Abstract
Animals have developed the means for supporting complex and dynamic consortia of microorganisms during their life cycle. A transcendent view of vertebrate biology therefore requires an understanding of the contributions of these indigenous microbial communities to host development and adult physiology. These contributions are most obvious in the gut, where studies of gnotobiotic mice have disclosed that the microbiota affects a wide range of biological processes, including nutrient processing and absorption, development of the mucosal immune system, angiogenesis, and epithelial renewal. The zebrafish (Danio rerio) provides an opportunity to investigate the molecular mechanisms underlying these interactions through genetic and chemical screens that take advantage of its transparency during larval and juvenile stages. Therefore, we developed methods for producing and rearing germ-free zebrafish through late juvenile stages. DNA microarray comparisons of gene expression in the digestive tracts of 6 days post fertilization germ-free, conventionalized, and conventionally raised zebrafish revealed 212 genes regulated by the microbiota, and 59 responses that are conserved in the mouse intestine, including those involved in stimulation of epithelial proliferation, promotion of nutrient metabolism, and innate immune responses. The microbial ecology of the digestive tracts of conventionally raised and conventionalized zebrafish was characterized by sequencing libraries of bacterial 16S rDNA amplicons. Colonization of germ-free zebrafish with individual members of its microbiota revealed the bacterial species specificity of selected host responses. Together, these studies establish gnotobiotic zebrafish as a useful model for dissecting the molecular foundations of host-microbial interactions in the vertebrate digestive tract.
Keywords: Danio rerio, symbiosis, host-microbial cross-talk, mice, DNA microarrays
Vertebrates have developed the means to support large societies of microbial partners during their life cycles. While considerable attention has been devoted to studying the molecular mechanisms that underlie pathogenic host-microbial relationships, relatively little is known about the molecular foundations of commensal or symbiotic host-microbial relationships and their contributions to normal animal development and adult physiology. Moreover, what we do know comes from a very limited number of model organisms (1).
The gut contains the vast majority of the mammalian microbiota (2). Most microorganisms in the intestine belong to Bacteria, although there are also members of Eukarya and Archaea (3). As with most complex ecosystems, it appears that the majority of species cannot be cultured when removed from their niches (4, 5). Although a full definition of biodiversity awaits systematic application of molecular enumeration techniques, such as genotyping DNA encoding 16S rRNA (rDNA) genes (6-8), it appears likely that at least 500-1,000 different species are distributed along the length of the adult human gastrointestinal tract (9). Colonization begins at birth and is followed by progressive assembly of a complex and dynamic microbial society (10). Assembly is presumably regulated by elaborate and combinatorial microbial-microbial and host-microbial interactions predicated on principles refined over the course of animal evolution.
Comparisons of rodents raised without exposure to any microorganisms [“germ-free” (GF)], rodents that have assembled a microbiota since birth [“conventionally raised” (CONR)], or rodents that have been colonized with components of the microbiota during or after completion of postnatal development [“conventionalized” (CONV)] have revealed a range of host functions affected by indigenous microbial communities. For example, the microbiota directs assembly of the gut-associated lymphoid tissue (GALT; ref. 11), helps educate the immune system (12, 13), affects the integrity of the intestinal mucosal barrier (14-16), modulates proliferation and differentiation of its epithelial lineages (17, 18), regulates angiogenesis (19), modifies the activity of the enteric nervous system (20), and plays a key role in extracting and processing nutrients consumed in the diet (21).
Despite these important effects, the mechanisms by which the mammalian gut microbial community influences host biology remain almost entirely unknown. Deciphering the pathways through which microbial signals operate promises to provide new chemical entities and host targets for enforcing health, and perhaps treating diseases affecting both the intestine and extra-intestinal tissues.
The zebrafish, Danio rerio, has several unique features that make it an attractive model organism for analyzing these pathways. First, zebrafish larvae and their digestive tracts are transparent from the time of fertilization through early adulthood, allowing in vivo observation of the developing gut (22, 23) and its resident microorganisms (24, 25). Second, zebrafish development occurs rapidly. Larvae hatch from their chorions at ≈3 days postfertilization (dpf). By 5 dpf, the yolk is largely absorbed and gut morphogenesis has proceeded to a stage that supports feeding and digestion (22, 23). Third, the organization of the zebrafish gut is similar to that of mammals. As in mice and humans, the intestinal epithelium undergoes renewal throughout life. A proliferative compartment, analogous to the mammalian crypt of Lieberkühn, is located at the bases of intestinal villi (26, 27). Epithelial progenitors give rise to cell types encountered in other vertebrates, including absorptive enterocytes, mucus-producing goblet cells, and an enteroendocrine lineage (22, 28). Fourth, methods for the derivation of GF larvae from other teleost species have been developed (29-31). These methods are based on the fact that embryos develop in an axenic environment that is protected by their chorions. Finally, the capacity to perform forward genetic analyses in a vertebrate that is transparent in the postembryonic period has already led to the identification of mutants with defects in gut development (22, 32, 33) and digestive physiology (23). Reverse genetic analyses using antisense morpholino oligonucleotides (34) or target-selected mutagenesis (35), as well as chemical screens (36, 37), provide additional means for identifying molecular mediators of host-microbial interactions. The imminent completion of the zebrafish genome will facilitate many of these approaches (www..sanger.ac.uk/Projects/D_rerio/).
In this report, we describe methods for raising GF zebrafish. Although our protocols support viability only through late juvenile stages, we were able to use the 6-dpf time point for morphological and functional genomic comparisons of GF, CONR, and CONV zebrafish digestive tracts. These results reveal a broad range of zebrafish host responses to the microbiota, including a number that are conserved between fish and mice. Because the zebrafish gut microbiota has not been characterized previously, we performed sequence-based 16S rDNA enumeration studies at various developmental time points. Monoassociation of GF fish with individual members of the microbiota demonstrated the bacterial specificity of selected conserved host responses. These findings provide a rationale for using gnotobiotic wild-type and/or genetically manipulated Danio rerio as a model organism for deciphering the molecular foundations of symbiotic/commensal host-bacterial relationships in the vertebrate digestive tract.
