094g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lim RKV, Lin Q. Acc. Chem. Res. 2011;44:828–839. doi: 10.1021/ar200021p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Song W, Wang Y, Qu J, Madden MM, Lin Q. Angew. Chem. Intl. Ed. 2008;47:2832–2835. doi: 10.1002/anie.200705805. [DOI] [PubMed] [Google Scholar]
- 72.Song W, Wang Y, Qu J, Lin Q. J. Am. Chem. Soc. 2008;130:9654–9655. doi: 10.1021/ja803598e. [DOI] [PubMed] [Google Scholar]
- 73.Wang Y, Song W, Hu WJ, Lin Q. Angew. Chem. Intl. Ed. 2009;48:5330–5333. doi: 10.1002/anie.200901220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Song W, Wang Y, Yu Z, Vera CIR, Qu J, Lin Q. ACS Chem. Biol. 2010;5:875–885. doi: 10.1021/cb100193h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang J, Zhang W, Song W, Wang Y, Yu Z, Li J, Wu M, Wang L, Zang J, Lin Q. J. Am. Chem. Soc. 2010;132:14812–14818. doi: 10.1021/ja104350y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu Z, Lim RK, Lin Q. Chemistry. 2010;16:13325–13329. doi: 10.1002/chem.201002360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zheng S-L, Wang Y, Yu Z, Lin Q, Coppens P. J. Am. Chem. Soc. 2009;131:18036–18037. doi: 10.1021/ja9094523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Y, Lin Q. Org. Lett. 2009;11:3570–3573. doi: 10.1021/ol901300h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu Z, Pan Y, Wang Z, Wang J, Lin Q. Angew. Chem. Intl. Ed. 2012;51:10600–10604. doi: 10.1002/anie.201205352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu Z, Ho LY, Lin Q. J. Am. Chem. Soc. 2011;133:11912–11915. doi: 10.1021/ja204758c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Y, Hu WJ, Song W, Lim RKV, Lin Q. Org. Lett. 2008;10:3725–3728. doi: 10.1021/ol801350r. [DOI] [PubMed] [Google Scholar]
- 82.Yu Z, Ho LY, Wang Z, Lin Q. Bioorg. Med. Chem. Lett. 2011;21:5033–5036. doi: 10.1016/j.bmcl.2011.04.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.An P, Yu Z, Lin Q. Chem Commun (Camb) 2013;49 doi: 10.1039/c3cc45752d. 10.1039/C1033CC45752D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ning X, Temming RP, Dommerholt J, Guo J, Ania DB, Debets MF, Wolfert MA, Boons GJ, van Delft FL. Angew. Chem. Intl. Ed. 2010;49:3065–3068. doi: 10.1002/anie.201000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Temming RP, Eggermont L, van Eldijk MB, van Hest JC, van Delft FL. Org. Biomol. Chem. 2013;11:2772–2779. doi: 10.1039/c3ob00043e. [DOI] [PubMed] [Google Scholar]
- 86.McKay CS, Moran J, Pezacki JP. Chem. Comm. 2010;46:931–933. doi: 10.1039/b921630h. [DOI] [PubMed] [Google Scholar]
- 87.McKay CS, Blake JA, Cheng J, Danielson DC, Pezacki JP. Chem. Comm. 2011;47:10040–10042. doi: 10.1039/c1cc13808a. [DOI] [PubMed] [Google Scholar]
- 88.McKay CS, Chigrinova M, Blake JA, Pezacki JP. Org. Biomol. Chem. 2012;10:3066–3070. doi: 10.1039/c2ob07165g. [DOI] [PubMed] [Google Scholar]
- 89.Kodama K, Fukuzawa S, Nakayama H, Sakamoto K, Kigawa T, Yabuki T, Matsuda N, Shirouzu M, Takio K, Yokoyama S, Tachibana K. Chembiochem. 2007;8:232–238. doi: 10.1002/cbic.200600432. [DOI] [PubMed] [Google Scholar]
