Abstract
Myelin is a dynamic, functionally active membrane necessary for rapid action potential conduction, axon survival, and cytoarchitecture. The number of debilitating neurological disorders that occur when myelin is disrupted emphasizes its importance. Using high-resolution 2D gel electrophoresis, mass spectrometry, and immunoblotting, we have developed an extensive proteomic map of proteins present in myelin, identifying 98 proteins corresponding to at least 130 of the ≈200 spots on the map. This proteomic map has been applied to analyses of the localization and function of selected proteins, providing a powerful tool to investigate the diverse functions of myelin.
Myelin is a dynamic, functionally active membrane (1), the loss or damage of which results in serious neurological disorders including leukodystrophies, central and peripheral neuropathies, and inflammatory demyelinating diseases such as multiple sclerosis (2-4). Rapid and efficient action potential conduction in the nervous system depends on myelin, which traditionally has been viewed as a passive contributor to conduction by increasing internodal membrane resistance and decreasing membrane capacitance (5). However, recent work has revealed additional active roles for myelin in nervous system development and function. For example, myelin regulates axon diameter and the formation of axon microtubular networks, and it is a key player in ion channel clustering at nodes of Ranvier (6-11). In return, the axon regulates myelin gene expression (12) and oligodendrocyte survival (13). The functional coupling of myelin and axons is further illustrated by the transfer of phospholipids (14) and N-acetylaspartate (15) from the axon to the myelin sheath.
A major impediment to understanding the active biological functions of myelin is the relative lack of myelin proteins that have been described and characterized. Thus, we have undertaken an extensive proteomic analysis of central nervous system myelin, developed a 2D PAGE map of myelin proteins, identified 98 of these proteins, and illustrated the power and utility of this approach through specific applications of comparative proteomics. For example, a comparison of CNS and peripheral nervous system (PNS) myelin proteins has revealed a previously undescribed component of Schwann cell microvilli, a proteomic matching of myelin from normal and genetically modified mice lacking key myelin lipids has demonstrated a dramatic reduction of two newly recognized myelin protein kinases, and an application of the map to a neuroimmunological analysis of demyelinating disease has identified a signaling mechanism (16).
Materials and Methods
Unless otherwise stated, materials were obtained from Sigma.
Antibodies. Rabbit polyclonal anti-Gβ (Santa Cruz Biotechnology), CRMP-2 (S. Strittmater, Yale University, New Haven, CT), Nm23 (Santa Cruz Biotechnology), Gαo (Santa Cruz Biotechnology), N-ethylmaleimide-sensitive fusion protein (NSF) (Santa Cruz Biotechnology), EF-2 (A. Nairn, Yale University), MnSOD (Upstate Biotechnology, Lake Placid, NY), ERp57 (StressGen Biotechnologies, Victoria, BC, Canada), VDAC-1 (Oncogene Science), OSP/claudin-11 (J. Bronstein, University of California, Los Angeles), and CNP (17) IgGs were used. Mouse monoclonal anti-myelin oligodendrocyte glycoprotein (C. Linington, Max Planck Institute for Neurobiology, Martinsried, Germany), cytochrome c oxidase (Molecular Probes), contactin (BD Biosciences, San Diego), Pan NaCh (18), GLNS (BD Biosciences), Munc-18 (BD Biosciences), NCAM (BD Biosciences), flotillin-1 (BD Biosciences), moesin (BD Biosciences), β-actin, and β-tubulin IgGs were used. Rat monoclonal anti-proteolipid protein (M. Lees, Shriver Center, University of Massachusetts, Waltham) IgG was used. Goat polyclonal anti-HSC70, CK-B, and Ezrin (all from Santa Cruz Biotechnology) IgGs were used. Horseradish peroxidase-conjugated secondary antibodies goat anti-mouse (Chemicon), goat anti-rat (Chemicon), donkey anti-rabbit (Santa Cruz Biotechnology), and donkey anti-goat (Santa Cruz Biotechnology) IgGs were used.
Immunoblotting. SDS/PAGE gels were transferred onto polyvinylidene difluoride membranes (Amersham Pharmacia Biosciences) and incubated with primary antibody at room temperature for 1 h or overnight at 4°C, followed by incubation with horseradish peroxidase-conjugated secondary antibodies at room temperature for 30 min. Antibodies were diluted in blocking solution (5% nonfat dry milk in Tris-buffered saline with 0.2% Tween 20). Blots were visualized by using enhanced chemiluminescence.
Myelin Purification. Myelin was purified from postnatal-day-35 C57BL/6J wild-type and ceramide galactosyl transferase (CGT)-null mouse brains as described (18). This method uses a combination of homogenization, hypotonic shock, and sucrose gradient centrifugation to produce a series of myelin-axolemmal complex fractions of increasing density. The quantitatively most abundant and most myelin-enriched fraction, or “main band,” was used for this study. In addition, PNS myelin was purified from adult Sprague-Dawley rat sciatic nerves. All solutions were used at pH 7.5 and 4°C. Purified myelin was resuspended in a minimal volume of 50 mM Tris/protease inhibitor mixture (10 μg/ml leupeptin/10 μg/ml aprotinin/1 mM phenylmethylsulfonyl fluoride). Aliquots were frozen on dry ice and stored at -80°C.
