Abstract
Explosion of disaster health information results in information overload among response professionals. The objective of this project was to determine the feasibility of applying semantic natural language processing (NLP) technology to addressing this overload. The project characterizes concepts and relationships commonly used in disaster health-related documents on influenza pandemics, as the basis for adapting an existing semantic summarizer to the domain. Methods include human review and semantic NLP analysis of a set of relevant documents. This is followed by a pilot-test in which two information specialists use the adapted application for a realistic information seeking task. According to the results, the ontology of influenza epidemics management can be described via a manageable number of semantic relationships that involve concepts from a limited number of semantic types. Test users demonstrate several ways to engage with the application to obtain useful information. This suggests that existing semantic NLP algorithms can be adapted to support information summarization and visualization in influenza epidemics and other disaster health areas. However, additional research is needed in the areas of terminology development (as many relevant relationships and terms are not part of existing standardized vocabularies), NLP, and user interface design.
Keywords: Disaster Information Management, Natural Language Processing, Semantic Predications, Online Information Extraction Application, Semi-Automated Ontology Development
INTRODUCTION
The United States and the international community are becoming increasingly concerned with building an infrastructure for disaster preparedness and response, and considerable effort is being directed at public health and medical aspects of disasters, particularly for influenza pandemic. There has been an exponential growth in relevant information. For example, the PubMed query “influenza pandemic” retrieves 299 MEDLINE citations for 1999–2000, and 2,166 for 2008–2009. An even larger increase has occurred in the number of non-academic information sources, which span a wide range of document types and sources, from lessons learned to policy development documents, frequently including updated federal guidelines and fact sheets, news items, and lately, social media postings.
Information explosion results in information overload among professionals who need to keep abreast of developments in the field, and several approaches to alleviating this overload have been proposed. One approach involves developing advanced information management tools to help the user find potentially useful documents. The output of such tools can provide practitioners with a basis for further, manual and human expert-based, steps in information management (e.g., human review, collaborative tagging). Semantic technology has been used to summarize and visualize information in clinical medicine, genomics, and research administration (Ahlers, Fiszman, Demner-Fushman, Lang, & Rindflesch, 2007; Fiszman, Demner-Fushman, Kilicoglu, & Rindflesch, 2009; Fiszman, Ortiz, Bray, & Rindflesch, 2008; Hurdle, Botkin, & Rindflesch, 2007). This technology is dependent upon domain knowledge in the Unified Medical Language System© (UMLS) © to represent concepts and relationships needed for natural language processing. However, at the present, the UMLS does not provide adequate coverage for disaster health applications.
This project makes use of semantic natural language processing (NLP), automatic summarization, and visualization as potential tools for addressing information overload in disaster health information management. The objective is to characterize the language used in describing concepts and relationships commonly used in disaster health-related documents on influenza pandemics, thus determining technical feasibility and potential usefulness of semantic NLP to contributing to an information overload solution. This work was the basis for extending existing semantic technology to the domain (Rosemblat et al., submitted). We also tested the application with health information specialists in order to better understand circumstances and requirements for potential real-life use and further inform the development of automatic methods for health disaster information management.
BACKGROUND
Information overload
Users of disaster health information are not a homogeneous group. They include public health professionals, emergency responders (e.g. firefighters, HAZMAT teams), response managers, healthcare providers, researchers, librarians, media representatives, and the lay public. Information sources are as diverse as the users. In addition to the academic research literature, users also need access to the so-called “grey literature” – “information produced on all levels of government, academics, business and industry in electronic and print formats not controlled by commercial publishing i.e. where publishing is not the primary activity of the producing body” (GreyNet International, retrieved March 2010). Additionally, the role of Twitter in propagating information about the public’s concerns points to the importance of monitoring the social media.
In a recently prepared report about information seeking behavior of professionals working in disaster health-related areas, Turoff and Hiltz (2008b) interviewed emergency coordinators, health professionals, academic researchers, and librarians about their preferred disaster health information sources, information needs, and behaviors. One of the most prominent findings was the diversity of sources (e.g. journals, Web Sites, books, reports, etc.) that various participants considered “highly useful.” Given the volume and diversity of information resources, information overload has become a major concern in the field of disaster health information management. The volume and diversity of information can be problematic for two reasons. On one level, it suggests that in most situations, the number of potentially relevant documents may be much greater than what one busy professional can read, causing information overload. On another level, it reflects a situation where the relevant information is likely to be distributed across a range of documents. In many cases, no single document may be the exact match to a particular professional’s information needs, with the useful information distributed across a range of sources and tangential to each. In this case, the overload is that of “suboptimal” information, from which one needs to somehow extract what is useful.
Disaster health information concepts and relationships
Disaster information seeking is further complicated by the lack of a universally accepted terminology for relevant concepts (Birnbaum, 2007; Mudalige, Carley, & Mackway-Jones, 2006). For example, some articles may refer to “disaster response,” while others may describe the same actions as “disaster recovery” or “mitigation.” While for many professionals specializing in emergency management each of these terms has a distinct meaning, much of the more general literature uses them interchangeably. The problem of vagueness also applies to more specialized areas of disaster medicine. For example, Braga and colleagues describe the variation in definitions and usage of terms such as “stress” and “trauma” in relationship to PTSD (Braga, Fiks, Mari, & Mello, 2008). Preferred terminology may also change over time in response to social factors, as in the case of “swine flu” vs. “H1N1.” Finally, many of the terms used are non-unique abbreviations, which introduce further ambiguity.
In addition to terminology, semantic NLP requires ontological knowledge about relationships in a domain, and how they are expressed in relevant text. In clinical medicine, for example, it is important to know that there is a “TREATS” relationship between disease concepts and drug concepts, which may be syntactically expressed in different ways.
The Unified Medical Language System (UMLS) provides extensive knowledge in the clinical domain. The Metathesaurus comprises more than 100 controlled vocabularies, such as SNOMED-CT and MeSH. Each concept contains synonymous terms in the constituent terminologies. They are also assigned one or more semantic types, or semantic super-categories. For example, the concept “Influenza” has the semantic type ‘Disease or Syndrome’, with synonyms “Flu” and “Grippe,” among others. In addition, the UMLS Semantic Network encodes ontological relationships in the biomedical domain. Each such relationship consists of a predication in which arguments are semantic types from the Metathesaurus. For example, the relationship “Pharmacologic Substance TREATS Disease or Syndrome” indicates that any Metathesaurus concept with the semantic type ‘Pharmacologic Substance’ is allowed to appear as the first argument in a TREATS relationship in which ‘Disease or Syndrome’ is the second argument. Although the UMLS was not devised for disaster health, its extensive coverage and formal structure serve as a valuable basis for developing terminology and ontological relationships needed to support semantic processing in the influenza epidemic domain.
