Abstract
Infection with Mycobacterium tuberculosis causes the illness tuberculosis with an annual mortality of ≈2 million. Understanding the nature of the host-pathogen interactions at different stages of tuberculosis is central to new strategies for developing chemotherapies and vaccines. Toward this end, we adapted microarray technology to analyze the change in gene expression profiles of M. tuberculosis during infection in mice. This protocol provides the transcription profile of genes expressed during the course of early tuberculosis in immune-competent (BALB/c) and severe combined immune-deficient (SCID) hosts in comparison with growth in medium. The microarray analysis revealed clusters of genes that changed their transcription levels exclusively in the lungs of BALB/c, SCID mice, or medium over time. We identified a set of genes (n = 67) activated only in BALB/c and not in SCID mice at 21 days after infection, a key point in the progression of tuberculosis. A subset of the lung-activated genes was previously identified as induced during mycobacterial survival in a macrophage cell line. Another group of in vivo-expressed genes may also define a previously unreported genomic island. In addition, our analysis suggests the similarity between mycobacterial transcriptional machinery during growth in SCID and in broth, which questions the validity of using the SCID model for assessing mycobacterial virulence. The in vivo expression-profiling technology presented should be applicable to any microbial model of infection.
Between infection and the appearance of the first symptoms of the disease, bacteria interact with different microenvironments within the host. The outcome of such host-pathogen interactions are in large part due to selective gene expression at different phases of infection (1). Consequently, understanding bacterial gene expression in vivo is central to our understanding of how bacteria colonize, invade, and interact with or disrupt the normal host cell functions and eventually produce disease. In this regard, Mycobacterium tuberculosis (Mtb) has a proven record for adaptation to different human microenvironments such as macrophages and epithelial cells (2-4). M. tuberculosis has claimed >2 million lives annually for the past decade (5). Several mycobacterial species, including the human pathogen, have developed resistance to several antibiotics (6). In addition, the historical vaccine against tuberculosis (Mycobacterium bovis bacillus Calmette-Guérin) has a questionable record of protecting against infection and can cause disease in immune-compromised individuals (7). A clear understanding of the molecular events responsible for establishing and maintaining tuberculosis will likely lead to improved drug and vaccine design. With the release of the M. tuberculosis genome sequence (8), large-scale genomics analysis of tuberculosis has become possible. This report describes the expression profile of M. tuberculosis during early infection by using DNA microarrays and the mouse model of tuberculosis.
The technology of DNA microarrays was introduced (9) to measure gene expression levels on a genome-wide scale. Questions related to the genome content of different species within mycobacteria (10) or its transcriptional regulation after exposure to specific inducers have been addressed with DNA microarray technology (11, 12). Recently, we used DNA microarrays for M. tuberculosis to profile the change of gene expression during transition from logarithmic to stationary phases in vitro (13). However, it has been a challenge to adapt this technology to measure the expression of a pathogen's genome in the host because of the low abundance of bacterial RNA in the host tissues and the nature of prokaryotic mRNA (short half-lives and lack of polyadenylation) (14, 15). To avoid this “sampling” problem, researchers studying Vibrio cholera have used bacterial cells extracted from human feces as a substrate for their DNA microarrays analysis (16), whereas others have used the rabbit ileal loop model to collect bacterial cells in cholera (17). In our hands, we have found that the use of genome-directed primers, a strategy that uses a minimal number of short oligonucleotides to preferentially transcribe bacterial mRNA in mixed RNA samples, is part of the solution to the sampling problem (18). A second technological improvement was the use of genomic DNA to normalize the signal generated from each transcript, which tended to improve the signal-to-noise ratio (13).
In this study we compared the expression profile of bacilli growing in standard medium (Middlebrook 7H9 broth) vs. those growing in a well characterized host (mice) during the first 28 days after infection. We also compared the expression profiles of bacterial growth in the immune-competent (BALB/c) vs. the severe combined immune-deficient (SCID) mice to identify mycobacterial genes influenced by the host immune responses. Genes identified from our search strategy could present the basis for diagnostic or vaccine antigens.
Materials and Methods
Animals. BALB/c and BALB/cSCID/SCID mice were purchased from Charles River Breeding Laboratories and were infected at 8 weeks of age by intranasal inoculation by allowing anesthetized mice to inhale 50 μl of PBS containing 103 colony-forming units (cfu) of M. tuberculosis H37Rv. Animals sampled at 7 days after infection were inoculated with 105 cfu per mouse to extract enough RNA for microarray analysis. The effective in vivo infectious dose was determined by harvesting the lungs from three mice at 1 h after inoculation to determine the actual deposition. Lungs from 50 mice per group (100 mice per group for day 7 samples) were rapidly harvested and placed on ice at each time point after initial infection. Each pair of lungs was added into a 50-ml conical tube containing 20 ml of 0.01% SDS (Sigma). Lungs were immediately homogenized and passed through loosely packed nylon-wool columns to remove large tissue debris. Tissue suspensions were then centrifuged at 4°C (4,000 × g) for 10 min to further eliminate lung tissues followed by resuspending bacterial pellets in Tri Reagent (Molecular Research Center, Cincinnati, OH) for RNA extraction as outlined (13). For each time point, total RNA used for DNA microarrays was extracted from at least two independent animal groups. Tuberculous bacilli were enumerated by plating serial dilutions of tissue homogenates on Middlebrook 7H11 agar (Microbio, Tempe, AZ). Tissue sections from a smaller number of infected animals (n = 4) were examined by using standard H&E and acid-fast staining.