- 90.Li N, Lim RK, Edwardraja S, Lin Q. J. Am. Chem. Soc. 2011;133:15316–15319. doi: 10.1021/ja2066913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cheng G, Lim RK, Li N, Lin Q. Chem Commun (Camb) 2013;49:6809–6811. doi: 10.1039/c3cc43479f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li J, Lin S, Wang J, Jia S, Yang M, Hao Z, Zhang X, Chen PR. J. Am. Chem. Soc. 2013;135:7330–7338. doi: 10.1021/ja402424j. [DOI] [PubMed] [Google Scholar]
- 93.Windt W, Boon N, Bulcke J, Rubberecht L, Prata F, Mast J, Hennebel T, Verstraete W. Antonie van Leeuwenhoek. 2006;90:377–389. doi: 10.1007/s10482-006-9088-4. [DOI] [PubMed] [Google Scholar]
- 94.Ojida A, Tsutsumi H, Kasagi N, Hamachi I. Tetrahedron Lett. 2005;46:3301–3305. [Google Scholar]
- 95.Brustad E, Bushey ML, Lee JW, Groff D, Liu W, Schultz PG. Angew. Chem. Intl. Ed. 2008;47:8220–8223. doi: 10.1002/anie.200803240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chalker JM, Wood CSC, Davis BG. J. Am. Chem. Soc. 2009;131:16346–16347. doi: 10.1021/ja907150m. [DOI] [PubMed] [Google Scholar]
- 97.Wang Y-S, Russell WK, Wang Z, Wan W, Dodd LE, Pai P-J, Russell DH, Liu WR. Molecular BioSystems. 2011;7:714–717. doi: 10.1039/c0mb00217h. [DOI] [PubMed] [Google Scholar]
- 98.Spicer CD, Davis BG. Chem. Comm. 2011;47:1698–1700. doi: 10.1039/c0cc04970k. [DOI] [PubMed] [Google Scholar]
- 99.Spicer CD, Triemer T, Davis BG. J. Am. Chem. Soc. 2012;134:800–803. doi: 10.1021/ja209352s. [DOI] [PubMed] [Google Scholar]
- 100.Spicer CD, Davis BG. Chem. Comm. 2013;49:2747–2749. doi: 10.1039/c3cc38824g. [DOI] [PubMed] [Google Scholar]
- 101.Dumas A, Spicer CD, Gao Z, Takehana T, Lin YA, Yasukohchi T, Davis BG. Angew. Chem. Intl. Ed. 2013;52:3916–3921. doi: 10.1002/anie.201208626. [DOI] [PubMed] [Google Scholar]
- 102.Blettner CG, König WA, Stenzel W, Schotten T. J. Org. Chem. 1999;64:3885–3890. [Google Scholar]
- 103.Li JH, Liu WJ, Xie YX. J. Org. Chem. 2005;70:5409–5412. doi: 10.1021/jo050353m. [DOI] [PubMed] [Google Scholar]
- 104.Han W, Liu C, Jin ZL. Org. Lett. 2007;9:4005–4007. doi: 10.1021/ol701709q. [DOI] [PubMed] [Google Scholar]
- 105.Razler TM, Hsiao Y, Qian F, Fu R, Khan RK, Doubleday W. J. Org. Chem. 2008;74:1381–1384. doi: 10.1021/jo802277z. [DOI] [PubMed] [Google Scholar]
- 106.Lercher L, McGouran JF, Kessler BM, Schofield CJ, Davis BG. Angew. Chem. Intl. Ed. 2013 doi: 10.1002/anie.201304038. DOI: 10.1002/anie.201304038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yusop RM, Unciti-Broceta A, Johansson EMV, Sánchez-Martín RM, Bradley M. Nat. Chem. 2011;3:239–243. doi: 10.1038/nchem.981. [DOI] [PubMed] [Google Scholar]
- 108.Lin YA, Chalker JM, Floyd N, Bernardes GaJL, Davis BG. J Am Chem Soc. 2008;130:9642–9643. doi: 10.1021/ja8026168. [DOI] [PubMed] [Google Scholar]
- 109.Chalker JM, Lin YA, Boutureira O, Davis BG. Chem Commun (Camb) 2009:3714–3716. doi: 10.1039/b908004j. [DOI] [PubMed] [Google Scholar]
- 110.Ai Hw, Shen W, Brustad E, Schultz PG. Angewandte Chemie International Edition. 2010;49:935–937. doi: 10.1002/anie.200905590. [DOI] [PubMed] [Google Scholar]
- 111.Lin YA, Chalker JM, Davis BG. J Am Chem Soc. 2010;132:16805–16811. doi: 10.1021/ja104994d. [DOI] [PubMed] [Google Scholar]
- 112.Lin YA, Davis BG. Beilstein J. Org. Chem. 2010;6:1219–1228. doi: 10.3762/bjoc.6.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cochrane SA, Huang Z, Vederas JC. Org Biomol Chem. 2013;11:630–639. doi: 10.1039/c2ob26938d. [DOI] [PubMed] [Google Scholar]