Advanced information management tools for disaster health preparedness
Informatics response to the challenges of disaster information management spans many types of initiatives, focusing on surveillance and threat detection (Chapman et al., 2005; Weiner & Trangenstein, 2008), portability of patients’ EHRs and tracking of victims during disasters (Gill et al., 2010), support of real-time data management in Emergency Operations Centers (Andersson & Pilemalm, 2008; Netten & van Someren, 2008) and virtual world simulations for education and training (Kamel Boulos, Ramloll, Jones, & Toth-Cohen, 2008).
Efforts to manage and organize textual documents are less common, but essential to mitigating information overload. For example, Roman et al. (2008) and Collins et al. (2009) developed an algorithm for identifying common themes in various California Wildfire documents and federal and states’ pan flu plans. In a different approach, Song and Chang (2008) used text mining to create an ontology of abbreviations used in disaster information literature. Our approach is based on Semantic MEDLINE (Kilicoglu et al., 2008), an application that helps users manage the results of PubMed searches. The tool uses concepts and relationships in the UMLS to summarize clinically significant information found in MEDLINE citations. Information is viewed as a network of predications, where a predication is a pair of concepts and a relationship that connects them (e.g., “Oseltamivir TREATS Influenza”). Such information is then visually represented in a graph which maintains links to the original documents.
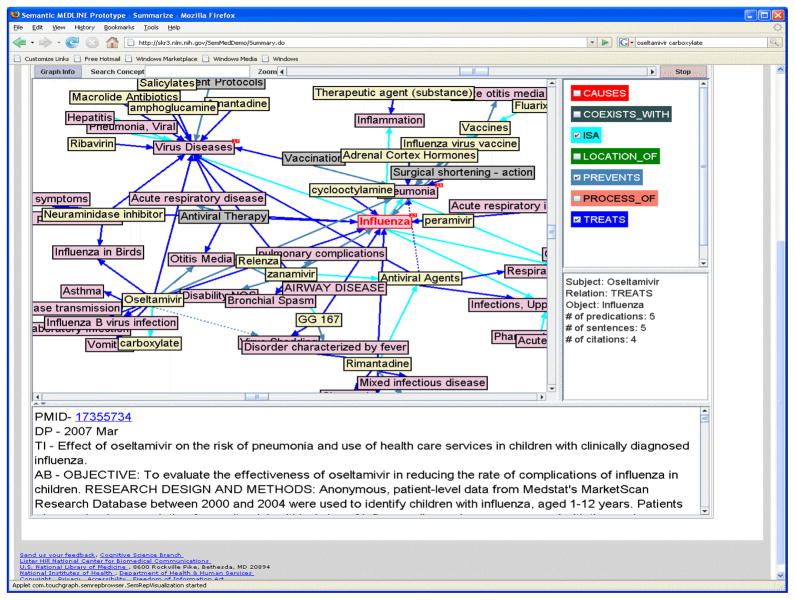
Figure 1 shows the results of summarizing over 3000 citations retrieved using the PubMed query “influenza AND drug therapy” and illustrates the ability to focus on specific information in a large number of documents. Concepts are represented by text boxes, and relationships by directional arrows. Clicking on the arrow of a relationship links to the sentences that contain that relationship. In this figure, the user has clicked on the arrow representing TREATS in the relationship “Oseltamivir TREATS Influenza.” One of the sentences from which this relation was extracted (from citation with PMID 17355734, “Effect of oseltamivir on the risk of pneumonia and use of health care services in children with clinically diagnosed influenza”) is displayed in [a]:
Figure 1.
Summary of the 3000 citations retrieved with the PubMed query “influenza AND drug therapy” (with Oseltamivir TREATS Influenza selected).
-
a
“Overall, children who received oseltamivir for the treatment of physician-diagnosed influenza were 51.7% less likely to be clinically diagnosed with pneumonia at a subsequent medical encounter.”
Semantic MEDLINE depends on semantic predications (or relationships) extracted from input text by SemRep (Rindflesch & Fiszman, 2003). The processor recognizes “oseltamivir” and “influenza” as concepts contained in the UMLS Metathesaurus, and identifies the former as a drug and the latter as a disease. SemRep further uses a rule that applies to this syntactic context and maps to the UMLS Semantic Network relationship TREATS, which represents the semantic relationship between the two concepts just mentioned. In previous work (Kilicoglu et al, 2008), Semantic MEDLINE was shown to be applicable to a variety of practical problems, from generating drug profiles to providing access to a medical encyclopedia (Fiszman, Rindflesch, & Kilicoglu, 2004; 2006). The thrust of the work reported here is to describe the enhancements needed to extend Semantic MEDLINE to the influenza epidemic domain by augmenting core terminology and relationships in the UMLS Metathesaurus and Semantic Network (see Figure 2 for a sample output of the enhanced tool). Further, we report on a preliminary user study that investigates the potential for exploiting semantic NLP in the disaster health domain.
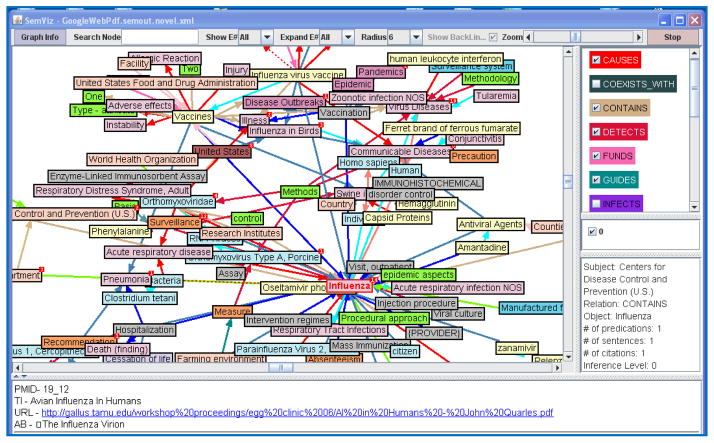
Figure 2.
Summary of pdf documents (selected relationships), obtained via “H1N1” Google query.
METHODS
Overview
Our methodology follows a bottom-up approach that combines manual and machine processing. We first analyzed disaster health-related documents on influenza pandemics in order to characterize common concepts and relationships in this domain. Computer assisted methods were then used to bring the results of this analysis into clearer focus, as the basis for the formal enhancements to the UMLS (Rosemblat et al., submitted). Finally, we ran Semantic MEDLINE extended with these concepts and relationships on documents in the domain and conducted a pilot test with users to assess its ability to assist in realistic tasks.