Bacteria. The virulent M. tuberculosis strain H37Rv was obtained from American Type Culture Collection. The organism was grown as a pellicle in Proskauer and Beck medium (Difco) passaged to obtain a working stock that was in log phase and then frozen as working stocks at -70°C. For inoculation, the frozen stock was thawed, diluted, sonicated to disperse clumps, and then diluted to the appropriate concentration in PBS containing 0.01% Tween 80 (Sigma). For in vitro samples, cultures for preparing RNA samples were grown in Middlebrook 7H9 broth (Difco) at 37°C and harvested by centrifugation at 4°C (4,000 × g for 20 min) at 7, 14, 21, or 28 days, respectively. Total RNA was extracted from cultures using the Tri Reagent according to the manufacturer's recommendations (13).
Construction of the DNA Microarrays and Sample Hybridization. Oligonucleotides (70-mers) representing 100% of the possible coding sequence of the Mtb strain H37Rv genome (http://genolist.pasteur.fr/TubercuList) were purchased from Operon Technologies (Alemeda, CA), resuspended in 3 × SSC buffer (final concentration, 40 μM) before robotic arraying onto poly-l-lysine-coated glass slides (75 × 25 mm). RNA samples were labeled by using FairPlay kits (Stratagene) as described in the manufacturer's manual, with the exception of using 6 μl of genome-directed primers (18) (250 ng/μl) to prime the transcription reactions instead of using random primers. The amount of total RNA used for each hybridization from in vitro (7 μg) and in vivo (10 μg) samples were determined empirically to give reproducible signals (>95% of the genes with <2-fold change in expression ratios from repeated samples). Mycobacterial genomic DNA was labeled as described (13). The labeled cDNA was adjusted to 4 × SSC/0.1% SDS and cohybridized with 0.5 μg of the labeled genomic DNA to the microarray glass slides overnight at 67°C. The slides were washed for 5 min at room temperature in low-stringency wash buffer (1 × SSC/0.1%SDS) followed by a 3-min wash in high-stringency buffer (0.1 × SSC).
Data Acquisition and Statistical Analysis. Hybridized microarray slides were scanned (GenePix4000, Axon Instruments, Inc.) with independent excitation of the fluorophores Cy3 and Cy5 at the 10 μm resolution. The signal and background fluorescence intensities were calculated for each DNA spot by using the segmentation method of the genpixpro software (Axon Instruments, Foster City, CA). The ratios of intensity for Cy3- to Cy5-labeled probes were determined for each DNA spot. The Cy5-labeled probe was dedicated for labeled genomic DNA, whereas all the cDNA samples were labeled with Cy3, and a standard statistical scoring system (Z) (19) was used to stratify the expression levels from low- to high-level “standard expression level” (13). We used only genes with Z scores ≥ ±2 in at least one time point (7, 14, 21, or 28 days) for hierarchical cluster analysis (20). The average of expression levels from a set of samples was compared with other samples to identify up- or down-regulated genes during mycobacterial growth at different time points and in different environments. The coefficient of variation for each gene was estimated between all hybridization replicates from two repeated biological samples and used as a filter to exclude genes with low hybridization reproducibility as suggested (21). Most of the genes had a coefficient of variation of <0.5. An average of six to eight replicate hybridizations from two independent biological replicates for each time point (total of 108 hybridizations) was used to calculate gene expression levels. Kruskall-Wallis rank test for multiple groups with the false discovery rate correction protocol was used to evaluate the differential expression levels between samples with genespring 5.1 software (Silicon Genetics, Redwood City, CA).
Real-Time, Quantitative PCR (qPCR). A randomly selected group of genes (n = 65) were subjected to real-time qPCR) as described (22) with the SYBR green dye and QunatiTect kit (Qiagen, Valencia, CA) according to the manufacture's protocol. The same cDNA samples (before fluorescence labeling) subjected to microarray analysis but from single animal groups were used as templates for qPCR with the iCycler thermocycler (Bio-Rad). For each amplification run, the calculated threshold cycle (Ct) for each gene amplicon was normalized to Ct of the 16S rRNA gene amplified from the corresponding sample before calculating gene fold change as described (22).
Results
In Vivo DNA Microarrays. We adapted the DNA microarray technology to profile the mycobacterial transcriptome during mouse infection. In these experiments, the virulent strain of M. tuberculosis was grown in broth, BALB/c or SCID mice. Lung tissues from infected BALB/c or SCID mice were harvested at different time points for RNA extraction and cDNA labeling and were subjected to DNA microarray analysis. Tuberculous bacilli growing in lung tissue were assayed because lungs are the site of bacterial entry and represent the first line of defense that Mtb must overcome to establish an infection. Oligonucleotide microarrays representing 100% of the predicted mycobacterial ORFs were used to interrogate the mycobacterial transcription profiles under in vivo and in vitro growth conditions at 7, 14, 21, and 28 days. These time points were chosen to reflect key points of early infection according to previous reports of the progression of tuberculosis in the mouse model of tuberculosis after respiratory infection (refs. 23 and 24 and supporting information, which is published on the PNAS web site).