2D PAGE. 2D PAGE was performed according to the method of Taylor and Pfeiffer (19). Purified myelin proteins were solubilized in lysis buffer [1% ASB-14 (Calbiochem)/25 mM Tris/5 mM EDTA/protease inhibitor mixture] at 37°C for 30 min, then precipitated with two volumes of 100% ethanol at -20°C overnight. The precipitated pellet was suspended in 20 μl of 10% ASB-14 and sonicated. Samples were then suspended up to 370 μl with rehydration buffer [7 M urea (ICN)/2 M thiourea (Fisher Scientific)/2% ASB-14/0.5% carrier ampholytes/100 mM DTT (Fisher Scientific)/0.001% bromophenol blue] and kept at room temperature for 1 h with occasional mixing. The sample was loaded into 18-cm IPGphor strip holders and overlaid with immobilized pH gradient (IPG) strips (Amersham Pharmacia). IPG strips were covered with 3.5 ml of mineral oil and rehydrated actively (30 V, 20°C) overnight by using an IPGphor (Amersham Pharmacia) isoelectric focusing (IEF) unit. After rehydration, hydrated filter wicks were placed under the gel. The cathodic filter wick was hydrated by using 100 mM DTT, whereas the anodic filter wick was hydrated by using water (19). IEF proceeded as follows: 200 V for 1 h, 500 V for 1 h, 1,000 V for 1 h, ramp to 6,000 V for 30 min, and hold at 6,000 V for 20,000 volt-hours. IPG strips were incubated in equilibration buffer (50 mM Tris, pH 8.8/6 M urea/30% glycerol/2% SDS) with 1% DTT for 15 min, then in equilibration buffer with 2% iodoacetamide for 15 min. IPG strips were placed on SDS/PAGE gels and sealed in place with 0.5% agarose. 2D SDS/PAGE was performed by using a Hoefer DALT tank (Amersham Pharmacia). Gels were transferred onto polyvinylidene difluoride for immunoblotting or stained with ammoniacal silver nitrate or colloidal Coomassie blue (Invitrogen).
Immunofluorescence Microscopy. Optic and sciatic nerves from adult and neonatal rats and adult mice were dissected and immediately fixed in ice-cold 4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.2, for 30 min. Fixed tissue was transferred to ice-cold 20% (wt/vol) sucrose in 0.1 M phosphate buffer until equilibrated, then transferred again to 30% sucrose. The tissue was then frozen in Tissue-Tek OCT mounting medium (Miles Scientific). Sections were cut, placed in 0.1 M phosphate buffer, spread on gelatin-coated coverslips, and allowed to air dry. Immunofluorescence staining was performed as described (10). Secondary antibodies were Alexa Fluor 488 or Alexa Fluor 594-conjugated to goat anti-rabbit or goat anti-mouse antibodies. Digital images were collected on a fluorescence microscope (Axioskop 2, Zeiss) fitted with a Hamamatsu ORCA-ER charge-coupled device camera (Hamamatsu Photonics, Hamamatsu City, Japan). A z-stack of images was collected at 0.2-μm intervals, and the resulting stacks were then deconvolved by iterative restoration using the software package volocity (Improvision, Lexington, MA). All digital images (immunofluorescence, immunoblots, and stained gels) were prepared by using photoshop (Adobe Systems, Mountain View, CA) and coreldraw (Corel, Ottawa).
Protein Identification. Coomassie blue-stained gel spots were excised and digested overnight with trypsin. Tryptic peptides were separated by microcapillary liquid chromatography coupled to a tandem mass spectrometer (LCQ-DECA; Finnigan-MAT, San Jose, CA; Center for Proteomics and Biological Mass Spectrometry at the University of Connecticut Health Center). Peptide fragmentation spectra were analyzed against the OWL database by using the SEQUEST algorithm (20) followed by further analysis using interact (21) and peptide prophet (22). In a few cases, tryptic peptides were analyzed by matrix-assisted laser desorption ionization time-of-flight (Micromass M@LDI-R; W. M. Keck Facility, Yale University), and peptide masses were analyzed by using peptide search (European Molecular Biology Laboratory) and profound (The Rockefeller University, New York) software.
Results
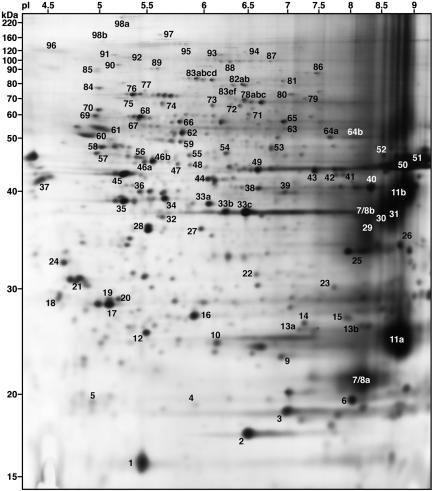
Identification and Mapping of Myelin Proteins. Proteins from highly enriched mouse myelin (18) were analyzed by 2D PAGE by using an enhanced method for resolving transmembrane and glycosylphosphatidylinositol-anchored proteins from lipid-rich membranes (19). The resulting pattern consists of ≈200 well resolved protein spots of varying intensities (Fig. 1). Higher-resolution gels corresponding to the pH ranges 4-7 and 6-9 are shown in Figs. 5 and 6, which are published as supporting information on the PNAS web site.