Selecting a representative document set
For training, we selected a set of public health and administrative influenza documents, of interest to public health professionals and administrators, as well as the lay public, from the Resource Guide for Public Health Preparedness, created by the New York Academy of Medicine (NYAM) and sponsored by the National Library of Medicine. The Guide is designed to provide “a single point of access to essential information resources in public health and disaster preparedness selected and cataloged by information specialists” (NYAM, 2010). It contains a broad range of document types, all highly relevant to disaster health information management, including but not limited to, expert guidelines, factsheets, and research and administrative reports.
The database was queried using a number of pandemic influenza and avian influenza related terms (e.g. pandemic influenza, avian influenza, highly pathogenic influenza, HPAI, H5N1). As this work was conducted before the 2009 H1N1 outbreak, H1N1-related terms were not used. We further selected a set of documents which consisted of documents 1) longer than five pages in length, containing abstracts or executive summaries and 2) five pages or fewer in length. Long documents without abstracts or summaries were not selected, because we wanted to limit extracted information that is minimally relevant to the main ides of the documents. Following the original NYAM document classification scheme, the final set of 106 documents was classified into the following types:
Research article or research report: 54
Administrative report: 15
Guideline or protocol: 12
Fact sheet: 6
Web site: 4
Journalistic article: 4
Presentation and conference paper: 3
Legislative document: 2
Speech or testimony; Commentary or editorial; Executive summary; Web cast; Training tool; Press release: 1 each
Discovering core semantic relationships
In order to identify important relationship classes and concepts relevant to pandemic influenza management, we read all the documents in the set, marking expressions that appeared to be important in the domain. For each relationship, concepts implicated in it were noted. For example, a guidance relationship between “Crisis Management Center” and “response” was extracted from “The United States is providing expertise and funding to assist the FAO to develop a Crisis Management Center that will facilitate their ability to mount and coordinate international rapid response to AI [avian influenza] outbreaks worldwide.” Resulting relationships were grouped into conceptual clusters, with potential concept synonyms noted. This clustering was discussed in several research group meetings, and a preliminary ontological schema of pandemic flu relationships was created. Finally, the schema was discussed with a public health disaster management expert, who confirmed the validity of the resulting representation and proposed some additions.
This preliminary work formed the basis for further, computer-assisted analysis, which brought into clearer focus the details needed to represent crucial concepts and relationships for the influenza epidemic domain consistently in the UMLS.
Computer-assisted analysis
Computer assisted processing proceeded in two phases, which are explained in more detail elsewhere (Rosemblat et al., submitted); we give an overview here. In the first phase (sentence selection), we exploited a small set of 344 sentences manually assigned to one of 6 predetermined core topics in disaster management (control, detection, information, sponsorship, surveillance, supervision). These sentences were used to train two separate classifiers for automatically assigning sentences to one of the 6 core topics. Two well-known supervised machine learning techniques, linear SVM (Vapnik, 1995) and C4.5 decision tree (Quinlan, 2001), underlie these classifiers. Sentences are represented as bag-of-words. We used svmlight (http://svmlight.joachims.org/) implementation of linear SVM and Weka J48 implementation of C4.5 decision tree (http://www.cs.waikato.ac.nz/ml/weka/). We evaluated the trained models on a set of 89 sentences, also manually labeled. With the linear SVM, we obtained precision of 0.79 and recall of 0.69. The classifier based on C4.5 decision tree performed better with 0.89 precision and 0.87 recall. We then applied these models to 2715 unlabeled sentences occurring in documents drawn from various government Web sites. For the second phase of the computer-assisted processing, we only considered those sentences on which both classifiers agreed, as these are most likely to be useful as the basis for ontology development.
In the second phase, for each relationship, labeled sentences were subjected to a program which attempted to map each noun phrase to concepts in the Metathesaurus and subsequently separated noun phrases that mapped to a concept from those that did not. The program also extracted all verbs from the sentences expressing each relationship. The output from this program was used for preliminary determination of UMLS coverage of the influenza epidemic domain. Noun phrases which mapped to the Metathesaurus were noted. Non-mapping noun phrases important in this domain were assigned a semantic type based on the informal analysis discussed in the subsection on “Discovering core semantic relationships”. These results served as the basis for formally modifying and extending the UMLS in support of Semantic MEDLINE processing of influenza epidemic documents (Rosemblat et al., submitted).
Testing enhanced Semantic MEDLINE with users
Finally, we ran a test to gain insight into the potential of Semantic MEDLINE summarization and visualization to help users find documents in the influenza epidemic domain that fulfilled their specific information needs. The query “H1N1” was issued at two Web sites: 1) NYAM Resource of Public Health Preparedness and b) the Center for Disease Control (CDC). Documents retrieved were submitted to Semantic MEDLINE enhanced for influenza epidemic, and the summarized graphs were examined by potential users.
Two information specialists from the Division of Specialized Information Services of the National Library of Medicine were given access to the semantic graphs produced by Semantic MEDLINE and asked to imagine developing a Web page with links to H1N1 2009 resources for health professionals and the lay public. Both participants had educational backgrounds in science and extensive experience with information technology; one of the participants had a Master’s Degree in biology specializing in bio-defense. Participants were told that the summaries were created by a preliminary version of the tool and were asked to imagine that the tool could extract documents with greater recall and precision. The task was described as reviewing the summaries and commenting about potential ways to integrate them into the task of collecting information for the Web page. Each session lasted approximately one hour and was video-recorded for analysis via Morae, TechSmith software.
RESULTS
Relationships and their organization: influenza epidemic themes
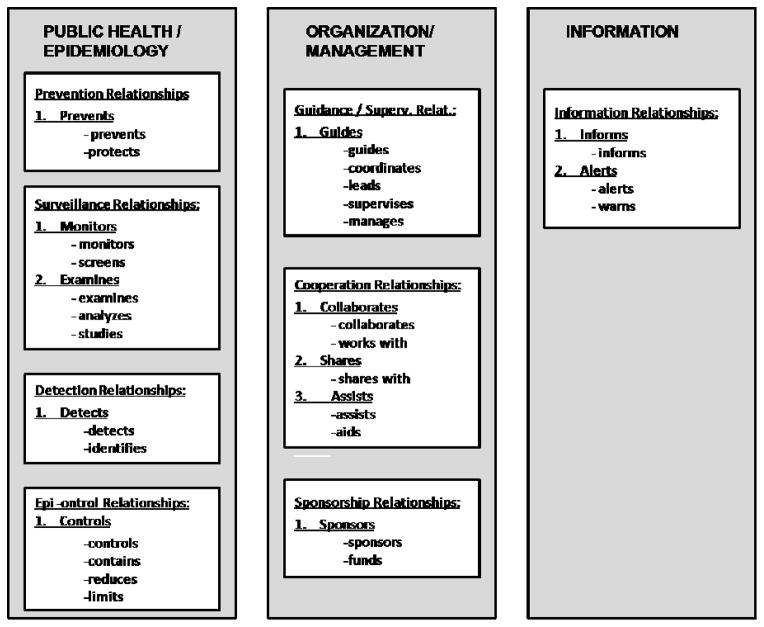
Three major themes emerged from analyzing the training documents on influenza epidemic; each theme can be characterized with a cluster of relationships. The first, A1, which we call Public Health and Epidemiology, pertains to actions most directly connected with preventing disease and preserving the health of the population: conducting surveillance, along with preventing and controlling disease. The second cluster, A2, Organization and Management, refers to actions of supervision, cooperation, and sponsorship among the organizations and entities involved in epidemics management. The third involves relationships related to dissemination of Information during epidemics (A.3). On the basis of this informal work, we conducted additional, computer-assisted analysis. The following specific relationships and concepts are involved in these semantic areas.