To test the use of microarrays for in vivo samples, we hybridized DNA microarrays to cDNA generated by mycobacterial genome-directed primers from RNA samples extracted from normal mouse lung tissues. Only 1.3-6.4% of mycobacterial genes were affected by cross hybridization (18). Stringent hybridization conditions were used throughout this study. To further reduce the level of cross hybridization from the host RNA, we introduced a centrifugation step to pellet mycobacterial cells (on average 5 × 107-108 cfu) before RNA extraction. All in vivo-extracted RNA samples were analyzed by running on agarose gel or by using LabChip technology (Agilent Technologies, Palo Alto, CA) and proved to be of a quality similar to RNA extracted from cultures growing in vitro (supporting information).
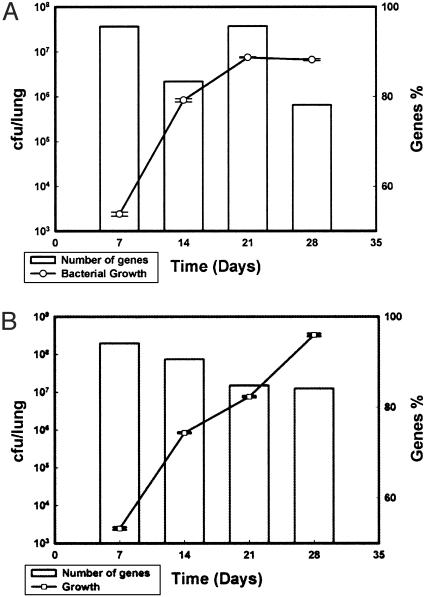
We also compared the number of genes with measurable expression levels (ratio of cDNA to genomic DNA for each gene of >1) from in vivo and in vitro RNA samples to the bacterial load of each sample. As shown in Fig. 1, >75% of the genes have detectable expression levels (R > 1) under all environments, regardless of the time of sample collection. Even as the number of bacteria per mouse lungs increased over time, no corresponding increase of the number of genes expressed occurred. In fact, RNA samples extracted at 28 days after infection from either BALB/c or SCID mice had a lower number of genes with measurable expression levels than samples extracted at 7 days after infection (Fig. 1 A and B). Variation in the percent of the detected transcriptome therefore is likely to represent changes in gene expression levels rather than any technological limitation. The expression of 3,882 genes (98.9%) have been reproducibly measured (R > 1) under at least one condition (in vitro or in vivo samples), whereas the expression of 42 genes was not detected (supporting information). The reliability of the microarray expression data were further assessed determining the fold expression change by quantitative, real time-PCR, (qPCR) (22) for a set of 62 randomly chosen genes. Overall, in 86% of the genes examined, agreement was in the direction of change in gene levels between both microarray and qPCR technologies (supporting information).
Fig. 1.
Microarray analysis of Mtb growing in vivo. Histograms show the number of genes with measurable expression levels during Mtb growth in either BALB/c (A) or SCID (B) mice. The left scale represents the total number of mycobacterial colony-forming units (cfu/lung) cultured before RNA extraction, and the right scale represents the percentage of the expressed genes.
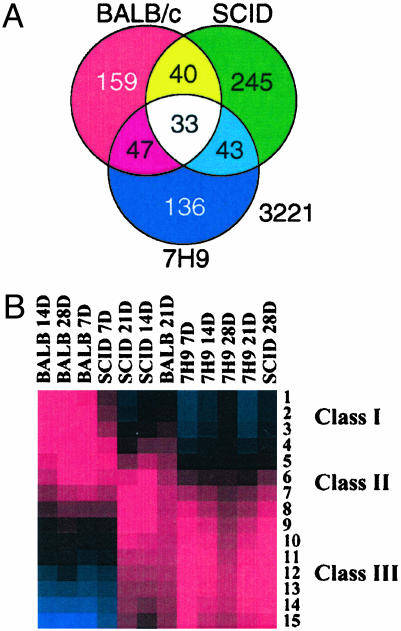
Temporal Changes in Gene Expression Levels During in Vivo Growth. We used two modes of analyses on the data. In one set, the expression values of the four time points (7, 14, 21, and 28 days) were compared within each condition (BALB/c, SCID mice, or broth) to identify genes that altered their expression levels during progression in different host microenvironments (growth-dependent genes). Alternatively, the expression levels from BALB/c and SCID samples were compared with those of the in vitro samples to identify unique genes regulated during different stages of early tuberculosis. For the former analysis, statistical tests revealed a group of 703 growth-dependent genes that was differentially expressed when Mtb were growing in BALB/c over the four time points (supporting information). Within this group of genes (Fig. 2A), we were able to identify growth-dependent genes that were significantly changed only in BALB/c (n = 159), only in SCID mice (n = 245), or only in the broth (n = 136). An additional group of 33 genes demonstrated significant changes in expression levels under the three environments. Two of these, aceA and sseB, have been implicated in Mtb pathogenesis (25, 26). A group of 40 genes significantly changed only during growth in BALB/c and SCID mice. This set of 40 genes could be part of a core set of genes regulated during in vivo infection, regardless of the host immune status reflecting the hostile microenvironment that Mtb faces inside the host. Genes such as rubB, dinF, and fdxA [induced at low pH and DNA damage stress (27)] were among this core of in vivo-regulated genes. A recent analysis of the Mtb growing in macrophages revealed the induction of the same three genes 24 h after macrophage infection (28). The complete list of the additional 30 genes that were induced in macrophage at 24 h (28) and BALB/c at 21 days of infection are summarized in the supporting information. Comparative analysis of the Mtb expression data after either macrophage (28) or BALB/c infections suggests that the host immune response at 21 days after infection is characterized by macrophage activation including lower pH and other stressors.