Fig. 1.
2D PAGE mapping of myelin proteins. Myelin proteins were separated by using a pH 3-10 IPG strip and 12% SDS/PAGE gels, and the gels stained with silver nitrate. Proteins have been assigned numbers corresponding to the spot numbers listed in Table 1. Approximate molecular mass (kDa) and pI values are listed.
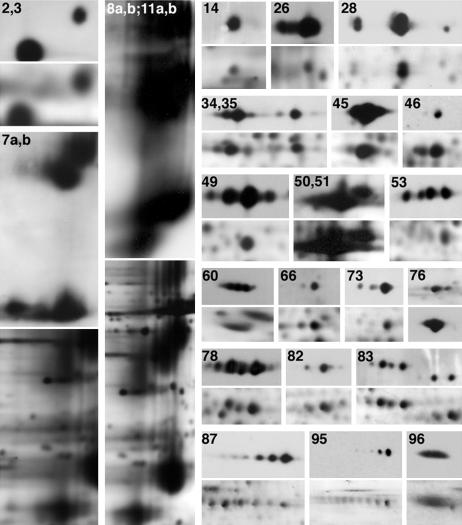
We next set out to identify and determine molecular correlates for these proteins. Proteins were identified by mass spectrometry and/or immunoblotting (see also ref. 23) and are summarized in Table 1, with their predicted molecular mass and pI. Most of the identified proteins migrate on the 2D PAGE in a manner consistent with their predicted molecular mass/pI values; exceptions are due to posttranslational modifications such as glycosylation and phosphorylation (24). We confirmed the identities of many proteins originally determined by mass spectrometry by using 2D immunoblotting (Fig. 2). Not surprisingly, this method often proved more sensitive than silver staining and led to the identification of additional spots on the 2D map. In some instances, this allowed for the identification of proteins for which the protein levels were too low for mass spectrometric identification [e.g., flotillin-1 (spot 53)]. In other cases, posttranslational modifications resulted in multiple immunoreactive spots for a single protein. For example, contactin (spot 95) appears on the map as a train of spots of increasing negative charge as a consequence of polysialylation (25). Combining mass spectrometry and immunoblotting, we have identified 98 proteins, corresponding to at least 130 of ≈200 spots on the map.
Table 1. Summary of protein identifications.
| Spot | Protein name | GenBank accession no. |
|---|---|---|
| 1 | MVP17* | O09198 |
| 2 | Nm23A†‡ | P15532 |
| 3 | Nm23B†‡ | Q01768 |
| 4 | ADP ribosylation factor 1† | P32889 |
| 5 | Cytochrome c oxidase† | P12787 |
| 6 | Cofilin† | P18760 |
| 7 | OSP/claudin-11‡ | Q60771 |
| 8 | DM2O‡ | P06905-2 |
| 9 | α Crystalline B chain† | P23927 |
| 10 | Rap-1A† | P10113 |
| 11 | PLP‡ | P06905 |
| 12 | ATP synthase D-subunit† | Q9DCX2 |
| 13 | Peroxiredoxin-1† | P35700 |
| 14 | MnSOD†‡ | P09671 |
| 15 | GST-pi† | P46425 |
| 16 | RhoA† | Q9QU10 |
| 17 | RhoB† | P01121 |
| 18 | Phosphoglycerate mutase 1† | Q9DBJ1 |
| 19 | RhoGDI 1† | Q99PT1 |
| 20 | Ubiquitin thiolesterase L1† | Q9R0P9 |
| 21 | 14-3-3 protein γ† | P35214 |
| 22 | Carbonic anhydrase II† | P00920 |
| 23 | Enoyl coenzyme A hydratase† | Q8BH95 |
| 24 | Elongation factor 1-β† | O70251 |
| 25 | V-ATPase E subunit† | P50518 |
| 26 | VDAC1†‡ | Q60932 |
| 27 | Malate dehydrogenase† | P14152 |
| 28 | Transducin β chain 1†‡ | P04901 |
| 29 | Lactate dehydrogenase A† | P06151 |
| 30 | GAPDH† | P16858 |
| 31 | Malate dehydrogenase (mito)† | P08249 |
| 32 | Lactate dehydrogenase B† | P16125 |
| 33 | SIR2L2 homolog† | Q9CXS5 |
| 34 | G(o) α-subunit 2†‡ | P18873 |
| 35 | G(o) α-subunit 1†‡ | P18872 |
| 36 | G(q) α-subunit† | P21279 |
| 37 | GAP-43† | P06837 |
| 38 | V-ATPase C-subunit† | Q9Z1G3 |
| 39 | Aldolase 3† | P05063 |
| 40 | Aldolase 1† | P05064 |