Public Health and Epidemiology
In the influenza epidemic literature this theme comprises four relationships: prevention, surveillance, detection, and epidemic control. There is considerable interconnectedness in the concepts involved in these relationships.
Prevention
Discussion about recommendations for preventing epidemics plays an important role in the preparedness discourse. Prevention is typically performed by interventions (e.g. hand washing), procedures (vaccinations), and drugs (neuraminidase inhibitors). The thing prevented is typically a disease or syndrome (influenza), but it can also be a less tangible concept (the spread or threat of a disease or virus).
Surveillance
Surveillance is the basis of epidemic control and is critical to public health and epidemic management. In our scheme, this cluster has two relationships: monitors and examines. Monitoring concerns populations, is conducted on a large scale, and involves tracking a large number of events, occurrences, or agents. For example, a health department can monitor the number of sales of over-the-counter medications. Examining involves analyzing or studying on a smaller scale. In the texts, actors performing surveillance are divided into public health actions (surveillance), devices (test kits), and organizations (health departments, national and international organizations, as well as local, state, and federal governmental departments). Surveillance sentences often refer to specific virus subtypes (H1N1) or disease subcategories (seasonal influenza). In the language of the documents, recipients of surveillance are also things that need to be examined, (hospitalizations, migratory birds, points of entry).
Detection
A less frequent, but important relationship is detects. Determining or discovering presence of disease is essential to epidemic management. Detection can be performed by organizations, public health actions (screening), as well as devices (test kit). Things that need to be detected include virus, outbreak, disease, pandemic, infection, drug resistance. As with surveillance, detection sentences often refer to specific virus subtypes or disease subcategories.
Epidemic control
Actions related to controlling epidemics are highly important in the non-academic influenza literature. The crux of these actions is managing the spread of the disease. Epidemic control is performed by drugs (either classes such as antiviral agent or neuraminidase inhibitor or specific drugs such as oseltamivir) and procedures (prophylaxis). In a few rare cases actors exercising epidemic control were professional groups (public health officials) or population groups (first responders, community members). For the most part, however, these actors were public health actions (surveillance, detection) and drugs and interventions (hand washing), the latter playing a critical role in epidemic control. Things to be controlled overlap with those that need to be detected: virus, outbreak, disease, pandemic, infection, drug resistance. The object of epidemic control is typically a disease or syndrome (influenza) or a virus (H1N1), in addition to a spread or a threat. Sentences that deal with control focus on the broad rhetoric of mitigating epidemics, as opposed to specifics (H5N1, seasonal influenza) typical of surveillance and detection.
Organization and Management
Guidance and supervision
Supervisory relationships are critical in administrative reports, guidelines and protocols, and define the chain of command in epidemics management. The main relationship of this cluster is guides. Both guidance and supervision can be done by an organization (Department of Homeland Security [DHS], Food and Agriculture Organization [FAO], Center for Disease Control [CDC]), professional or occupational group (healthcare providers, Secretary of Homeland Security, leaders, governors, facilitators), population group (experts) or information resources (National Avian Influenza Response Plan). Guidance or supervision is exercised over a less authoritative organization (local authorities), professional or occupational or population groups (residents, participants, the public), or response action (messaging, preparedness, operations).
Cooperation
While supervision is an important component of influenza pandemics management, so is cooperation, as many organizations work together, coordinating efforts and sharing resources. Our cooperation cluster has three relationships: collaborates, shares (defined as giving out a portion or using jointly), and assists. Collaboration typically occurs between organizations or professional, occupational or population groups. Sharing also occurs between organizations or groups, and it involves sharing of resources.
Sponsorship
Another important aspect of epidemics management is sponsorship, characterized by the funds relationship. Organizations or geopolitical entities (countries) sponsor other organizations or geopolitical entities and activities.
Information
Informing and alerting
Communication among responders, health professionals, the public, and the media is essential during epidemics. Two information relationships noted in the texts are informs and alerts. Representing these relationships helps summarize information about the communication flow during epidemics. Informs concerns disseminating information; alerts is related to it, but implies an urgent nature and a brief message. Informing and alerting can be performed by organizations (Departments of Health), professional groups, geopolitical entities (Utah), population groups, and information resources (guideline, action plan). The objects of these actions are, similarly, organizations and professional groups, geopolitical entities, and population groups.
The relationships seen in the training documents are summarized in Figure 3, along with examples of the terms that expressed them. On the basis of this informal work we conducted additional, computer-assisted analysis.
Figure 3.
Relationships in the training documents, with some examples of the terms that expressed them (the relationships are numbered; the verbs under them are the terms that expressed them)
Sentences classified for influenza epidemic relationships
Table 1 provides examples of sentences from the training data automatically assigned as expressing relationships by the machine-leaning classifiers. The sentences retrieved by the classifiers then served as the basis for further computer-assisted analysis. The results of determining which noun phrases mapped to concepts in the Metathesaurus are given in the next section, followed by verbs extracted from the sentences for each relationship. Some of the relationships were not submitted to the machine classifier because they were part of the original UMLS, or because their frequency in the training set was low.
Table 1.
Sentences assigned to relationships by automatic classifiers
| Theme | Relationship | Sentence |
|---|---|---|
| Public Health and Epidemiology | Detects | Surveillance preparedness to date has emphasized early detection of an outbreak |
| (Epidemic) controls | The use of social mitigation measures may represent the most effective means for reducing transmission of virus in the fall when it is spreading most efficiently. | |
| Organization and Management | Supervises | The federal government must take a lead in providing guidelines to states on surge capacity planning. |
| Sponsors | Also, the country must make a sustained commitment to pandemic preparedness by providing consistent federal funding for stockpiling medicines and medical supplies, training, and planning activities. | |
| Information | Informs | Can local or state health officials determine and share information with other decision-makers about the following? |
UMLS Terminology and relationship indicators
Below, we give an overview of the UMLS Metathesaurus concepts deemed important for the influenza epidemic domain, and of those, which concepts had a domain-appropriate semantic type. The concepts were generated through automated analysis of the training documents, as detailed in the Computer-assisted analysis section.