Materials and Methods
Zebrafish were from the C32 inbred line (a gift from Stephen Johnson, Department of Genetics, Washington University School of Medicine). All experiments involving these animals were conducted using protocols approved by the Animal Studies Committee of Washington University.
Fish Husbandry. Conventionally raised (CONR) zebrafish. CONR zebrafish were reared through 14 dpf at a density of ≈0.4 individual per milliliter of static water that had been harvested from tanks in a recirculating zebrafish aquaculture facility. Animals were subsequently maintained at ≈0.03 individual per milliliter of static water through 28 dpf and then moved to recirculating tanks. Zebrafish were fed rotifers (Aquatic Biosystems) beginning at 3 dpf, followed by brine shrimp (Aquafauna Bio-Marine) beginning at 14 dpf, and then advanced to a diet of brine shrimp, TetraMin flakes (Tetra), and Hikari micropellets (Hikari) at 28 dpf.
Germ-free (GF) zebrafish. To generate and rear GF zebrafish, adult male and female CONR zebrafish were collected, killed in 3-aminobenzoic acid ethyl ester (Sigma; final concentration 1 mg/ml; 10-min exposure), and then immersed in a bath of 10% polyvinylpyrrolidone (PSS Select) for 2 min at room temperature. After the abdominal walls of the males had been carefully opened to avoid rupturing their intestines, testes were removed, placed in a sterile 1.5-ml Eppendorf tube containing 500 μl of sterile Hanks' solution (4°C), and dissociated with a sterile pestle. The abdominal walls of gravid females were opened in a similar fashion, ovaries were ruptured, and eggs were removed from the body cavity with a sterile Pasteur pipette. Eggs were fertilized in vitro with the collected sperm in sterile plastic 60-mm-diameter Petri dishes (10-min incubation at room temperature). Fertilized eggs were subsequently washed three times in sterile water (3 min per cycle at room temperature) and incubated for 6 h at room temperature in ≈10 ml of a sterile solution of 0.3 mg/ml marine salt (Coralife), 100 μg/ml ampicillin, 5 μg/ml kanamycin, and 250 ng/ml amphotericin B. Embryos were then washed at room temperature in 0.1% polyvinylpyrrolidone for 2 min, rinsed three times with sterile water at room temperature, immersed in 0.003% sodium hypochlorite (Novel Wash) for 20 min at room temperature, and simultaneously transferred into plastic gnotobiotic isolators (Standard Safety Equipment; ref. 21). Once inside the gnotobiotic isolators, zebrafish embryos were rinsed three times with sterile water and then reared in these isolators in a static solution of sterile (autoclaved) gnotobiotic zebrafish medium [GZM; 0.3 g/liter marine salt (Coralife); neutral pH buffer (Bullseye 7.0, Wardley)] at a density of ≈0.4 individual per milliliter of GZM, in 400-ml glass beakers. Each day, 50% of the GZM in each beaker was replaced with fresh medium. Water temperature was maintained at 28°C by using an external K-MOD 107 heating system (Allegiance Healthcare). Beginning on 3 dpf, the solution was supplemented with dissolved autoclaved chow (ZM000, ZM Ltd; 20 mg of dry weight per liter). To ensure that the isolators were free of contaminating bacteria or fungi, their inside surfaces were routinely swabbed, and aliquots of GZM containing dissolved food were removed from beakers and cultured aerobically and anaerobically at 28°C and 37°C in three different media (nutrient broth, brain/heart infusion broth, and Sabouraud dextrose broth).
Conventionalized (CONV) zebrafish. To generate these animals, water was collected from recirculating tanks in a conventional zebrafish aquaculture facility and passed through a 5-μm pore filter (Millipore). Microbial density in the filtrate was defined by culture under aerobic and anaerobic conditions at 28°C on brain/heart infusion blood agar; 104 colony-forming units of bacteria were added per milliliter of GZM containing 3-dpf GF zebrafish.
Monoassociations. In some experiments, GF animals were colonized at 3 dpf with a single bacterial species. Aeromonas hydrophila (ATCC 35654) and Pseudomonas aeruginosa (strain PA01; a gift from Stephen Lory, Department of Microbiology, Harvard Medical School) were grown overnight under aerobic conditions in tryptic soy broth (TSB) at 30°C and in nutrient broth at 37°C, respectively, and then added to beakers containing 3-dpf GF zebrafish at final concentrations of 104 colony-forming units per milliliter of sterile GZM.
Supporting Materials and Methods. Supporting Materials and Methods, which is published as supporting information on the PNAS web site, presents methods used for (i) light and transmission electron microscopy (EM) analyses of the effects of the microbiota on gut morphology; (ii) functional genomic studies of the impact of the microbiota on gene expression in the zebrafish digestive tract (RNA isolation, fabrication of zebrafish DNA microarrays, synthesis of cDNA probes, data collection and analysis, plus SYBR-green-based real-time quantitative RT-PCR assays); and (iii) sequencing 16S rDNA amplicons generated from the digestive tract microbiota of CONR and CONV animals.
Results and Discussion
Generation of GF Zebrafish. To investigate the impact of indigenous microbial communities on zebrafish biology, we developed procedures for producing and rearing GF zebrafish (see Supporting Materials and Methods). GF and CONR zebrafish belonging to the C32 inbred strain started to feed at 5 dpf and were indistinguishable macroscopically through ≈8 dpf (Fig. 1 A and B). At 9 dpf, GF animals began to develop a stereotyped, rapidly progressive epidermal degeneration phenotype manifested by epidermal opacity, loss of epidermal integrity, and sloughing of epidermal cells (Fig. 1 D and E). Mortality was 100% by 20 dpf (n = 824 zebrafish scored). The phenotype was rescued by exposing 3-dpf or 6-dpf GF animals to the microbiota contained in water obtained from a conventional zebrafish aquaculture facility (Fig. 1F and data not shown). This finding indicates that the degenerative changes observed in late larval stage GF animals are not due to irreversible insults acquired earlier in development. Our observations that (i) animals conventionalized at 3 dpf and fed the same autoclaved diet can live to adulthood (≥42 dpf), and (ii) unfed GF animals do not develop this phenotype through 12 dpf (n = 44 scored) suggest that this phenomenon is due to undefined and deleterious effects of exposure to autoclaved chow, which are ameliorated by the presence of the microbiota (see below for further mechanistic evaluation).