| 41 | Citrate synthase† | Q9CZU6 |
| 42 | Phosphoglycerate kinase 1† | P09411 |
| 43 | Creatine kinase, ubiquitous† | P30275 |
| 44 | Septin 2† | P42208 |
| 45 | β-Actin‡ | P02570 |
| 46 | Creatine kinase B chain†‡ | Q04447 |
| 47 | NDRG1 protein† | Q62433 |
| 48 | NADH-coenzyme Q reductase† | Q91WD5 |
| 49 | Glutamine synthetase† | P15105 |
| 50 | CNP1‡ | P16330-2 |
| 51 | CNP2‡ | P16330 |
| 52 | Septin 7† | O55131 |
| 53 | Flotillin-1‡ | O08917 |
| 54 | α Enolase† | P17182 |
| 55 | Rab GDI 3† | Q61598 |
| 56 | Ubiquinol-cytochrome c reductase§ | Q9CZ13 |
| 57 | γ Enolase† | P17183 |
| 58 | ATP synthase β chain† | P56480 |
| 59 | Vesicle amine transport protein 1† | Q8R3G0 |
| 60 | Tubulin β-2 chain‡§ | P05217 |
| 61 | Tubulin α-2 chain§ | P05213 |
| 62 | Sep2 fragments† | Q9ESF7 |
| 63 | CAP 1† | P40124 |
| 64 | ATP synthase α chain† | Q03265 |
| 65 | Pyruvate kinase M2 isozyme† | P52480 |
| 66 | ERp57† | P27773 |
| 67 | HSP60† | P19226 |
| 68 | Neurofilament-66† | P46660 |
| 69 | Rab GDI α† | P50396 |
| 70 | Protein phosphatase 2† | Q8C2E1 |
| 71 | MPAST1† | Q9WVK4 |
| 72 | Stress-induced phosphoprotein† | Q99L66 |
| 73 | CRMP-2†‡ | O08553 |
| 74 | DLAT fragment† | Q91ZB1 |
| 75 | Annexin VI† | P14824 |
| 76 | HSC71†‡ | P08109 |
| 77 | GRP75§ | P38647 |
| 78 | Munc-18† | O08599 |
| 79 | Transketolase† | P40142 |
| 80 | Calpain 5† | O08688 |
| 81 | Transferrin† | Q92111 |
| 82 | Vesicular-fusion protein NSF†‡ | P46460 |
| 83 | Ezrin†‡ | P26040 |
| 84 | GRP78† | P20029 |
| 85 | HSP84† | P11499 |
| 86 | Similar to aconitase† | Q99K10 |
| 87 | Elongation factor 2†‡ | P58252 |
| 88 | Dynamin-1 (isoform 5)† | P39053 |
| 89 | Gelsolin† | P13020 |
| 90 | TER ATPase† | Q01853 |
| 91 | APG-2† | Q61316 |
| 92 | Ubiquitin-activating enzyme E1† | Q02053 |
| 93 | Dipeptidylpeptidase 6† | Q9Z218 |
| 94 | Hexokinase, tumor isozyme† | P17710 |
| 95 | Contactin†‡ | P12960 |
| 96 | NCAM-120†‡ | P13594 |
| 97 | Phospholipase C β-1† | Q8K5A5 |
| 98 | Fodrin α chain† | P16086 |
MVP17 was sequenced by Edman degradation (23). Predicted molecular weight and pI values are available through the Swiss-Prot and TrEMBL databases located on the ExPASy proteomics server (http://us.expasy.org/sprot).
Liquid chromatography mass spectrometry.
Immunoblotting.
Matrix-assisted laser desorption ionization.
Fig. 2.
2D immunoblotting of myelin proteins. Each numbered immunoblot is shown above its corresponding region from a silver-stained gel. Numbers correspond to protein spots from Fig. 1 and Table 1. For display purposes, OSP/claudin-11 and proteolipid protein/DM20 immunoblots and silver-stained gel regions were minimized relative to other spots.
The proteins identified can be grouped into functional categories. These include cytoskeletal/scaffolding (e.g., spots 6, 45, 60, 61, 73, 83, 89, and 98), signaling (e.g., spots 4, 10, 16, 17, 19, 21, 28, 34, 35, 36, 55, 69, and 70), and vesicle transport, docking, and fusion (e.g., spots 1, 59, 78, 82, 88, and 90) proteins. One particularly interesting group included proteins traditionally associated with mitochondria and/or energy metabolism (e.g., spots 5, 12, 14, 26, 31, 39, 40, 43, 56, 58, and 86).