- 114.Lin YA, Boutureira O, Lercher L, Bhushan B, Paton RS, Davis BG. J Am Chem Soc. 2013;135:12156–12159. doi: 10.1021/ja403191g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stockmann H, Neves AA, Stairs S, Brindle KM, Leeper FJ. Org. Biomol. Chem. 2011;9:7303–7305. doi: 10.1039/c1ob06424j. [DOI] [PubMed] [Google Scholar]
- 116.Imming P, Mohr R, Müller E, Overheu W, Seitz G. Angew. Chem. Intl. Ed. 1982;21:284–284. [Google Scholar]
- 117.Stairs S, Neves AA, Stöckmann H, Wainman YA, Ireland-Zecchini H, Brindle KM, Leeper FJ. Chembiochem. 2013;14:1063–1067. doi: 10.1002/cbic.201300130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sletten EM, Bertozzi CR. J. Am. Chem. Soc. 2011;133:17570–17573. doi: 10.1021/ja2072934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Agarwal P, van der Weijden J, Sletten EM, Rabuka D, Bertozzi CR. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:46–51. doi: 10.1073/pnas.1213186110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wittig G, Schöllkopf U. Chemische Berichte. 1954;87:1318–1330. [Google Scholar]
- 121.Alam J, Keller TH, Loh TP. Chem. Comm. 2011;47:9066–9068. doi: 10.1039/c1cc12926k. [DOI] [PubMed] [Google Scholar]
- 122.Han MJ, Xiong DC, Ye XS. Chem. Comm. 2012;48:11079–11081. doi: 10.1039/c2cc35738k. [DOI] [PubMed] [Google Scholar]
- 123.Li Q, Dong T, Liu X, Lei X. J Am Chem Soc. 2013 doi: 10.1021/ja401989p. [DOI] [PubMed] [Google Scholar]
- 124.Willis NJ, Bray CD. Chemistry. 2012;18:9160–9173. doi: 10.1002/chem.201200619. [DOI] [PubMed] [Google Scholar]
- 125.van Kasteren SI, Kramer HB, Jensen HH, Campbell SJ, Kirkpatrick J, Oldham NJ, Anthony DC, Davis BG. Nature. 2007;446:1105–1109. doi: 10.1038/nature05757. [DOI] [PubMed] [Google Scholar]
- 126.Brustad EM, Lemke EA, Schultz PG, Deniz AA. J. Am. Chem. Soc. 2008;130:17664–17665. doi: 10.1021/ja807430h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lawrence AL, Adlington RM, Baldwin JE, Lee V, Kershaw JA, Thompson AL. Org. Lett. 2010;12:1676–1679. doi: 10.1021/ol100138k. [DOI] [PubMed] [Google Scholar]
- 128.Wu B, Wang Z, Huang Y, Liu WR. Chembiochem. 2012;13:1405–1408. doi: 10.1002/cbic.201200281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Willems LI, Verdoes M, Florea BI, van der Marel GA, Overkleeft HS. Chembiochem. 2010;11:1769–1781. doi: 10.1002/cbic.201000280. [DOI] [PubMed] [Google Scholar]
- 130.Wan W, Huang Y, Wang Z, Russell WK, Pai PJ, Russell DH, Liu WR. Angew. Chem. Intl. Ed. 2010;49:3211–3214. doi: 10.1002/anie.201000465. [DOI] [PubMed] [Google Scholar]
- 131.Karver MR, Weissleder R, Hilderbrand SA. Angew. Chem. Intl. Ed. 2012;51:920–922. doi: 10.1002/anie.201104389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Niederwieser A, Spate AK, Nguyen LD, Jungst C, Reutter W, Wittmann V. Angew. Chem. Intl. Ed. 2013;52:4265–4268. doi: 10.1002/anie.201208991. [DOI] [PubMed] [Google Scholar]
- 133.Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. Science. 2008;320:664–667. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Greiss S, Chin JW. J. Am. Chem. Soc. 2011;133:14196–14199. doi: 10.1021/ja2054034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bianco A, Townsley FM, Greiss S, Lang K, Chin JW. Nat. Chem. Biol. 2012;8:748–750. doi: 10.1038/nchembio.1043. [DOI] [PubMed] [Google Scholar]