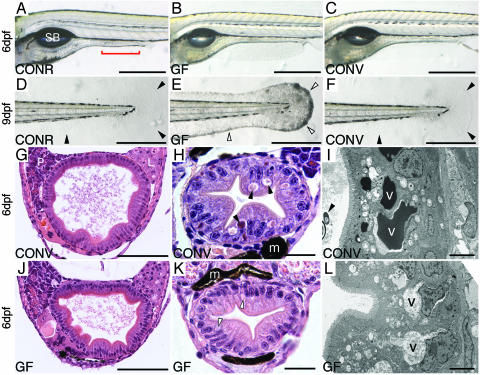
Fig. 1.
Morphologic studies of CONR, CONV, and GF zebrafish. (A-C) Whole-mount preparations of 6-dpf zebrafish. Rostral is to the left, dorsal is to the top. A shows the position of the swim bladder (SB) and the boundary of intestinal segment 2 (red bracket). Segments 1 and 3 lie rostral and caudal to segment 2, respectively. (D-F) Whole mounts of the caudal regions of 9-dpf CONR, GF, and CONV (conventionalized at 3 dpf) animals, showing onset of epidermal degeneration phenotype in GF fish. This phenotype is manifested by loss of transparency and integrity of the epidermis in fin folds (the edges of these fin folds are highlighted with open arrowheads in E). CONR and CONV fin folds remain transparent (edges indicated by filled black arrowheads in D and F). (G, H, J, and K) Hematoxylin- and eosin-stained transverse sections showing intestinal segment 1 (G and J) and segment 2 (H and K) in 6-dpf CONV and GF zebrafish. There are no detectable epithelial abnormalities in intestinal segment 1, whether judged by light microscopy (G and J) or by transmission EM (data not shown). In contrast, enterocytes in segment 2 contain prominent supranuclear vacuoles filled with eosinophilic material in CONV (and CONR) fish (e.g., black arrowheads in H). These vacuoles appear clear in GF animals (e.g., open arrowheads in K). Pigmented melanocytes (m) lie adjacent to the intestine in H and K. (I and L) EM study of 6-dpf intestines, showing electron-dense material in the supranuclear vacuoles (v) of segment 2 CONV enterocytes, and electron-lucent material in GF enterocytes. The filled black arrowhead in I points to a bacterium in the intestinal lumen. (Bars: 500 μm in A-F; 100 μmin G and J;20 μmin H and K; 5 μmin I and L.)
GF zebrafish harvested at 6 dpf, and animals conventionalized at 3 dpf and killed 3 days later (CONV), have a similar gross morphology (Fig. 1 B and C). Additionally, GF zebrafish at 6 dpf exhibit no statistically significant differences in their average body length compared with age-matched CONV and CONR larvae [4.06 ± 0.11 mm (GF); 4.09 ± 0.11 mm (CONV); and 4.02 ± 0.15 mm (CONR); P > 0.3 for each comparison based on Student's t test]. Given the phenotype observed in GF fish ≥9 dpf, we focused our analysis of the effects of the microbiota on host biology by using 6-dpf animals.
The Zebrafish Microbiota Affects Epithelial Renewal and Enterocyte Morphology. The zebrafish is a stomachless teleost: its esophagus is continuous with the proximal intestine (segment 1), which is largely responsible for lipid absorption. Segment 2 of the intestine (Fig. 1A) is involved in absorption of other macromolecules, whereas a short distal domain (segment 3) is postulated to participate in water and ion transport (26, 38, 39).
The proximal intestine, liver, pancreas, and gallbladder of GF and CONV animals were indistinguishable, whether judged by examination of whole-mount preparations (Fig. 1 B and C), serial hematoxylin- and eosin-stained sections (e.g., Fig. 1 G and J; n = 20-34 animals per treatment), or transmission EM (data not shown).
Enterocytes are the principal epithelial lineage in the mammalian intestinal epithelium. In larval and adult stomachless teleosts, macromolecules taken up from the segment 2 lumen are transported through a network of superficial invaginations and vesicles in enterocytes to large supranuclear vacuoles, where they are stored and/or degraded (26, 38, 39). A similar tubulovesicular network exists in neonatal mammalian enterocytes (40-42) but disappears at the time of initiation of protein digestion. Light microscopic and EM studies of serially sectioned GF zebrafish revealed a consistent morphologic phenotype in their segment 2 enterocytes: the large supranuclear vacuoles were filled with clear electron-lucent material, in contrast to CONV and CONR animals, where the material was eosinophilic and electron-dense (Fig. 1 H, I, K, and L plus data not shown).
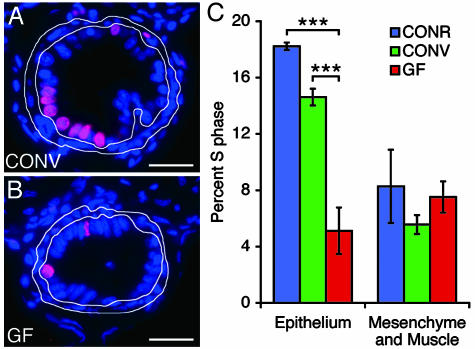
GF mice have reduced rates of epithelial proliferation in their intestinal crypts of Lieberkühn compared with their CONR or CONV counterparts (17). A similar situation occurs in zebrafish. Quantitative 5-bromodeoxyuridine labeling studies disclosed that the fractional representation of S-phase cells in the intestinal epithelium was significantly greater in 6-dpf CONV and CONR zebrafish compared with GF animals (P < 0.0001 in each case based on Student's t test; n ≥ 12 animals per condition; Fig. 2). No significant differences were observed in the underlying mesenchyme/muscle (Fig. 2C). The increase in epithelial proliferation was not accompanied by a statistically significant change in apoptosis, as judged by terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) assays of epithelium and underlying mesenchyme/muscle in the same animals (data not shown; P > 0.3 for all comparisons).