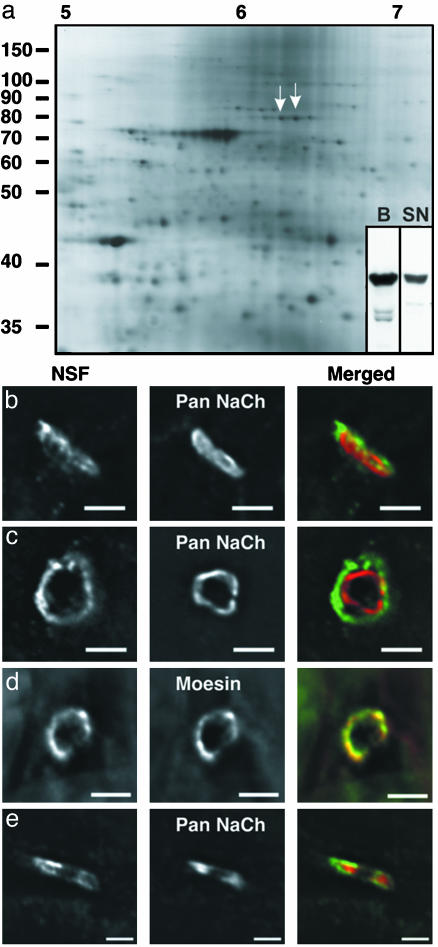
Functional Applications. The database generated by our analysis serves as a powerful resource for developmental and functional studies of myelin. Two examples emanating from this study are instructive. Although CNS and PNS myelin serve many of the same functions, there are interesting differences in its synthesis, cell biology, and protein composition (26). Nevertheless, because there is a substantial overlap in protein composition, the proteomic map of CNS myelin presented here also provides an extensive map of many PNS myelin proteins as well (Fig. 3a; unpublished data). Further, it has become clear that the cellular location and function of a protein are inextricably linked. Therefore, as one approach for placing the identified proteins into functional contexts, we are carrying out an unbiased immunohistochemical analysis on protein localization in optic nerve, spinal cord, and sciatic nerve. Among a number of interesting staining patterns (data not shown), immunostaining with antibodies against NSF [spot 82; also present in PNS myelin (Fig. 3a)] is particularly intriguing. In CNS optic nerve longitudinal sections, NSF staining is diffusely present along myelinated fibers (data not shown). In contrast, in the PNS sciatic nerve, high levels of NSF are concentrated at nodes of Ranvier (Fig. 3b). When examined in cross section, double immunostaining of voltage-gated sodium channels and NSF showed that NSF immunoreactivity formed a halo around the sodium channel clusters (Fig. 3c). Because PNS nodes of Ranvier are contacted by Schwann cell microvilli, we hypothesized that NSF is present in these structures. Indeed, double immunostaining of NSF with known markers of Schwann cell microvilli, such as moesin (27, 28), demonstrated their colocalization (Fig. 3d). Finally, because Schwann cell microvilli have been implicated in efficient node of Ranvier formation in the PNS, we examined its localization during early development and found that NSF is present in high densities and overlaps with sodium channels at forming nodes (Fig. 3e). As in other cells, vesicle transport from the trans-Golgi network to sites of newly forming myelin membrane is regulated in part by soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins at sites of vesicle docking and fusion (29). A number of these molecules have been identified in oligodendrocytes (OLs) and myelin (30), several of which are represented in this map. NSF plays an important role in the regulation of vesicle priming and recycling (31). Therefore, its localization in the microvilli of Schwann cells in sciatic nerve suggests that microvilli might be localized sites of membrane deposition. Because microvilli have been implicated in sodium channel clustering during development at nodes of Ranvier (27, 28), these data suggest that membrane synthesis could be involved in this process.
Fig. 3.
NSF is a previously undescribed component of Schwann cell microvilli. (a) 2D PAGE of adult PNS myelin proteins shows the presence of NSF (arrows). (Inset) 1D immunoblot of NSF in brain homogenate (B) and sciatic nerve (SN). (b and c) Immunostaining of NSF (Left and Right in green) and voltage-gated sodium channels (Center and Right in red) in adult sciatic nerve transverse section (b) and cross section (c). (Bar = 3 μm.) (d) Immunostaining of NSF (Left and Right in green) and moesin (Center and Right in red) in adult sciatic nerve cross section. (Bar = 3 μm.) (e) Immunostaining of postnatal day 1 sciatic nerve transverse section. (Bar = 2 μm.) Axon orientation in b is from bottom left to upper right. Axon orientation in e is from left to right.
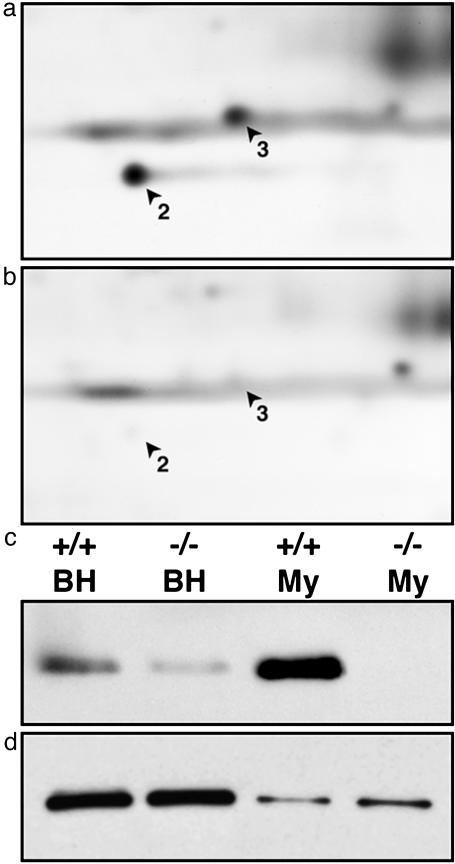
Additionally, a comparative proteomic approach was used to compare the expression of proteins in myelin from wild-type and genetically modified CGT-null mice (32, 33). These mice are unable to express two key myelin glycosphingolipids, galactocerebroside and sulfatide. In particular, we noted that two newly identified myelin proteins, Nm23A and -B (Fig. 1, spots 2 and 3, respectively), are dramatically reduced in CGT-null brain and virtually absent from CGT-null myelin (Fig. 4 a and b). Consistent with this, Nm23A and -B are highly enriched in myelin compared with brain homogenate. Nm23A and -B are nucleoside diphosphate kinases involved in the transfer of the γ-phosphate from ATP to other diphosphates (e.g., GDP) as well as protein kinases (34). Among the substrates of Nm23 is the β-subunit of heterotrimeric GTP-binding proteins (35), one of the proteins that undergoes altered phosphorylation status in an antibody-mediated model of demyelinating disease (16) (see Discussion). Further, Nm23 is reduced in the brains of patients afflicted with Alzheimer's disease or Down's syndrome (36). Thus, a direct comparison between two myelin proteomic maps has allowed a rapid identification of an interesting mutation-related regulation. Similar studies can be performed for numerous myelin mutants, with the identities of misregulated proteins often directly ascertained from the master 2D map (Fig. 1).