Public Health and Epidemiology
Methods and procedures for surveillance and detection are well represented in the UMLS, exemplified by the following terms: “Test” (semantic type ‘Laboratory Procedure’), “Test kit” (‘Medical Device’), “Surveillance” (‘Health Care Activity’), “Screening method / protocol” (‘Health Care Activity’), “Assay” (‘Laboratory Procedure’), “Diagnostic device” (‘Medical Device’). Representation of organizations and professional or occupational groups, on the other hand, is less comprehensive. Federal agencies, such as the Department of Health and Human Services, and the Centers for Disease Control and Prevention, and well-known entities, such as the World Health Organization occur in the Metathesaurus; however, World Organization for Animal Health does not. Concepts for many professional groups, such as public health specialist and animal health specialist are not represented.
Medically-oriented subjects of control are largely represented in the UMLS: e.g. “Antiviral agent” is a ‘Pharmacological substance’, “Chemoprophylaxis” is a ‘Therapeutic or Preventive Procedure’, “Decontamination kit” (‘Medical Device’), “Quarantine” (‘Therapeutic or Preventive Procedure’). However, other agents of control either do not appear (non-pharmaceutical intervention, social distancing) or their semantic types are not useful for the influenza epidemic domain. For example, “Hand washing” has semantic types ‘Daily or Recreational Activity’, ‘Healthcare Activity’, and ‘Finding’. Of the three, only ‘Healthcare Activity’ falls within the scope of this domain.
The objects of mitigation are well-represented in the UMLS, with appropriate semantic types ‘Disease or Syndrome’, ‘Virus’, or ‘Phenomenon or Process’ (e.g. “Outbreak” and “Pandemic”).
Organization and Management
Things that can be guided, supervised, or funded are diverse. Often, these are organizations or population groups (e.g. residents, public, participants) or specific activities (e.g. communication campaign, vaccination program), but they can be highly general concepts, such as efforts, decisions, actions, response, steps, preparedness, and activities. These general concepts are in the UMLS, but are denoted by very diverse semantic types, which are not usually useful in the influenza epidemic domain. For example, ‘Decision” is a ‘Mental Process’, while “Step” is a ‘Conceptual entity’. A wide range of relevant concepts are not in the UMLS, including the following: Center for Biologics Evaluation and Research, border patrol, governor, local leader, and Secretary of Homeland Security.
Information
Actors and recipients of information relationships are often organizations and population groups, similar to the organizations and professional and population groups involved in the other relationships. They are only sparsely present in the UMLS, especially in the case of local organizations. For example, both the organization ‘State Emergency Operation Center’ (EOC) and the expression ‘managers’ are missing.
Verbs serving as possible indicators of relationships
Table 2 contains verbs automatically extracted from sentences classified by relationship. Such verbs are used by Semantic MEDLINE to map to relationships in the ontology.
Table 2.
Verbs extracted from classified sentences.
| Theme | Relation | Verbs |
|---|---|---|
| Public Health and Epidemiology | MONITORS | monitors, screens, conducts surveillance |
| EXAMINES | inspects, examines | |
| EPIDEMIC CONTROLS | reduces, limits, controls, mitigates | |
| Organization and Management | GUIDES | coordinates, leads, guides, manages, supervises |
| COLLABORATES | coordinates, collaborates, works with | |
| SPONSORS | funds, sponsors, invests, contributes, allocates, provides | |
| Information | INFORMS | informs, provides information |
| ALERTS | alerts, warns |
Running enhanced Semantic MEDLINE and user testing
Based on the results discussed above, both the UMLS Metathesaurus and the Semantic Network were modified to support Semantic MEDLINE processing in the influenza epidemic domain (See Rosemblat, et al., submitted, for details). Searches were conducted at the NYAM and CDC Web sites, as well as via general Google search and a wide range of documents were retrieved. For example, the 46 most relevant documents retrieved from the NYAM site included documents from the CDC Web site, the Trust for America’s Health (www.healthyamericans.org), a research article from the BMC Public Health journal, a report prepared by the U.S. House of Representatives Committee on Homeland Security about pandemic preparedness, and a report issued by several health care workers unions concerned about protecting union members during an influenza epidemic. Semantic MEDLINE extracted a total of 2468 predications from these documents, including many in the influenza epidemic domain. For example, 60 DETECTS predications included “United States DETECTS Disease Outbreaks,” and EPI_CONTAINS predications (for epidemiologically contains) included “Quarantine EPI_CONTAINS public health emergency.” All predications were summarized and visualized with Semantic MEDLINE.
Finally, we conducted two case studies in which users were given access to Semantic MEDLINE results. Our goal was to generate pilot data about the potential usefulness of the application to professionals seeking information on influenza epidemics. Due to the small sample of testers, the information seeking behavior of the participants should be seen as a source of hypotheses, rather than conclusions.
Types of inquiries and the impact of searcher’s background knowledge
While using the tool, both participants engaged in three types of information seeking behavior. The first can best be described as goal-driven, when the participant searches the graph for information answering a specific, predefined question. For example, both participants at some point in their session attempted to find articles about antiviral medications that treat influenza. This involved making sure “TREATS” relationship was selected, locating names of known antiviral drugs (e.g., oseltamivir, zanamavir), and looking up the relationship between these medications and “influenza.” The second type of inquiry is best described as opportunity-driven. It occurs when the user, while searching the graph for a particular purpose, notices something that is not related to the purpose, but appears to be a valuable path to pursue. For example, while looking for treatment and prevention relationships, the first participant noticed “Employer INSPECTS workers,” which prompted her to follow-up with searches for business guidelines for continuity of operations. Inquiries of the third type, curiosity-driven, were similarly unplanned and also elicited surprise (as in “I wonder what this is doing here”). For example, at one point in her search, the first participant noticed “Vaccination TREATS ascending hemiplegia”, frowned in puzzlement, and decided to look up the article that contained this relationship. Ascending hemiplegia was not part of her mental model of the domain, and puzzlement was the primary reason for following up, in order to either fill a gap in her domain knowledge or identify an NLP error.
The use of various inquiries seemed to depend on the depth of background knowledge of the subject. The first participant, who had extensive knowledge of influenza and epidemic management, relied on a combination of goal-driven and opportunity-driven inquiries. Without prompting, she started the exercise by developing an outline for the Web page she wanted to create. The outline included the following sections: 1) Overview, 2) Signs and symptoms, 3) Treatment, 4) Global biosurveillance and 5) Diagnostics and tests. “Prevention” was added later. She then attempted to search for information on each section of the outline in turn. Her background knowledge and familiarity with the terminology guided her to the right relationships, when they were included in the summary, as in the case of treatment and prevention. Difficulties arose when the relationship was not in the summary (as in the case of information on signs and symptoms), because she had difficulty recognizing the absence and spent time looking for non-included information.