Fig. 2.
The microbiota stimulates intestinal epithelial proliferation. (A and B) Sections prepared from the intestines of 6-dpf CONV and GF zebrafish after a 24-h exposure to bromodeoxyuridine in their environmental water. Sections were stained with antibodies to bromodeoxyuridine (magenta) and the nuclear stain bisbenzimide (blue). The mesenchyme and muscle surrounding the intestinal epithelium are outlined in white. (C) Quantitation of S-phase cells in the intestinal epithelium and mesenchyme. The percentage of cells in S phase in GF intestinal epithelium is significantly lower than in CONR or CONV animals (P < 0.0001, indicated by brackets with three asterisks). Data are expressed as the mean of two independent experiments ± SEM (n = 19-31 sections scored per animal; ≥6 animals per experiment). (Bars: 25 μm in A and B.)
Host Transcriptional Responses to the Microbiota. To gain additional insights about the mechanisms underlying these microbiota-associated phenotypes, as well as other aspects of host physiology affected by gut microbes, we conducted a broad, functional genomics-based analysis of gene expression in the digestive tracts of 6-dpf GF, CONV, and CONR zebrafish. Comparisons were performed by using DNA microarrays containing 16,228 65-mer oligonucleotides representing zebrafish genes and ESTs. RNA was isolated from the pooled digestive tracts of 30 animals per treatment group. Two independently generated cohorts of animals were evaluated for each condition (i.e., a total of 60 animals). These “biological duplicates,” together with Cy3- and Cy5-labeled probe dye swap controls, produced a total of four DNA microarray datasets for each of the two comparisons performed (i.e., CONV versus GF; CONR versus GF; see Fig. 4, which is published as supporting information on the PNAS web site).
Using criteria described in Supporting Materials and Methods, we found 212 genes to exhibit differential expression in both GF versus CONV and GF versus CONR comparisons. Table 1, which is published as supporting information on the PNAS web site, provides a complete gene list, as well as fold-differences in their relative expression. Table 2, which is published as supporting information on the PNAS web site, provides an overview of the functions of all genes for which Gene Ontology (GO) term assignments could be made (www.geneontology.org).
In addition, we referenced zebrafish genes culled from comparisons of GF versus CONV and/or GF versus CONR animals to our previous DNA microarray datasets of genes differentially expressed in the gastrointestinal tracts (small intestine, colon, or liver) of adult GF mice versus ex-GF mice colonized with components of the normal mouse intestinal microbiota (F. Bäckhed, L. Hooper, M. Lecuit, and J.I.G., unpublished data; ref. 15). Sixty-six homologous genes were identified as responsive to the microbiota in both fish and mice (Tables 3 and 4, which are published as supporting information on the PNAS web site). Expression of 54 of these changed in the same direction (up or down) in both species (Table 3). Moreover, 59 of the 66 genes were identified in our analysis of the response of the mouse intestine and did not occur in mouse liver datasets (Tables 3 and 4). Several groups of these zebrafish genes, named according to their mouse homologs, are discussed below.
Conserved Transcriptional Responses to the Microbiota. Epithelial proliferation. The increased epithelial proliferation associated with the microbiota was manifested by the increased expression of 15 genes involved in DNA replication and cell division. They include thymidylate kinase (Dtymk), four minichromosome maintenance genes (Mcm2, Mcm3, Mcm5, and Mcm6), origin-recognition complex subunit 4 (Orc4l), proliferating cell nuclear antigen (Pcna), and ribonucleotide reductase subunit M2 (Rrm2; Tables 1 and 3). Nutrient metabolism. Developmental studies of Ppara (peroxisome proliferator-activated receptor α) expression in the GF mouse gastrointestinal tract have revealed a marked induction of this ligand-activated transcription factor during the suckling-weaning transition, with a continued rise in mRNA levels until animals reach adulthood. In contrast, expression of these genes in CONR mice is markedly lower and remains constant throughout postnatal development and adulthood (21). These findings indicate that adult GF mice manifest a fasting-like phenotype even though they consume more chow than their CONR counterparts (43). This phenomenon likely reflects their inability to efficiently use energy sources in their diet that are normally liberated by microbe-dependent processing (e.g., degradation of complex plant polysaccharides by the myriad of glycosylhydrolases produced by Bacteroides spp.; refs. 9 and 44).
A zebrafish homolog of Ppara, as well as homologs of several of its target genes [Fiaf (fasting-induced adipose factor, also known as angiopoietin-like 4, a secreted protein that inhibits lipoprotein lipase; ref. 45), and Cpt1a and Ctp1b (two carnitine palmitoyltransferases involved in mitochondrial fatty acid oxidation)] and Fbp1 (encodes the gluconeogenic enzyme fructose-1,6-bisphosphatase) are also up-regulated in GF fish (Tables 1 and 3). These changes indicate that, as in mice, the absence of a microbiota in zebrafish is associated with a compromised ability to use nutrients and the assumption of a metabolic state that shares features associated with fasting.
Colonization of GF mice with components of the intestinal microbiota also has a pronounced effect on the expression of genes involved in various aspects of lipid metabolism (15). Expression of farnesyl-diphosphate synthetase (Fdps), a key enzyme in isoprenoid biosynthesis that produces C15 precursors for various sterols, dolichols, and ubiquinones, is increased in the distal intestine of CONV versus GF mice, as well as in CONV versus GF zebrafish (Table 3). Apolipoprotein B (Apob), a protein secreted from enterocytes and hepatocytes that plays a pivotal role in intra- and extracellular cholesterol trafficking, is up-regulated by the microbiota in the mouse intestine and in the zebrafish digestive tract, whereas expression of the liver-specific cholesterol 7α-hydrolase (Cyp7a1), which catalyzes the first step in cholesterol catabolism and bile acid biosynthesis, and its transcriptional activator, Nr5a2, are reduced (Tables 1 and 3). Together, these findings reveal that the microbiota modulates cholesterol metabolism and trafficking in both mice and zebrafish.