Fig. 2.
Expression profiles for Mtb in different environments. (A) Venn diagram showing the number of overlapping and unique set of growth-dependent genes. (B) Self-organizing maps of gene expression profiles of Mtb under different conditions. Genes with Z scores ≥ ±2 in at least one time point from BALB/c mice, SCID mice, or medium samples were included in the analysis. The means of each cluster of genes (15 clusters) are represented with red, blue, or black boxes indicating up-regulation, down-regulation, or no change of gene expression levels, respectively.
The change in the growth-dependent gene levels could reflect the adaptation to the host lung environment and to the nutrient depletion (in both in vivo and in vitro) at the same time with different sets of genes but from the same functional group. Therefore, genes identified as activated based on in vitro models only could be misleading to the real change in gene expression during infection. For example, genes regulated only in BALB/c mice included genes that could contribute to mycobacterial survival in vivo, such as proZ (transport system permease protein), aceAa (probable isocitrate lyase involved in lipid metabolism) and genes encoding regulatory proteins such as sigK, sigE (RNA polymerase σ-factors), and kdpE (transcriptional regulatory protein). However, for survival in broth other genes were regulated such as cstA (carbon starvation-induced stress-response protein), cysW (sulfate transport system permease protein), Rv3383c (transferase involved in lipid biosynthesis), and gene-encoding regulatory proteins such as sigJ (σ-factor) and Rv1167c or Rv1994c (transcriptional regulators). Finally, although more genes showed a significant change in SCID mice (n = 245) than in BALB/c mice (n = 159), >80% of these genes displayed a modest fold change (< ±2.5). Several of the genes encoding ribosomal protein subunits (such as rpsL, rpsN, and rpsS) and the extracytoplasmic alternative σ-factor (sigI) were included in this set of genes, reflecting the exponential increase of tuberculous bacilli in SCID mice.
Host Influence on the Expression Levels of Tuberculosis Genes. In the second approach for gene expression analysis, we organized the gene expression levels collected from different samples (BALB/c, SCID mice, and broth) into gene groups by using the self-organizing map algorithm (29). The goal behind such analysis is to determine the influence of different growth environments on the Mtb gene expression levels. A set of 578 highly expressed genes (Z scores ≥ ±2) was grouped into 15 subclusters (Fig. 2B and supporting information). Among these, we were able to identify three classes of expression profiles: class 1, a profile composed of three clusters (186 genes) with high expression levels in BALB/c at 7, 14, and 28 days and SCID mice at 7 days; class 2, another profile composed of five clusters (162 genes) with high expression levels in BALB/c at 21 days and SCID at 14 and 21 days; and class 3, a third profile composed of seven clusters (230 genes) with high expression levels in SCID mice at 28 days and in all in vitro samples. Putative genes for virulence, protein, and peptide secretion and cytochrome P450 were significantly regulated only in classes 1 and 2 of genes that were predominantly regulated during growth in BALB/c or SCID mice (supporting information). Genes involved in cell division and chelatase expression were predominant in class 3. Overall, self-organizing map analysis classified gene groups according to the host environment with the exception of SCID mice at 28 days and BALB/c at 21 days. The expression profile of Mtb in SCID mice at 28 days resembled the expression profile in culture probably reflecting the logarithmic increase of the number of bacteria (Fig. 1B) that eventually kill the immune-compromised host by 35 days after infection. However, the expression profile of bacilli in BALB/c at 21 days did not cluster with the rest of BALB/c profiles, indicating an inflection of the expression profile during the course of infection.