In addition to goal-driven inquiries, this participant conducted many opportunity-driven look-ups, using the results to expand her outline. Both goal-driven and opportunity-driven lookups frequently served to jog her memory, prompting several “Aha!” moments. For example, seeing the concept “isolation” in the graph prompted her to add “non-pharmaceutical interventions” bullet to the treatment and prevention section of her outline.
The second participant did not have the background knowledge to provide a skeleton for goal-driven inquiries, and conducted more opportunity driven inquiries. In his own words, he was using the tool to “get the lay of the land” and to develop “a framework of what is important” and “a reading list.” Yet, limited background knowledge may restrict one’s information schema and make it difficult to come up with a meaningful sequence of inquiries. While the second participant viewed the documents with predications relating a) influenza and vaccination and b) influenza and specific antiviral drugs, he did not view the documents with predications about non-pharmaceutical interventions for influenza treatment.
Both participants conducted some curiosity-driven look-ups, or look-ups of predications that did not seem to make sense to them. Most often, predications that appeared surprising, such as “vaccine CAUSES influenza”, were due to inaccurate extractions, which are a concern for using NLP tools in a practical setting. Like any NLP tool, Semantic MEDLINE produces an output that has less than 100% precision, extracting some predications that are not accurate. Neither participant, however, had difficulty recognizing them as system imperfections and moving on with the task. They would typically select the predication, choose to view the sentence in the text that the predication was extracted from, and dismiss the document.
Match between system’s relationships and participants’ inquiries
Both participants understood the idea of predicational representation and were able to use it, but not with equal ease. The second participant stated that he would have preferred being able to view all the articles concerning a group of related concepts. He pointed to the three concepts: “Surveillance”, “Preparedness,” and “Measure,” all color-coded gold (because they are activities that can act as subject arguments of PREVENTS), and said, “I like gold items and I want to see all the gold items.”
Both participants also paid attention to clusters, or concepts that were connected to many other concepts, and often chose to investigate by cluster, rather than by relationship type. A predication is not the most intuitively accessible unit of information seeking for human information seekers. Relationships are often implied, but not explicitly specified, in queries. For example, a searcher conducting “zanamivir AND influenza” inquiry is likely to be interested in how zanamivir treats influenza, but the focus is on these two argument noun, rather than on the verb or part of speech that connects them. Yet, both participants in the study selected and deselected relationships and manipulated them in order to gain different perspectives on the information in the dataset.
The participants focused on clinical and public health and epidemiology relationships rather than on the organizational and management relationships. The first participant explained this by the nature of her work and the task, saying that the Division of Specialized Information Services [NIH] was less likely to cover organization and management in its Web page. However, she also stated her interest in searching for business continuity of operation guidelines, which involved management relationships.
Both participants viewed predications that involved TREATS, PREVENTS and LOCATION_OF relationships. The first participant also viewed predications containing ISA, CONTAINS, INSPECTS and MANAGES, while the second participant also viewed predications containing INFECTS relationship. The first participant explored three relationships that are not part of the standard UMLS Semantic Network – CONTAINS, INSPECTS and MANAGES, while the second participant only viewed standard UMLS relationships.
Recognizing which relationships are important may be a function of background knowledge. The first participant also activated relationships in groups that resembled our relationship clusters. For example, she started out by deactivating FUNDS, ISA, MANAGES, COORDINATES, and COEXISTS_WITH, thus isolating and removing the management-organization cluster. The second participant did not use such strategies, possibly because the system did not provide him with a legend for interpreting relationships (he commented that he would like to have a way to manage relationships).
Tool improvement comments and suggestions
Both participants felt that the tool was useful for previewing a new domain with rapidly developing information topics, and for triggering one’s recall in a familiar domain. The current test version of the tool could only process a limited number of documents, thus affecting the scope of the results displayed to the participants. Both agreed that with greater completeness of coverage, the tool would be useful to a variety of disaster health information seeking tasks. Occasional retrieval errors seemed to be less of a concern, because they were recognized and filtered out by human users.
The participants made some suggestions for organizing the information for the viewers and enabling users to work together. The first participant suggested a set of filters that would allow viewing information from specific sources, such as federal government, local government, academia, and commercial sources. The second participant envisioned a social networking version of the tool, where each user could identify predications considered important, share graphs, and compare them with other users.
Both participants were able to use the tool’s basic functions (e.g., controlling the view by zooming and dragging boxes). When asked about the tool’s usability, the first participant said that she found the tool easy to use. The second participant wished for a way to understand and manage relationships (currently displayed as a list), grouping them and moving aside those he had already viewed. He also wished for a three-dimensional representation of the graph, supplemented with a legend explaining the graph’s color codes. This participant also suggested that the tool should have a way to bookmark and manage articles linked to the tool’s predications.
DISCUSSION
Ontology, terminology, NLP and computing issues
The project demonstrates the feasibility of applying semantic NLP to address information overload in disaster health information management. For influenza management, the domain could be represented with a limited set of relationships, and the actors and recipients of the relationships belong to a manageable number of semantic types. We expect that organization management and information relationships added for influenza management may be applied to other areas of disaster health (e.g., chemical or radiological attack, biological attack, natural disaster). Public health and epidemiology relationships are likely to apply to some, though not all, disaster health situations. Future efforts, applied to broader range of document collections, will identify the relationships relevant to the other areas of disaster health, as well as some additional relationships of influenza management.
Our work also demonstrates that significant research and development effort is needed before semantic natural language processing can be applied to the practical task of disaster literature summarization. Disaster health is not a uniform domain. The type of response depends on the specific nature of the disaster, and so does the language that describes it. While we expect some overlap among the relationships in various domains, domain-specific relationships are likely to be common. The difference among the domains is bound to be even greater in the realm of terminology, with some domains requiring many more additions to the UMLS than influenza epidemics management. Developing disaster health terminology is a challenging process for a number of reasons. One challenge has to do with the task of collecting the terms. This task can be partly ameliorated by machine methods. Machine methods may be used to extract frequently occurring n-grams (noun strings of n terms) from a disaster / emergency literature text corpus, but the final term selection requires human review. Defining useful terms also requires domain expertise, which would allow terminology developers to extract terms at the level of granularity useful for summarization. For example, “immunization safety” is an important concept in epidemics management. It consists of two tokens, “immunization” and “safety”, both of them currently in the UMLS, but the complete 2-gram concept is not, and would have to be added to ensure coverage.