All three of the solute carrier family genes in our datasets that are involved in the transport of amino acids or peptides are expressed at higher levels in GF compared with CONV and/or CONR zebrafish: Slc7a3, Slc38a4, and Slc15a2 (Tables 1 and 3). One possibility is that 6-dpf GF animals up-regulate these transporters to compensate for an inability to efficiently process ingested proteins. Xenobiotic metabolism. Components of the microbiota of other fish species have been shown to assist in the breakdown of ingested fish feed binders (46) and xenobiotics (47). The mouse gut microbiota also degrades xenobiotics, providing an explanation for the observed increased expression of genes involved in their metabolism in GF animals (15). We identified six members of the cytochrome P450 monooxygenase family that are modulated by the zebrafish microbiota. Among those with increased expression in the GF zebrafish digestive tract were homologs of mammalian Cyp1a1, Cyp2b6, and Cyp2c19, which are known to participate in xenobiotic metabolism (48-50). These results indicate that the GF zebrafish digestive tract may be less competent to detoxify dietary constituents and other components of the environment. The findings are consistent with the hypothesis that xenobiotic compounds present in the autoclaved chow (e.g., fish feed binders), may be toxic to larvae without a microbiota, and therefore contribute to the early lethality observed in fed GF zebrafish.
Innate immune responses. Functional genomic studies indicate that conventionalization of adult GF mice with an unfractionated microbiota harvested from CONR animals produces a series of responses involving genes expressed in various lymphocyte populations (F. Bäckhed, L. Hooper, M. Lecuit, and J.I.G., unpublished data). Although zebrafish develop an adaptive immune system (51), they do not appear to be capable of mounting an adaptive immune response until 4 weeks postfertilization (52). Consistent with this observation, comparisons of 6-dpf GF versus CONV or CONR zebrafish digestive tracts did not reveal remarkable differences in expression of the wide range of lymphocyte-specific genes represented on our microarrays (Tables 1-4). Nonetheless, our dataset of conserved microbiota-associated responses in the zebrafish digestive tract included genes involved in innate immunity. For example, there was induction of the zebrafish homologs of mouse serum amyloid A1 (Saa1), C-reactive protein (Crp), complement component 3 (C3), angiogenin 4 (Ang4, a microbiota-regulated antibiotic protein produced by members of the Paneth cell lineage in the mouse small intestinal epithelium that has species-selective bactericidal activity against gut pathogens; ref. 16), and suppressor of cytokine signaling 3 (Socs3, a member of a family of feedback inhibitors of JAK/STAT pathways). We also documented a microbiota-associated increase in expression of a myeloperoxidase homolog (Mpo; also known as Mpx in zebrafish, where it is a granulocyte-specific marker; ref. 53). In the mouse intestine, the microbiota induces expression of the oxidative stress response gene Gpx2 (glutathione peroxidase 2; ref. 54). Similarly, the zebrafish microbiota induces expression of a gene displaying sequence homology to both mouse Gpx2 and its constitutively expressed paralog, Gpx1 (the induced zebrafish gene is referred to as Gpx2 in Tables 1 and 3).
Shared Characteristics of the Zebrafish and Mammalian Digestive Tract Microbiota. While these studies reveal a wide range of conserved responses of the zebrafish digestive tract to the presence of a microbiota, the nature of this microbiota, and its degree of similarity to microbial communities that reside in the mouse or human gut, had not been previously defined. Therefore, we generated and sequenced libraries of bacterial 16S rDNA amplicons produced by PCR of DNA prepared from the microdissected digestive tracts of CONR 6-, 10-, 20-, and 30-dpf and adult animals (see Supporting Materials and Methods for details concerning the size of each pool, the number of libraries produced from various sibships at each time point, and how sequences were referenced against ribosomal databases for genus and species identification). Because a number of variables can affect the composition of a microbiota (e.g., nutrient supply, aquaculture conditions, and developmental stage), we used our sequence data only to identify bacterial genera/species that can occur within the zebrafish digestive tract.
The only genera found at all time points surveyed were Aeromonas and Pseudomonas. Vibrio and Lactococcus spp. were also commonly encountered (Table 5, which is published as supporting information on the PNAS web site). Comparisons of the digestive tract microbiotas of CONV versus CONR 6-dpf zebrafish indicated an enrichment of Aeromonas in the former (61% of all sequenced clones in CONV versus 0.3% in CONR), and of Vibrio in the latter (57% in CONR versus 12% in CONV; Table 5). This observation may help explain why our datasets of differentially expressed genes identified from comparisons of CONV versus GF and CONR versus GF digestive tracts were not completely overlapping (see http://gordonlab.wustl.edu/).
Some of the species we identified in the zebrafish microbiota have been shown to help control the growth and establishment of pathogens in other fish species. For example, Lactococcus lactis and Pseudomonas spp. can inhibit growth of Vibrio anguillarum (55, 56). Because of the absence of an adaptive immune response during the larval-juvenile stages in zebrafish, incorporation of these bacterial species into the gut microbiota may be important for maintaining their health during this free-living period.
Our results not only are consistent with earlier culture-based analyses of other freshwater and marine teleosts (57-59) but also reveal some limited similarities to the mammalian gut microbiota. For example, the zebrafish microbiota contains members of Bacteroidetes (e.g., Flavobacterium and Flexibacter), a major phylum in mice, humans, and other mammals (2), components of Ralstonia and Plesiomonas genera (5, 60), as well as a number of lactic acid bacteria (Lactococcus lactis, Lactobacillus fermentum, Leuconostoc citreum, and Weissella confusa).