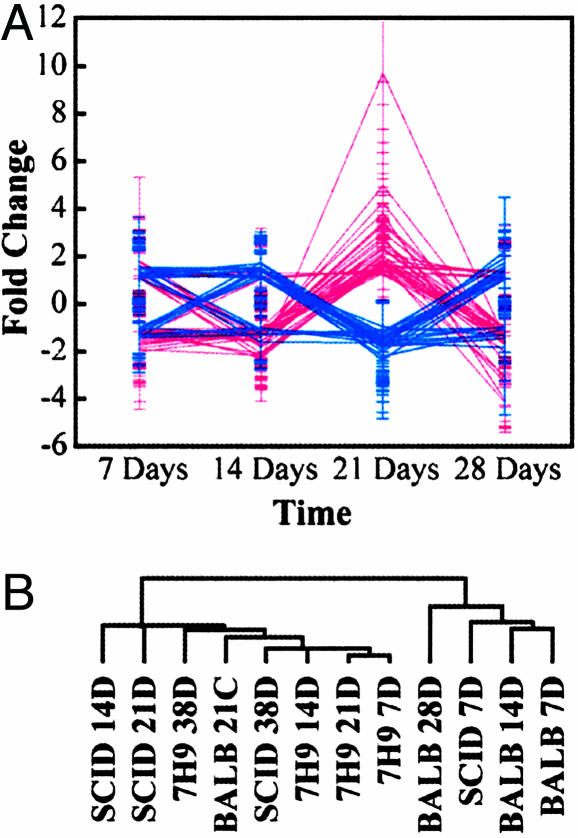
A more detailed analysis of the bacilli expression levels in BALB/c relative to SCID mice (Fig. 3A) identified a group of 122 genes that displayed a significant change in their expression levels in BALB/c vs. SCID mice. The “surge” in gene expression levels at 21 days after infection is responsible for the clustering of BALB/c at 21 days profile with Mtb growing in log phase. However, this surge of gene expression might reflect the Mtb response to the host microenvironments affected by the start of mounting an increased immune response against the infection at 21 days (23, 24). The increase in host responses to the Mtb infection was also confirmed with our histopathological examination of mice infected at different time points after infection (supporting information). Genes up-regulated in samples collected at 21 days after infection from BALB/c compared with SCID mice included three genes (atpE, atpF, and atpH) of the ATP synthase operon. The ATP synthase operon encodes the Mg2+-dependent, proton-translocating membrane protein that could be involved in maintaining the intracellular pH homeostasis required for optimal growth of mycobacterial bacilli as suggested (30, 31). Genes involved in iron metabolism and regulation (fdxA, ferredoxin A; mbtD, mycobactin synthesis D; and hupB, iron-regulated protein) (32, 33) were also up-regulated in this gene group reflecting the essential role of iron for bacilli growth in the immune-competent host.
Fig. 3.
Identification of immune-responsive genes by using DNA microarrays. (A) The fold change in genes with significant change in expression levels when Mtb bacilli were grown in BALB/c (red) vs. SCID (blue) mice at 7, 14, 21, and 28 days after infection (n = 122, 67 up- and 55 down-regulated in BALB/c relative to 7H9 samples). (B) The dendogram tree of hierarchical clustering analysis of genes expressed in BALB/c or SCID mice and broth over the first 28 days after infection. List of genes is provided in the supporting information.
Other genes that might be involved in pathogenicity of Mtb in vivo (lipF, lipase and clpX, atp-dependent protease) were also included in the 21-day BALB/c sample. With signature-tagged mutagenesis, LipF was also identified as important for mycobacterial persistence in mice (34). The fact that expression of a particular gene group was induced in BALB/c but not in SCID mice strongly indicates modulation of the bacterial gene expression in response to host immune responses. Moreover, analysis of BALB/c expression levels relative to in vitro samples indicate an additional 143 genes regulated at 21 days after infection (the supporting information), again indicating the bacterial response to the host's immune microenvironment.
Functional Analysis of in Vivo-Expressed Genes. We also applied an agglomerative hierarchical clustering algorithm (20) to group genes by expression patterns that may reflect similar function once the bacilli establish an infection. Hierarchical clustering supported the gene classes derived by self-organizing map analysis (Fig. 3B). It also identified several gene groups as differentially expressed between in vivo and in vitro (the supporting information). In one cluster, most of the genes were highly expressed in both broth and SCID mice samples, confirming the logarithmic character of the bacilli growing in SCID mice as noted before. This gene cluster included the transcriptional regulators, sigK, Rv0744c, and the possible two-component transcriptional regulator, Rv1626 indicating their importance in regulating gene expression during the log phase of growth. Genes encoding the IS1539 transposase (Rv2885c), the mutator protein (mutT3), and a possible resolvase (Rv0921) were also present in the same cluster, which may indicate that genetic rearrangements occur during mycobacterial growth at these time points. Several other clusters displayed genes highly expressed in in vivo vs. in vitro samples, including genes that were shown to be involved in cell wall biosynthesis (e.g., ald), transcriptional regulation (e.g., pvdS), purine biosynthesis (e.g., pure), or biotin metabolism (e.g., birA) and may contribute to the establishment of the infection inside the host. Moreover, we searched differentially expressed genes for the presence of secreted antigens that were characterized by using a bioinformatics approach (35). Among highly expressed mycobacterial genes, a set of 11 genes (supporting information) were expressed in BALB/c mice compared with only four and six genes in SCID and broth, respectively. The BALB/c-secreted antigens included the abundant antigen, Ag 85C, which was shown to play an important role in the permeability and integrity of the mycobacterial cell wall (36).