While machine methods can speed up term extraction, assigning semantic types is a laborious manual process. Another challenge has to do with the difficulty separating disaster health terms from often synonymous general clinical terms. For example, the concepts of “exposure” and “contamination” are critical to the medical management of radiation events. While both of these terms are currently in the UMLS, it is desirable for a disaster health summarizer to distinguish radiation exposure and contamination from other kinds of exposure and contamination, so separate concepts would need to be created. An additional difficulty stems from the international nature of disaster health, where each geographic area, group and organization brings to the table its unique language, yet each could really benefit from the ability to communicate seamlessly. A decision also needs to be made about where to draw the line between health-related and non-health-related aspects of disaster management. To address these difficulties, disaster health terminology needs to be conducted as a large-scale, sustainable process, by a group with continuous funding and large technical capabilities.
Since the development of the all-encompassing disaster health information summarizer is not a quick task, it can be undertaken via a modular approach, in which some domains are developed and released for practical use before the others. Influenza epidemics management is a good candidate for a priority release, because of partial coverage in the existing version of UMLS, and because of influenza epidemics’ relatively high likelihood of occurring. Increase terminology coverage will lead to improved accuracy of the underlying natural language processing, with both higher precision and recall, and that will translate into a more accurate, practical tool.
Usefulness and usability
Our pilot study suggested that a summarization tool can be useful for information specialists and health practitioners trying to keep abreast of the literature on rapidly evolving topics. Turoff and Hiltz describe three categories of users of emergency health information: 1) Emergency Professionals and Coordinators, 2) Health Related Professionals and 3) Researchers / Academics and Librarians. We envision this tool as more useful for the second and third categories, as these professionals work in settings that are more conducive to dealing with the unstructured texts that the tool is prepared to handle. One potential use case may involve a librarian in a large academic health center, collecting information on H1N1 vaccination and care of pediatric patients during a pandemic. The librarian, who is not the domain expert, may use the tool to organize the information to match queries from various professionals. This may include findings on drug / dose effectiveness for academic researchers, as well as storage capacity and safety monitoring for public health practitioners. Other use cases may involve an academic researcher using the tool to visualize information on the development of new therapies for H1N1, or a public health official using the tool to represent various aspects of establishing a large immunization program. A separate usability evaluation will be necessary for each use case and group of users, which should be conducted following additional ontology, terminology, NLP and computing work on the tool.
Specific usability recommendations for the tool are beyond the scope of this paper, but making the tool as intuitive as possible for busy professionals with no background in linguistics and NLP is essential. Informal discussion about the tool suggests that while the participants in our pilot study did not have trouble viewing information in terms of predications rather than individual concepts, other inexperienced users might, so a training module would be helpful. At the present time, the relationships in the control panel are not organized into clusters. Concepts are color-coded by semantic type, but there is no legend explaining the coding. These aspects of the tool need to be enhanced.
A less trivial enhancement, suggested by the information specialists in our pilot testing, involves introducing filters that would allow distinguishing between document types (e.g., guidelines, lessons learned, continuity of operations plans) and source domain (e.g., federal, state, academic, commercial). Recognizing the domain of the documents is not technically challenging, and neither is handling document types, provided that information is specified in the source database. For example, the NYAM Resource Guide for Public Health Preparedness tagged documents by type, allowing an easy distinction. For databases that do not include such tags, the issue becomes conceptual rather than computational.
Like any information management resource, a summarization tool aids human judgment rather than replace it. Users apply their background knowledge and the purpose of the specific task they are performing to select and follow the most valuable leads in the visual summary. In our pilot test exercise, participants with different level of background knowledge directed their attention to different predications. It appears that some general domain knowledge is beneficial for making the best use of the tool, because it helps one to group related propositions into a single search strategy (e.g., reviewing articles with propositions of the DRUG_treats_influenza type, followed by a review of non-pharmaceutical interventions for influenza). The tool may also provide support to users with less background knowledge by including semantic types of concepts in the graph along with the concepts (e.g., DRUG: oseltamivir) and by incorporating elements of social media (e.g., displaying what relationships were viewed and “approved” by other users).
Automation vs. expert-led approach
As mentioned above, NLP summarization is not a replacement of human judgment, and neither is it the only technological approach to alleviating information overload. Disaster information organization is a moving target, due to constantly evolving terminology, rapid emergence of new topics and changing policies. Under the circumstances, human expertise is essential for information management. The optimal expertise combines deep knowledge of specific disaster and health domains with information management expertise, and is often beyond the grasp of a single individual. As one potential solution, Turoff (2008a, 2009) advocates for establishing communities of practice and developing electronic tools to support information management within the communities. Vetted community members may contribute documents to databases and use collaborative (“folksonomy”) tagging to organize resources. While this is well beyond the scope of the pilot project we described, collaborative tagging and NLP can potentially be integrated in a tool that capitalizes on the strengths of each approach. For example, vocabulary development may combine automated terms extraction and collaborative tagging, thus relying on the wisdom of human experts while using automation to ease their work load.
Limitations of the study
While this study outlines and validates a method for reducing information overload, it does not present a finished product. The ontology of influenza epidemics management, presented in this paper, therefore, should not be viewed as a complete representation of the domain’s structure. Our goal was to propose a concept and outline the direction for promising future research, working with a manageable initial training data set. For this training set, we judiciously selected a resource that included documents from critical authoritative public health information sources (e.g., CDC, flu.gov, WHO). Yet, the documents were filtered via a single database. Additionally, while the extracted relationships were reviewed by an expert in public health disaster management, the initial extraction was done by the first and the last authors, who are not disaster management experts. It is, therefore, fully expected that the ontological coverage of the domain is not exhaustive. Another limitation of the study involves a small number of participants and the informal nature of the user study, which also stems from the proof-of-concept nature of the work. Once additional ontology, terminology and NLP work is conducted, a larger, more detailed evaluation will help finalize the interface and develop a tool for practitioners.
CONCLUSION
The goal of this project was to determine the feasibility of using semantic natural language processing, automatic summarization, and visualization to address information overload in disaster health information management. The work suggests that the ontology of the disaster health domain can be described via a manageable number of semantic relationships that involve concepts from a limited set of semantic types. With modifications that include the addition of (non-UMLS) relationships and concepts, Semantic MEDLINE could be extended to represent the domain of influenza epidemics management. Pilot testing suggests that the tool has the potential for reducing information overload for information specialists searching for documents in the domain. Additional work is needed in the areas of terminology building, natural language processing, and user interface design. Future efforts should focus on large-scale terminology development and NLP fine-tuning, applying the resulting tool to a variety of document sets, and testing the outcome with a range of professionals, including information specialists, public health workers and emergency managers.