Monoassociation Experiments Reveal Microbial Specificity for Some Host Responses. It is remarkable that responses to the gut microbiota are conserved between zebrafish and mice, given their great evolutionary distance and the compositional differences in their indigenous microbial communities. To determine whether some of the observed evolutionarily conserved host responses to the microbiota exhibit microbial species specificity, we colonized 3-dpf GF zebrafish with individual components of the digestive tract microbiota for 3 days. Two culturable and genetically manipulatable Gram-negative bacterial species were chosen for these monoassociation experiments as representatives of the Aeromonas and Pseudomonas genera that were consistently represented in the digestive tracts of 6-dpf-adult zebrafish: Aeromonas hydrophila and Pseudomonas aeruginosa. RNA was isolated from the pooled digestive tracts of 10 animals per condition at 6-dpf (n = two groups per condition), and host transcriptional responses were quantified by using real-time quantitative RT-PCR (see Table 6, which is published as supporting information on the PNAS web site). Two control RNAs were used as reference standards: 6-dpf GF and 6-dpf CONV digestive tracts (n = 30 per group; two independent groups per condition to generate biological duplicates). Importantly, the average number of viable organisms recovered from the digestive tracts of CONV or monoassociated animals was not significantly different (4.4-8.3 × 104 colony-forming units per digestive tract; P ≥ 0.26).
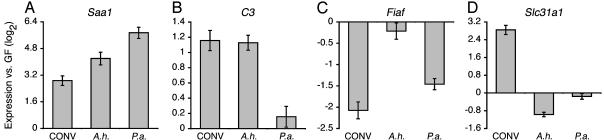
The subset of genes selected for real-time quantitative RT-PCR (qRT-PCR) assays represented a range of key host biological processes (e.g., innate immunity, nutrient uptake and metabolism, and xenobiotic metabolism). The qRT-PCR results showed that the response of some of these genes, Apob, Saa1, Mpo, and Arg-2 (arginase 2), was robust whether there was colonization with an unfractionated microbiota or with either of the two individual species (Fig. 3A and data not shown). In contrast, C3 responded to the presence of a normal microbiota and to A. hydrophila, but not to P. aeruginosa (Fig. 3B). Conversely, Fiaf responded to a normal microbiota and P. aeruginosa, but not to A. hydrophila (Fig. 3C). Expression of other genes, Gpx2, Ppara, Slc31a1 (solute carrier required for high-affinity uptake of Cu2+), Cyp27a1 (cytochrome P450 family 27 A1), Ddost (dolichyl-diphosphooligosaccharide protein glycotransferase), Gstp1 (glutathione S-transferase π 1), Dmbt1 (deleted in malignant brain tumors 1; also called crp-ductin), Itgb1bp3 (integrin β-1-binding protein 3), Wars (tryptophanyl-tRNA synthetase), and Yars (tyrosyl-tRNA synthetase), was regulated by the unfractionated microbiota but not dramatically altered in response to monoassociation with either A. hydrophila or P. aeruginosa (Fig. 3D and data not shown). These findings indicate that, as in mice (15), at least a subset of zebrafish genes are sensitive to factors represented in only a subset of bacterial components of the gut microbiota.
Fig. 3.
Real-time quantitative RT-PCR studies of the microbial species specificity of selected evolutionarily conserved zebrafish responses to the digestive tract microbiota. Expression levels of serum amyloid A1 (Saa1; A), complement component 3 (C3; B), fasting-induced adipose factor (Fiaf; C), and solute carrier family 31 member 1 (Slc31a1; D) in digestive tracts from 6-dpf conventionalized (CONV), A. hydrophila-monoassociated (A.h.), and P. aeruginosa-monoassociated (P.a.) larvae are shown relative to 6-dpf GF larval digestive tracts. Assays were performed in triplicate (n ≥ 4 assays per gene). Data were normalized to 18S ribosomal RNA and results are expressed as mean log2 values ± SEM.
Finally, DNA microarray-based comparisons of gene expression in the digestive tracts of conventionally raised 6-, 10-, and 20-dpf zebrafish revealed that a number of the genes found to be regulated by the microbiota exhibit changes in their relative expression levels during the larval to late juvenile stages of development (e.g., Fig. 5, which is published as supporting information on the PNAS web site). This observation underscores the importance of considering (i) the potential contributions of the microbiota when designing and interpreting genetic and/or chemical screens for factors that modulate zebrafish development, and (ii) that observed features of postembryonic animal development may represent formerly unappreciated manifestations of host-microbial interactions.
Prospectus. While coupling gnotobiotic mouse models with functional genomics has been valuable for documenting the wide range of host functions that are modulated by the microbiota, identification of the host and microbial signals and signaling pathways that mediate such interactions will be very challenging. The present study indicates that the zebrafish possesses several characteristics that should facilitate this discovery process: (i) they exhibit a number of responses to the gut microbiota that are shared with mammals; (ii) a subset of these responses show microbial specificity; (iii) the transparency of developing zebrafish could allow host-microbial interactions to be observed in their digestive tracts, through the use of fluorescent protein-expressing bacteria and/or fluorescent protein-expressing host transgenes (see below); and (iv) genetic and chemical screenings could be performed by using normal and genetically engineered gnotobiotic zebrafish raised in multiwell arrays (61).
Although the present study shows the feasibility of studying host-microbial interactions in larval gnotobiotic zebrafish, extension to adult stages will require continued refinement of animal husbandry protocols. Many of the issues faced with GF zebrafish, such as identifying a diet that allows survival to reproductive maturity, the capacity to breed strains of GF animals over multiple generations, and optimizing equipment for precise control of environmental variables, are similar to those overcome by gnotobiologists during the past 50 years as they have learned to effectively propagate GF mice.
We anticipate that in the near future transgenic GF zebrafish will be created containing fluorescent protein reporters under the control of transcriptional regulatory elements from biologically important genes that display evolutionarily conserved microbial regulation. These animals could then be used as instructive models for genetic and/or chemical analyses of the molecular foundations, and physiological significance, of mutually beneficial (symbiotic) or pathogenic relationships between microorganisms and their vertebrate hosts.
Supplementary Material
Acknowledgments
We are grateful to Maria Karlsson, David O'Donnell, and Janaki Guruge for assistance with fish husbandry; Michael Heinz, Chris Sawyer, Wes Warren, Rick Waterman, Stephen Johnson, and Yiyong Zhou for providing zebrafish DNA microarrays; Fredrik Bäckhed, Lora Hooper, and Marc Lecuit for sharing unpublished DNA microarray datasets from gnotobiotic mice; Peter Kang, Jian Xu, Donald Williams, Josh Conyers, Vince Magrini, and Elaine Mardis for help with 16S rDNA enumeration; and Jaime Dant for electron microscopy. This work was supported by the Ellison Medical Foundation. Studies that produced DNA microarray datasets from gnotobiotic mice were funded by the National Institutes of Health (DK30292). J.F.R. is the recipient of a National Research Service Award postdoctoral fellowship (DK62675). B.S.S. is a National Science Foundation Graduate Research Fellow.