Additional analysis of the hierarchical clusters identified a cluster (Fig. 4) of 49 in vivo-expressed genes that has 20 genes occupying a contiguous region of 34.1 kb of the Mtb genome sequence (8). Because this set of genes was highly expressed only in vivo, and not in vitro, we denoted this region as the in vivo-expressed genomic island (iVEGI). The GC content of the iVEGI is <55%, whereas that of the surrounding region and Mtb in general is on average >65% GC (8). The majority of the genes in this region are involved in cell wall biosynthesis (Rv0969, Rv0970, and Rv0985c--Rv0987) and lipid metabolism (Rv0971c-Rv0975c). Genes with putative virulence phenotypes that could facilitate the Mtb survival inside the host are also located in the iVEGI, including a serine protease (Rv0983) and acyl-CoA dehydrogenase (Rv0975c) that is similar to the human acyl-CoA dehydrogenase. Two genes (mprA and mprB) in the iVEGI belong to the family of two-component response regulators and were proved to be involved in mycobacterial persistence during infection (37). Additional analysis of the regions flanking the iVEGI did not identify sequences encoding prophages or transposable elements, hallmarks of transposed islands. However, a tRNA gene for alaV is located downstream of the iVEGI. All the iVEGI genes are found in the pathogenic strains analyzed (M. tuberculosis H37Rv and CDC1551, M. bovis, and Mycobacterium leprae), whereas only four genes are found in the nonpathogenic strain, Mycobacterium smegmatis. Specifically, genes of the iVEGI with GC content <55% (mscL and Rv0986-Rv0988) display only a weak sequence similarity (E-value <0.05) to any ORF in the M. smegmatis (http://www.tigr.org/tdb) genome. Moreover, all the genes constituting the iVEGI were present in M. bovis bacillus Calmette-Guérin and in Mtb H37Ra strain as demonstrated by PCR-based analysis (data not shown). Of note, not all pathogenicity islands fulfill all the criteria set for characterizing pathogenicity islands (38).
Fig. 4.
Genomic organization of the iVEGI where arrowheads indicate the direction of transcription as predicted from the genome sequence of Mtb strain H37Rv. Filled arrows represent mycobacterial genes with higher expression levels in mice samples compared with in vitro samples.
Discussion
We have adapted DNA microarray technology to assess Mtb gene expression profile during growth in the host. The expression patterns of all the genes of Mtb have now been assessed during the early time course of infection in both immune-competent and immune-compromised mice, with several profiles of note. First, the expression profile of Mtb in SCID mice most resembles the profile when grown in broth, rather than the profile when grown in BALB/c mice. Second, a group of 67 genes was significantly activated in BALB/c but not in SCID mice at 21 days after infection, supporting the notion that the growth of Mtb bacilli in an immunocompetent host is influenced by the host immune responses (23, 24) and its growth requirements. Third, a group of 33 genes reported as induced at 24 h after infection in an in vitro macrophage model (28) were also induced at 21 days after infection in mice reflecting the nature of the host immune response at this stage of tuberculosis. Fourth, at least 49 genes are only expressed in vivo and 20 of these are contiguous in a distinctively lower GC content area of the Mtb genome. We recognize that some of the genes expressed in vivo could reflect the normal transition of the tuberculous bacilli to different phases of growth regardless of the host environment or because of the handling procedure taken during RNA extraction from infected lungs. Nevertheless, by comparing relative gene expression of infected vs. in vitro samples handled similarly to in vivo samples, the identified expression profiles during infection provide a starting point for further studies to differentiate between host-responsive vs. phase-dependent genes.
Several of the genes we have identified as differentially expressed between growth in culture and in mice have also been identified by other technologies, supporting the validity of the amplified differential gene expression (ADGE) technology for the in vivo analysis. Genes considered to be differentially expressed (mostly of unknown function) by ADGE (such as Rv0024, mce1, Rv0592, Rv0594, Rv1200, Rv1968, and Rv3088) were also identified by sequence homology-based strategy used during the initial sequencing of Mtb. Additional genes identified by ADGE profiling as up-regulated in vivo (e.g., Rv0170, Rv0834c, sigE, tkt, sseA, uvrA, lipF, and ponA′) were also identified by other protocols based on genomic screening in macrophage cell cultures by selective capture of transcribed sequences (25), differential fluorescence induction (39) analysis, or recombinant-based in vivo expression technology (40). Another set of genes (pks6, fadB4, mtrA, and groEL1) identified by the selective capture of transcribed sequences and differential fluorescence induction protocols was down-regulated in vivo by the ADGE profiling. In addition, among 22 genes induced in BALB/c mice lung tissues as detected by qPCR (28) at 21 days, we characterized four genes (Rv1738, Rv2626c, aceA, and fadB2) as repressed in vivo vs. in vitro at the same time point. Discrepancies between ADGE profiling and other technologies (selective capture of transcribed sequences, recombinant-based in vivo expression technology, differential fluorescence induction, and qPCR) could arise from the difference in the model system, experimental conditions, or the criteria set by investigators for identifying differentially expressed genes. Nonetheless, ADGE profiling provides a more dynamic view of the changes of gene expression profiles over time with a higher resolution on a gene-by-gene basis and on a genome-wide level compared with other protocols.