Acknowledgments
This study was supported in part by the Intramural Research Program of the National Institutes of Health, National Library of Medicine.
References
- Ahlers CB, Fiszman M, Demner-Fushman D, Lang FM, Rindflesch TC. Extracting semantic predications from Medline citations for pharmacogenomics. Pacific Symposium on Biocomputing. 2007:209–220. [PubMed] [Google Scholar]
- Andersson D, Pilemalm S. Evaluation of crisis management operations using Reconstruction and Exploration. Proceedings of the 5th International Conference on Information Systems for Crisis Response and Management (5th ISCRAM); Washington, DC. 2008. pp. 118–125). [Google Scholar]
- Birnbaum ML. What’s in a word? Prehospital and Disaster Medicine. 2007;22(3):155–156. doi: 10.1017/s1049023x00004581. [DOI] [PubMed] [Google Scholar]
- Braga LL, Fiks JP, Mari JJ, Mello MF. The importance of the concepts of disaster, catastrophe, violence, trauma and barbarism in defining posttraumatic stress disorder in clinical practice. BMC Psychiatry. 2008;8:68. doi: 10.1186/1471-244X-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman WW, Christensen LM, Wagner MM, Haug PJ, Ivanov O, Dowling JN, Olszewski RT. Classifying free-text triage chief complaints into syndromic categories with natural language processing. Artificial Intelligence in Medicine. 2005;33(1):31–40. doi: 10.1016/j.artmed.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Collins LM, Blake M, Hoare G, Mane KK, Martinez MLB, Pittman J, Powell JE, Roman JH, Spearing S, White R, Yao X. Proceedings of the 6th International Conference on Information Systems for Crisis Response and Management (6th ISCRAM); Gothenburg, Sweden. 2009. Retrieved March 4, 2010, from http://www.iscram.org/ISCRAM2009/papers/ [Google Scholar]
- Fiszman M, Demner-Fushman D, Kilicoglu H, Rindflesch TC. Automatic summarization of MEDLINE citations for evidence-based medical treatment: a topic-oriented evaluation. Journal of Biomedical Informatics. 2009;42(5):801–13. doi: 10.1016/j.jbi.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiszman M, Ortiz E, Bray BE, Rindflesch TC. Semantic processing to support clinical guideline development. Proceedings of the AMIA 2008 Annual Symposium (AMIA’08); Washington, DC. 2008. pp. 187–191. [PMC free article] [PubMed] [Google Scholar]
- Fiszman M, Rindflesch TC, Kilicoglu H. Summarization of an online medical encyclopedia. Studies in Health Technology and Informatics. 107(Pt. 1):506–510. [PubMed] [Google Scholar]
- Fiszman M, Rindflesch TC, Kilicoglu H. Summarizing drug information in Medline citations. Proceedings of the AMIA 2006 Annual Symposium (AMIA’08); Washington, DC. 2006. pp. 254–258. [PMC free article] [PubMed] [Google Scholar]
- Gill M, Pearson G, Neve L, Miernicki G, Antani S, Thoma G. Lost Person Finder (LPF) 2010 Mar 4; Project description posted to http://archive.nlm.nih.gov/proj/lpf.php.
- GreyNet International. Mission Statement. Retrieved March 4, 2010, from http://www.greynet.org/
- Hurdle JF, Botkin J, Rindflesch TC. Leveraging semantic knowledge in IRB databases to improve translation science. Proceedings of the AMIA 2007 Annual Symposium (AMIA’07); Chicago, IL. 2007. pp. 349–353. [PMC free article] [PubMed] [Google Scholar]
- Kamel Boulos MN, Ramloll R, Jones R, Toth-Cohen S. Web 3D for public, environmental and occupational health: early examples from second life. International Journal of Environmental Research and Public Health. 2008;5(4):290–317. doi: 10.3390/ijerph5040290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilicoglu H, Fiszman M, Rodriguez A, Shin D, Ripple AM, Rindflesch TC. Semantic MEDLINE: A Web application to manage the results of PubMed searches. Proceedings of the Third International Symposium for Semantic Mining in Biomedicine; Turku, Finland. 2008. pp. 69–76. [Google Scholar]
- Mudalige M, Carley S, Mackway-Jones K. Who’s who at a major incident: standardising role titles for emergency planners. Emergency Medicine Journal. 2006;23(5):408–409. doi: 10.1136/emj.2005.030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netten N, van Someren M. Identifying segments for routing emergency response dialogues. Proceedings of the 5th International Conference on Information Systems for Crisis Response and Management (5th ISCRAM); Washington, DC. 2008. pp. 108–117. [Google Scholar]
- New York Academy of Medicine (NYAM) [accessed June 16, 2010];Resource Guide for Public Health Preparedness. http://www.phpreparedness.info/about.php.
- Quinlan R. C4.5: Programs for Machine Learning. Morgan Kaufmann; 1992. [Google Scholar]
- Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. Journal of Biomedical Informatics. 2003;36(6):462–477. doi: 10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Roman JH, Marks Collins L, Mane KK, Martinez MLB, Dunford CE, Powell JE., Jr Reducing information overload in emergencies by detecting themes in web content. Proceedings of the 5th International Conference on Information Systems for Crisis Response and Management (5th ISCRAM); Washington, DC. 2008. pp. 101–107. [Google Scholar]
- Rosemblat G, Keselman A, Kilicoglu H, Fiszman M, Owolabi Y, Jin H, Shin D, Rindflesch TC. Semantic predications for disaster information management. Manuscript submitted for publication.
- Song M, Chang P. Automatic extraction of abbreviation for emergency management websites. Proceedings of the 5th International Conference on Information Systems for Crisis Response and Management (5th ISCRAM); Washington, DC. 2008. pp. 93–100. [Google Scholar]
- Turoff M, Hiltz SR. Assessing the health information needs of the emergency preparedness and management community. Journal of Information Services and Use. 2008a;28:269–280. [Google Scholar]
- Turoff M, Hiltz SR. Information seeking behavior and viewpoints of emergency preparedness and management professionals concerned with health and medicine. for the National Library of Medicine; 2008b. Unpublished report. Available at http://is.njit.edu/turoff. [Google Scholar]
- Turoff M, Hiltz SR. future of professional communities of practice. In: Weinhardt C, Luckner S, Stößer J, editors. WeB 2008 Proceedings. LNBIP, Springer; Heidelberg: 2009. pp. 144–158. [Google Scholar]
- Vapnik VN. The Nature of Statistical Learning Theory. Springer-Verlag; New York: 1995. [Google Scholar]
- Weiner EE, Trangenstein PA. Informatics solutions for emergency planning and response. Studies in Health Technology and Informatics. 2007;129(Pt. 2):1164–1168. [PubMed] [Google Scholar]