Fig. 4.
Nm23A and -B are dramatically reduced in CGT-null myelin. (a and b) Myelin proteins purified from wild-type (a) and CGT-null mice (b) were separated by 2D PAGE and silver stained. Nm23A (2) and -B (3) levels were dramatically reduced in CGT-null myelin when compared with wild-type myelin. (c) Similar reductions also seen in brain homogenates (BH) were confirmed by immunoblotting. Note the enrichment of Nm23 in myelin (My) compared with brain homogenate. (d) Levels of actin remained unchanged in CGT-null mice compared with wild-type controls.
Discussion
Myelin is a complex community of proteins and lipids serving a variety of vital functions. This work offers a major advance in this regard by identifying a significant number of myelin proteins and displaying them on a 2D map, greatly facilitating subsequent direct comparisons and identifications of proteins in other experimental contexts. To realize its full potential, each protein must be placed into its specific biological context. Thus, we have organized the identified proteins into families with related metabolic and cellular function and demonstrated the utility of the map by providing examples of its application to functional analyses of nervous system physiology and pathology.
A current forefront of myelin biological research stems from the recognition of an extensive bidirectional communication between the myelin sheath and the axon. As a prelude to this analysis, we reexamined the classical methods of “purifying” myelin by biochemical means (18). It was noted that even myelin highly enriched by density centrifugation included low but significant levels of specific axolemmal proteins, particularly components of the paranodal and juxtaparanodal regions (see Materials and Methods). This and other observations led to the conclusion that there are high-affinity interactions between the inner lamella and paranodal loops of myelin and the axonal membrane (axolemma), suggesting an intercellular neuron/oligodendrocyte functional unit, the myelin-axolemmal complex (18). Therefore, some proteins displayed on the 2D map may be components of attached axolemmal. Further examination by immunohistochemical localization is expected to resolve this issue, as we have done for NSF (Fig. 3).
Despite our success in identifying and mapping a large number (at least 130 of ≈200 spots) of the myelin proteins displayed on the 2D map, there are proteins represented on the 2D map that remain unidentified. In most cases, the lack of protein identity is due to relatively low abundances below the sensitivity of tandem mass spectrometric analysis (37). Several options are available to overcome this. First, immunological detection of proteins can be more sensitive than silver staining or mass spectrometry. Thus, additional identifications will be made by immunoblotting for proteins whose presence in myelin is predicted from other studies. In this vein, Fountoulakis and colleagues (38) performed a 2D PAGE analysis of crude human brain homogenates; many spots identified in the mouse myelin proteome are consistent (with respect to pI and molecular mass) with their findings, making crossover identifications possible. A second approach recognizes that a cornerstone of proteomic research is prior fractionation (39). The high abundances of two myelin proteins, proteolipid protein (≈50%) and isoforms of myelin basic protein (≈30%), suggest a requirement for further enrichment of the low-abundance proteins. Because both of these proteins are soluble at 4°C in Triton X-100, whereas many other myelin proteins remain insoluble (40, 41), prior Triton X-100 fractionation can potentially increase the concentration of these insoluble proteins >5-fold. Third, immunoprecipitation of protein complexes using subfractions of the myelin-axolemmal complex (18) (see above) offers additional avenues of enrichment for subsets of proteins. Fourth, despite important technical advances, there are proteins that by virtue of their size, charge, posttranslational modifications, and/or hydrophobicity are not efficiently resolved by 2D analysis (19). This includes some proteins previously reported to be in myelin. For example, the glycoproteins myelin-associated glycoprotein and myelin oligodendrocyte glycoprotein are poorly resolved but can be identified by immunoblotting as dispersed signals; the very basic myelin basic proteins are beyond the range of the pI system used but have been previously resolved, along with other basic myelin proteins, by nonequilibrium pH gradient gel electrophoresis (42) (see below). These proteins can in principle be identified by batch-wise [“shotgun” (43)] mass spectrometric analysis. Thus, the present map is expanding and will become increasingly comprehensive as these approaches are used.
Included among the proteins identified in this study are GTP-binding proteins and their modulators/effectors, several of which are involved in membrane dynamics, SNARE proteins necessary for membrane biogenesis, and cytoskeletal and scaffolding proteins necessary for oligodendrocyte process extension and local actin dynamics (44). In addition, and initially surprising to us, a number of mitochondrial proteins were identified. However, it is important to note that early electron microscopy revealed thin elongated mitochondria in the cytoplasmic veins of PNS myelin (45). Consistent with these data, we have visualized mitochondria in the cytoplasmic veins of the myelin-like membrane (46) elaborated by mature oligodendrocytes in culture, as well as CNS and PNS myelin, by immunostaining (unpublished data). Further, during myelinogenesis, oligodendrocytes elaborate extensive membrane sheets at a dramatic rate (1), placing an enormous energetic demand on the cell. Mitochondria are often anchored by the cytoskeleton at regions of high ATP consumption (47). It is plausible that at least part of this demand is supplied by local “myelin mitochondria.” That mitochondria have been implicated in several neurodegenerative diseases (48) accompanied by loss of myelin adds additional interest to these findings.