Microarray analysis of the in vivo-expressed genes also provided evidence of coexpression of a group of genes that occupy the same genetic loci, named iVEGI, that may contribute to the in vivo survival of Mtb. Based on established criteria for characterizing genomic islands (41) and the expression profile of this particular cluster of genes, we can denote iVEGI as a genomic island encoding cell wall components and participating in lipid metabolism required for mycobacterial survival in vivo. Whether the iVEGI is also a pathogenicity island will require further characterization of genes included in the iVEGI region by both animal testing and further analysis of the avirulent mycobacterial strains. Analysis of gene expression at longer time points (>28 days) could extend our knowledge of the expression of iVEGI and contribute to our understanding of the state of bacilli during chronic infection.
Finally, the classification of genes in expression patterns may give insight into optimal drug and vaccine targets. Given that Mtb remains one of the most devastating human pathogens (5), this insight could be useful. For example, the genes that are significantly up-regulated at early time points may be good drug candidates, in particular, if they serve in critical, conserved functions in pathogenic bacteria. It may also be possible to gain insight into the best vaccine candidates. We have demonstrated a technology (expression library immunization; ref. 42) for screening the whole genome of any pathogen for genes that encode effective vaccines. If in vivo microarray analysis was able to restrict the scope of the genes to be tested, it would greatly facilitate this search. Regardless, the techniques used here to assess gene expression of M. tuberculosis in vivo should be applicable to other pathogens.
Supplementary Material
Acknowledgments
We thank Annie Alsup, Walker Hale, Quiha Sun, Tom Giesler, and Kristen Garrison for technical assistance; Preston Hunter; and Ross Chambers, Mike McGuire, and Vanessa Sperandio for helpful discussions. This work was supported by Defense Advanced Research Planning Agency and National Institutes of Health grants (to S.A.J. and R.L.) and University of Wisconsin, Madison, grants (to A.M.T.). A.M.T. was also supported in part by a Molecular Cardiology Training grant to University of Texas Southwestern Medical Center.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Mtb, Mycobacterium tuberculosis; iVEGI, in vivo-expressed genomic island; ADGE, amplified differential gene expression; qPCR, quantitative PCR; SCID, severe combined immune-deficient; cfu, colony-forming unit.
References
- 1.Krinos, C. M., Coyne, M. J., Weinacht, K. G., Tzianabos, A. O. & Kasper, D. L. (2001) Nature 414, 555-558. [DOI] [PubMed] [Google Scholar]
- 2.Sturgill-Koszycki, S., Schlesinger, P. H., Chakraborty, P., Haddix, P. L., Collins, H. L., Fok, A. K., Allen, R. D., Gluck, S. L., Heuser, J. & Russell, D. G. (1994) Science 263, 678-681. [DOI] [PubMed] [Google Scholar]
- 3.Garbe, T. R., Hibler, N. S. & Deretic, V. (1999) Infect. Immun. 67, 460-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang, M., Kim, K. J., Iyer, D., Lin, Y. G., Belisle, J., McEnery, K., Crandall, E. D. & Barnes, P. F. (1997) Infect. Immun. 65, 692-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dye, C., Scheele, S., Dolin, P., Pathania, V. & Raviglione, R. C. (1999) J. Am. Med. Assoc. 282, 677-686. [DOI] [PubMed] [Google Scholar]
- 6.Pablos-Mendez, A., Raviglione, M. C., Laszlo, A., Binkin, N., Rieder, H. L., Bustreo, F., Cohn, D. L., Lambregts-van Weezenbeek, C. S. B., Kim, S. J., Chaulet, P., et al. (1998) N. Engl. J. Med. 338, 1641-1649. [DOI] [PubMed] [Google Scholar]
- 7.Cohn, D. L. (1997) Am. J. Med. Sci. 313, 372-376. [DOI] [PubMed] [Google Scholar]
- 8.Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S. V., Eiglmeier, K., Gas, S., Barry, C. E., III, et al. (1998) Nature 393, 537-538. [DOI] [PubMed] [Google Scholar]
- 9.Schena, M., Shalon, D., Davis, R. W. & Brown, P. O. (1995) Science 270, 467-470. [DOI] [PubMed] [Google Scholar]