Abbreviations: rDNA, DNA encoding rRNA genes; GF, germ-free; CONR, conventionally raised; CONV, conventionalized; dpf, days postfertilization; EM, electron microscopy.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY536921-AY538099).
References
- 1.McFall-Ngai, M. J. (2002) Dev. Biol. 242, 1-14. [DOI] [PubMed] [Google Scholar]
- 2.Savage, D. C. (1977) Annu. Rev. Microbiol. 31, 107-133. [DOI] [PubMed] [Google Scholar]
- 3.Eckburg, P. B., Lepp, P. W. & Relman, D. A. (2003) Infect. Immun. 71, 591-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suau, A., Bonnet, R., Sutren, M., Godon, J. J., Gibson, G. R., Collins, M. D. & Dore, J. (1999) Appl. Environ. Microbiol. 65, 4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salzman, N. H., de Jong, H., Paterson, Y., Harmsen, H. J., Welling, G. W. & Bos, N. A. (2002) Microbiology 148, 3651-3660. [DOI] [PubMed] [Google Scholar]
- 6.Harmsen, H. J., Raangs, G. C., He, T., Degener, J. E. & Welling, G. W. (2002) Appl. Environ. Microbiol. 68, 2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashi, H., Sakamoto, M. & Benno, Y. (2002) Microbiol. Immunol. 46, 535-548. [DOI] [PubMed] [Google Scholar]
- 8.Blaut, M., Collins, M. D., Welling, G. W., Dore, J., van Loo, J. & de Vos, W. (2002) Br. J. Nutr. 87, Suppl. 2, S203-S211. [DOI] [PubMed] [Google Scholar]
- 9.Xu, J. & Gordon, J. I. (2003) Proc. Natl. Acad. Sci. USA 100, 10452-10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Favier, C. F., Vaughan, E. E., De Vos, W. M. & Akkermans, A. D. (2002) Appl. Environ. Microbiol. 68, 219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cebra, J. J. (1999) Am. J. Clin. Nutr. 69, 1046S-1051S. [DOI] [PubMed] [Google Scholar]
- 12.Braun-Fahrlander, C., Riedler, J., Herz, U., Eder, W., Waser, M., Grize, L., Maisch, S., Carr, D., Gerlach, F., Bufe, A., et al. (2002) N. Engl. J. Med. 347, 869-877. [DOI] [PubMed] [Google Scholar]
- 13.Kelly, D., Campbell, J. I., King, T. P., Grant, G., Jansson, E. A., Coutts, A. G., Pettersson, S. & Conway, S. (2004) Nat. Immunol. 5, 104-112. [DOI] [PubMed] [Google Scholar]
- 14.MacPherson, A. J., Gatto, D., Sainsbury, E., Harriman, G. R., Hengartner, H. & Zinkernagel, R. M. (2000) Science 288, 2222-2226. [DOI] [PubMed] [Google Scholar]
- 15.Hooper, L. V., Wong, M. H., Thelin, A., Hansson, L., Falk, P. G. & Gordon, J. I. (2001) Science 291, 881-884. [DOI] [PubMed] [Google Scholar]
- 16.Hooper, L. V., Stappenbeck, T. S., Hong, C. V. & Gordon, J. I. (2003) Nat. Immunol. 4, 269-273. [DOI] [PubMed] [Google Scholar]
- 17.Uribe, A., Alam, M., Midtvedt, T., Smedfors, B. & Theodorsson, E. (1997) Scand. J. Gastroenterol. 32, 691-699. [DOI] [PubMed] [Google Scholar]
- 18.Bry, L., Falk, P. G., Midtvedt, T. & Gordon, J. I. (1996) Science 273, 1380-1383. [DOI] [PubMed] [Google Scholar]
- 19.Stappenbeck, T. S., Hooper, L. V. & Gordon, J. I. (2002) Proc. Natl. Acad. Sci. USA 99, 15451-15455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Husebye, E., Hellström, P. M. & Midtvedt, T. (1994) Dig. Dis. Sci. 39, 946-956. [DOI] [PubMed] [Google Scholar]
- 21.Hooper, L. V., Midtvedt, T. & Gordon, J. I. (2002) Annu. Rev. Nutr. 22, 283-307. [DOI] [PubMed] [Google Scholar]
- 22.Pack, M., Solnica-Krezel, L., Malicki, J., Neuhauss, S. C., Schier, A. F., Stemple, D. L., Driever, W. & Fishman, M. C. (1996) Development (Cambridge, U.K.) 123, 321-328. [DOI] [PubMed] [Google Scholar]
- 23.Farber, S. A., Pack, M., Ho, S. Y., Johnson, I. D., Wagner, D. S., Dosch, R., Mullins, M. C., Hendrickson, H. S., Hendrickson, E. K. & Halpern, M. E. (2001) Science 292, 1385-1388. [DOI] [PubMed] [Google Scholar]
- 24.Davis, J. M., Clay, H., Lewis, J. L., Ghori, N., Herbomel, P. & Ramakrishnan, L. (2002) Immunity 17, 693-702. [DOI] [PubMed] [Google Scholar]
- 25.van der Sar, A. M., Musters, R. J., van Eeden, F. J., Appelmelk, B. J., Vandenbroucke-Grauls, C. M. & Bitter, W. (2003) Cell. Microbiol. 5, 601-611. [DOI] [PubMed] [Google Scholar]
- 26.Stroband, H. W. & Debets, F. M. (1978) Cell Tissue Res. 187, 181-200. [DOI] [PubMed] [Google Scholar]
- 27.Rombout, J. H. W. M., Stroband, H. W. J. & Taverne-Thiele, J. J. (1984) Cell Tissue Res. 236, 207-216. [DOI] [PubMed] [Google Scholar]
- 28.Harder, W. (1975) Anatomy of Fishes (Eschweizerbartsche, Stuttgart).