Comparative proteomics is a powerful tool for identifying changes in the expression pattern of proteins in response to changing environmental and physiological conditions. For example, changes in the protein profiles of oligodendrocytes as they enter terminal differentiation have been examined by comparative 2D gel electrophoresis. In one such study, [32P]GTP overlay identified a set of small GTP-binding proteins, many of which are also present in myelin, that are strongly up-regulated as the cells enter terminal differentiation (49). Similar studies using nonequilibrium pH gradient gel electrophoresis identified basic proteins that are either up- or down-regulated on differentiation (42). Last, a 2D PAGE study was conducted to detect the synthesis of proteins during myelination of rat optic nerve (50). In each of these examples, the identities of most proteins were not resolved. Application of the present myelin 2D proteomic map is expected to elucidate the identification of many of them.
We recently applied this proteomic paradigm to a signaling mechanism in mature oligodendrocytes with important implications for demyelinating disease. Shortly after antibody crosslinking of myelin oligodendrocyte glycoprotein, a molecule strongly implicated in multiple sclerosis, specific oligodendrocyte proteins undergo changes in their phosphorylation state followed by major changes in cell cytoarchitecture (16). Although the low quantities of proteins available from primary cultures of oligodendrocytes precluded direct mass spectrometric analysis, we were able to identify these proteins by comparing the silver-stained oligodendrocyte gels with the myelin 2D proteomic map.
Conclusion
The development of this proteomic map offers a major advance for investigations into the diverse biological roles of the myelin membrane and suggests a paradigm for similar studies in other neural systems.
Supplementary Material
Acknowledgments
We thank Drs. R. Bansal and K. Menon for advice and encouragement, and A. Taylor for technical assistance. This work was supported by National Institutes of Health Grants NS10861 (to S.E.P.), NS41078 (to S.E.P.), NS45440 (to C.M.T.), NS44916 (to M.N.R.), and HL70694 (to D.K.H.) and National Multiple Sclerosis Society Grants RG2181 (to S.E.P.) and FG1423 (to C.B.M.).
Abbreviations: IPG, immobilized pH gradient; PNS, peripheral nervous system; NSF, N-ethylmaleimide-sensitive fusion protein; CGT, ceramide galactosyl transferase.
References
- 1.Pfeiffer, S. E., Warrington, A. E. & Bansal, R. (1993) Trends Cell Biol. 3, 191-197. [DOI] [PubMed] [Google Scholar]
- 2.Keegan, B. M. & Noseworthy, J. H. (2002) Annu. Rev. Med. 53, 285-302. [DOI] [PubMed] [Google Scholar]
- 3.Koeppen, A. H. & Robitaille, Y. (2002) J. Neuropathol. Exp. Neurol. 61, 747-759. [DOI] [PubMed] [Google Scholar]
- 4.Zhou, L. & Griffin, J. W. (2003) Curr. Opin. Neurol. 16, 307-313. [DOI] [PubMed] [Google Scholar]
- 5.Morell, P. & Norton, W. T. (1980) Sci. Am. 242 (5), 88-90, 92, 96. [DOI] [PubMed] [Google Scholar]
- 6.Sanchez, I., Hassinger, L., Paskevich, P. A., Shine, H. D. & Nixon, R. A. (1996) J. Neurosci. 16, 5095-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin, X., Crawford, T. O., Griffin, J. W., Tu, P., Lee, V. M., Li, C., Roder, J. & Trapp, B. D. (1998) J. Neurosci. 18, 1953-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ching, W., Zanazzi, G., Levinson, S. R. & Salzer, J. L. (1999) J. Neurocytol. 28, 295-301. [DOI] [PubMed] [Google Scholar]
- 9.Boiko, T., Rasband, M. N., Levinson, S. R., Caldwell, J. H., Mandel, G., Trimmer, J. S. & Matthews, G. (2001) Neuron 30, 91-104. [DOI] [PubMed] [Google Scholar]
- 10.Rasband, M. N. & Trimmer, J. S. (2001) Dev. Biol. 236, 5-16. [DOI] [PubMed] [Google Scholar]
- 11.Peles, E. & Salzer, J. L. (2000) Curr. Opin. Neurobiol. 10, 558-565. [DOI] [PubMed] [Google Scholar]
- 12.Goto, K., Kurihara, T., Takahashi, Y. & Kondo, H. (1990) Neurosci. Lett. 117, 269-274. [DOI] [PubMed] [Google Scholar]