- 10.Behr, M. A., Wilson, M. A., Gill, W. P., Salamon, H., Schoolnik, G. K., Rane, S. & Small, P. M. (1999) Science 284, 1520-1523. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez, G. M., Voskuil, M. I., Gold, B., Schoolnik, G. K. & Smith, I. (2002) Infect. Immun. 70, 3371-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson, W., DeRisi, J., Kristensen, H. H., Imboden, P., Rane, S., Brown, P. O. & Schoolnik, G. K. (1999) Proc. Natl. Acad. Sci. USA 96, 12833-12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talaat, A. M., Howard, S. T., Hale, I. W., Lyons, R., Garner, H. & Johnston, S. A. (2002) Nucleic Acids Res. 30, E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lucchini, S., Thompson, A. & Hinton, J. C. D. (2001) Microbiology 147, 1403-1414. [DOI] [PubMed] [Google Scholar]
- 15.Lakey, D. L., Zhang, Y., Talaat, A. M., Samten, B., Desjardin, L. E., Eisenach, K. D., Johnston, S. A. & Barnes, P. F. (2002) Microbiology 148, 2567-2572. [DOI] [PubMed] [Google Scholar]
- 16.Merrell, D. S., Butler, S. M., Qadri, F., Dolganov, N. A., Alam, A., Cohen, M. B., Calderwood, S. B., Schoolnik, G. K. & Camilli, A. (2002) Nature 417, 642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu, Q., Dziejman, M. & Mekalanos, J. J. (2003) Proc. Natl. Acad. Sci. USA 100, 1286-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talaat, A. M., Hunter, P. & Johnston, S. A. (2000) Nat. Biotechnol. 18, 679-682. [DOI] [PubMed] [Google Scholar]
- 19.Thomas, J. G., Olson, J. M., Tapscott, S. J. & Zhao, L. P. (2001) Genome Res. 11, 1227-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tseng, G. C., Oh, M. K., Rohlin, L., Liao, J. C. & Wong, W. H. (2001) Nucleic Acids Res. 29, 2549-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmittgen, T. D., Zakrajsek, B. A., Mills, A. G., Gorn, V., Singer, M. J. & Reed, M. W. (2000) Anal. Biochem. 285, 194-204. [DOI] [PubMed] [Google Scholar]
- 23.Orme, I. M., Andersen, P. & Boom, W. H. (1993) J. Infect. Dis. 167, 1481-1497. [DOI] [PubMed] [Google Scholar]
- 24.Dunn, P. L. & North, R. J. (1996) J. Med. Microbiol. 45, 103-109. [DOI] [PubMed] [Google Scholar]
- 25.Graham, J. E. & Clark-Curtiss, J. E. (1999) Proc. Natl. Acad. Sci. USA 96, 11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKinney, J. D., Zu, B. K., Munoz-Elias, E. J., Miczak, A., Chen, B., Chan, WT, Swenson, D., Sacchettini, J. C., Jacobs, W. R., et al. (2000) Nature 406, 735-738. [DOI] [PubMed] [Google Scholar]
- 27.Fisher, M. A., Plikaytis, B. B. & Shinnick, T. M. (2002) J. Bacteriol. 184, 4025-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schnappinger, D., Ehrt, S., Voskuil, M. I., Liu, Y., Mangan, J. A., Monahan, I. M., Dolganov, G., Efron, B., Butcher, P. D., Nathan, C., et al. (2003) J. Exp. Med. 198, 693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamyo, P., Slonim, D., Mesirov, J., Zhu, Q., Kitareewan, S., Dmitrovsky, E., Lander, E. & Golub, T. (1999) Proc. Natl. Acad. Sci. USA 96, 2907-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piddington, D. L., Kashkouli, A. & Buchmeier, N. A. (2000) Infect. Immun. 68, 4518-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rao, M., Streur, T. L., Aldwell, F. E. & Cook, G. M. (2001) Microbiology 147, 1017-1024. [DOI] [PubMed] [Google Scholar]
- 32.De Voss, J. J., Rutter, K., Schroeder, B. G. & Barry, C. E. (1999) J. Bacteriol. 181, 4443-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohavy, O., Harth, G., Horwitz, M., Eggena, M., Landers, C., Sutton, C., Targan, S. R. & Braun, J. (1999) Infect. Immun. 67, 6510-6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Camacho, L. R., Ensergueix, D., Perez, E., Gicquel, B. & Guilhot, C. (1999) Mol. Microbiol. 34, 257-267. [DOI] [PubMed] [Google Scholar]
- 35.Gomez, L., Johnson, S. & Gennaro, M. L. (2000) Infect. Immun. 68, 2323-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson, M., Raynaud, C., Laneelle, M. A., Guilhot, C., Laurent-Winter, C., Ensergueix, D., Gicquel, B. & Daffe, M. (1999) Mol. Microbiol. 31, 1573-1587. [DOI] [PubMed] [Google Scholar]
- 37.Zahrt, T. C. & Deretic, V. (2001) Proc. Natl. Acad. Sci. USA 98, 12706-12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elliott, S. J., Wainwright, L. A., McDaniel, T. K., Jarvis, K. G., Deng, Y. K., Lai, L. C., McNamara, B. P., Donnenberg, M. S. & Kaper, J. B. (1998) Mol. Microbiol. 28, 1-4. [DOI] [PubMed] [Google Scholar]
- 39.Triccas, J. A., Berthet, F. X., Pelicic, V. & Gicquel, B. (1999) Microbiology 145, 2923-2930. [DOI] [PubMed] [Google Scholar]
- 40.Saviola, B., Woolwine, S. C. & Bishai, W. R. (2003) Infect. Immun. 71, 1379-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hacker, J. & Kaper, J. B. (2000) Annu. Rev. Microbiol. 54, 641-679. [DOI] [PubMed] [Google Scholar]
- 42.Barry, M. A., Lai, W. C. & Johnston, S. A. (1995) Nature. 377, 632-635. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.