- 29.Baker, J. A. & Ferguson, M. S. (1942) Proc. Soc. Exp. Biol. Med. 51, 116-119. [Google Scholar]
- 30.Trust, T. J. (1974) Appl. Microbiol. 28, 340-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lesel, R. & Lesel, M. (1976) Ann. Hydrobiol. 7, 21-25. [Google Scholar]
- 32.Chen, J. N., Haffter, P., Odenthal, J., Vogelsang, E., Brand, M., van Eeden, F. J., Furutani-Seiki, M., Granato, M., Hammerschmidt, M., Heisenberg, C. P., et al. (1996) Development (Cambridge, U.K.) 123, 293-302. [DOI] [PubMed] [Google Scholar]
- 33.Mayer, A. N. & Fishman, M. C. (2003) Development (Cambridge, U.K.) 130, 3917-3928. [DOI] [PubMed] [Google Scholar]
- 34.Nasevicius, A. & Ekker, S. C. (2000) Nat. Genet. 26, 216-220. [DOI] [PubMed] [Google Scholar]
- 35.Wienholds, E., van Eeden, F., Kosters, M., Mudde, J., Plasterk, R. H. & Cuppen, E. (2003) Genome Res. 13, 2700-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peterson, R. T., Link, B. A., Dowling, J. E. & Schreiber, S. L. (2000) Proc. Natl. Acad. Sci. USA 97, 12965-12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khersonsky, S. M., Jung, D. W., Kang, T. W., Walsh, D. P., Moon, H. S., Jo, H., Jacobson, E. M., Shetty, V., Neubert, T. A. & Chang, Y. T. (2003) J. Am. Chem. Soc. 125, 11804-11805. [DOI] [PubMed] [Google Scholar]
- 38.Noaillac-Depeyre, J. & Gas, N. (1976) Tissue Cell 8, 511-530. [DOI] [PubMed] [Google Scholar]
- 39.Stroband, H. W., van deer Meer, H. & Timmermans, L. P. (1979) Histochemistry 64, 235-249. [DOI] [PubMed] [Google Scholar]
- 40.Kraehenbuhl, J. P. & Campiche, M. A. (1969) J. Cell Biol. 42, 345-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hugon, J. S. (1971) Histochemie 26, 19-27. [DOI] [PubMed] [Google Scholar]
- 42.Worthington, B. B. & Graney, D. O. (1973) Anat. Rec. 175, 37-61. [DOI] [PubMed] [Google Scholar]
- 43.Wostmann, B. S., Larkin, C., Moriarty, A. & Bruckner-Kardoss, E. (1983) Lab. Anim. Sci. 33, 46-50. [PubMed] [Google Scholar]
- 44.Xu, J., Bjursell, M. K., Himrod, J., Deng, S., Carmichael, L. K., Chiang, H. C., Hooper, L. V. & Gordon, J. I. (2003) Science 299, 2074-2076. [DOI] [PubMed] [Google Scholar]
- 45.Yoshida, K., Shimizugawa, T., Ono, M. & Furukawa, H. (2002) J. Lipid Res. 43, 1770-1772. [DOI] [PubMed] [Google Scholar]
- 46.Gilmour, A., McCallum, M. F. & Allan, M. C. (1976) Aquaculture 7, 161-172. [Google Scholar]
- 47.Malone, T. C. (1970) Nature 227, 848-849. [DOI] [PubMed] [Google Scholar]
- 48.Denison, M. S. & Whitlock, J. P., Jr. (1995) J. Biol. Chem. 270, 18175-18178. [DOI] [PubMed] [Google Scholar]
- 49.Ekins, S. & Wrighton, S. A. (1999) Drug Metab. Rev. 31, 719-754. [DOI] [PubMed] [Google Scholar]
- 50.Desta, Z., Zhao, X., Shin, J. G. & Flockhart, D. A. (2002) Clin. Pharmacokinet. 41, 913-958. [DOI] [PubMed] [Google Scholar]
- 51.Traver, D., Herbomel, P., Patton, E. E., Murphey, R. D., Yoder, J. A., Litman, G. W., Catic, A., Amemiya, C. T., Zon, L. I. & Trede, N. S. (2003) Adv. Immunol. 81, 253-330. [PubMed] [Google Scholar]
- 52.Lam, S. H., Chua, H. L., Gong, Z., Lam, T. J. & Sin, Y. M. (2004) Dev. Comp. Immunol. 28, 9-28. [DOI] [PubMed] [Google Scholar]
- 53.Lieschke, G. J., Oates, A. C., Crowhurst, M. O., Ward, A. C. & Layton, J. E. (2001) Blood 98, 3087-3096. [DOI] [PubMed] [Google Scholar]
- 54.Esworthy, R. S., Binder, S. W., Doroshow, J. H. & Chu, F. F. (2003) Biol. Chem. 384, 597-607. [DOI] [PubMed] [Google Scholar]
- 55.Villamil, L., Tafalla, C., Figueras, A. & Novoa, B. (2002) Clin. Diagn. Lab. Immunol. 9, 1318-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spanggaard, B., Huber, I., Nielsen, J., Sick, E. B., Pipper, C. B., Martinussen, T., Slierendrecht, W. J. & Gram, L. (2001) Environ. Microbiol. 3, 755-765. [DOI] [PubMed] [Google Scholar]
- 57.Cahill, M. M. (1990) Microb. Ecol. 19, 21-41. [DOI] [PubMed] [Google Scholar]
- 58.Ringø, E., Strøm, E. & Tabachek, J.-A. (1995) Aquaculture Res. 26, 773-789. [Google Scholar]
- 59.Hansen, G. H. & Olafsen, J. A. (1999) Microb. Ecol. 38, 1-26. [DOI] [PubMed] [Google Scholar]
- 60.Arai, T., Ikejima, N., Itoh, T., Sakai, S., Shimada, T. & Sakazaki, R. (1980) J. Hyg. (London) 84, 203-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patton, E. E. & Zon, L. I. (2001) Nat. Rev. Genet. 2, 956-966. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.