- 13.Barres, B. A. & Raff, M. C. (1999) J. Cell Biol. 147, 1123-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ledeen, R. W., Golly, F. & Haley, J. E. (1992) Mol. Neurobiol. 6, 179-190. [DOI] [PubMed] [Google Scholar]
- 15.Chakraborty, G., Mekala, P., Yahya, D., Wu, G. & Ledeen, R. W. (2001) J. Neurochem. 78, 736-745. [DOI] [PubMed] [Google Scholar]
- 16.Marta, C. B., Taylor, C. M., Coetzee, T., Kim, T., Winkler, S., Bansal, R. & Pfeiffer, S. E. (2003) J. Neurosci. 23, 5461-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bansal, R. & Pfeiffer, S. E. (1985) J. Neurosci. Res. 14, 21-34. [DOI] [PubMed] [Google Scholar]
- 18.Menon, K., Rasband, M. N., Taylor, C. M., Brophy, P., Bansal, R. & Pfeiffer, S. E. (2003) J. Neurochem. 87, 995-1009. [DOI] [PubMed] [Google Scholar]
- 19.Taylor, C. M. & Pfeiffer, S. E. (2003) Proteomics 3, 1303-1312. [DOI] [PubMed] [Google Scholar]
- 20.Eng, J. K., McCormack, A. L. & Yates, J. R., III (1994) J. Am. Soc. Mass Spectrom. 5, 976-989. [DOI] [PubMed] [Google Scholar]
- 21.Han, D. K., Eng, J., Zhou, H. & Aebersold, R. (2001) Nat. Biotechnol. 19, 946-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keller, A., Nesvizhskii, A. I., Kolker, E. & Aebersold, R. (2002) Anal. Chem. 74, 5383-5392. [DOI] [PubMed] [Google Scholar]
- 23.Kim, T., Fiedler, K., Madison, D. L., Krueger, W. H. & Pfeiffer, S. E. (1995) J. Neurosci. Res. 42, 413-422. [DOI] [PubMed] [Google Scholar]
- 24.Resing, K. A. (2002) Ann. N.Y. Acad. Sci. 971, 608-614. [DOI] [PubMed] [Google Scholar]
- 25.Rios, J. C., Melendez-Vasquez, C. V., Einheber, S., Lustig, M., Grumet, M., Hemperly, J., Peles, E. & Salzer, J. L. (2000) J. Neurosci. 20, 8354-8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arroyo, E. J. & Scherer, S. S. (2000) Histochem. Cell Biol. 113, 1-18. [DOI] [PubMed] [Google Scholar]
- 27.Melendez-Vasquez, C. V., Rios, J. C., Zanazzi, G., Lambert, S., Bretscher, A. & Salzer, J. L. (2001) Proc. Natl. Acad. Sci. USA 98, 1235-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gatto, C. L., Walker, B. J. & Lambert, S. (2003) J. Cell Biol. 162, 489-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madison, D. L. & Pfeiffer, S. E. (1997) in Cell Biology and Pathology of Myelin: Evolving Biological Concepts and Therapeutic Approaches, eds. Juurlink, B. H. J., Devon, R. M., Doucette, J. R., Schreyer, D. J. & Verge, V. M. L. (Plenum, New York), pp. 145-155.
- 30.Trapp, B. D., Pfeiffer, S. E., Anitei, M. & Kidd, G. J. (2004) in Myelin Biology and Disorders, ed. Lazzarini, R. A. (Academic, New York), Vol. 1, pp. 29-55. [Google Scholar]
- 31.Lin, R. C. & Scheller, R. H. (2000) Annu. Rev. Cell Dev. Biol. 16, 19-49. [DOI] [PubMed] [Google Scholar]
- 32.Dupree, J. L., Coetzee, T., Suzuki, K. & Popko, B. (1998) J. Neurocytol. 27, 649-659. [DOI] [PubMed] [Google Scholar]
- 33.Coetzee, T., Fujita, N., Dupree, J., Shi, R., Blight, A., Suzuki, K. & Popko, B. (1996) Cell 86, 209-219. [DOI] [PubMed] [Google Scholar]
- 34.Roymans, D., Willems, R., Van Blockstaele, D. R. & Slegers, H. (2002) Clin. Exp. Metastasis 19, 465-476. [DOI] [PubMed] [Google Scholar]
- 35.Cuello, F., Schulze, R. A., Heemeyer, F., Meyer, H. E., Lutz, S., Jakobs, K. H., Niroomand, F. & Wieland, T. (2003) J. Biol. Chem. 278, 7220-7226. [DOI] [PubMed] [Google Scholar]
- 36.Kim, S. H., Fountoulakis, M., Cairns, N. J. & Lubec, G. (2002) Biochem. Biophys. Res. Commun. 296, 970-975. [DOI] [PubMed] [Google Scholar]
- 37.Aebersold, R. & Mann, M. (2003) Nature 422, 198-207. [DOI] [PubMed] [Google Scholar]
- 38.Langen, H., Berndt, P., Roder, D., Cairns, N., Lubec, G. & Fountoulakis, M. (1999) Electrophoresis 20, 907-916. [DOI] [PubMed] [Google Scholar]
- 39.Taylor, R. S., Wu, C. C., Hays, L. G., Eng, J. K., Yates, J. R., III, & Howell, K. E. (2000) Electrophoresis 21, 3441-3459. [DOI] [PubMed] [Google Scholar]
- 40.Taylor, C. M., Coetzee, T. & Pfeiffer, S. E. (2002) J. Neurochem. 81, 993-1004. [DOI] [PubMed] [Google Scholar]
- 41.Kim, T. & Pfeiffer, S. E. (1999) J. Neurocytol. 28, 281-293. [DOI] [PubMed] [Google Scholar]
- 42.Yamaguchi, Y. & Pfeiffer, S. E. (1999) J. Neurosci. Res. 56, 199-205. [DOI] [PubMed] [Google Scholar]
- 43.McDonald, W. H. & Yates, J. R., III (2003) Curr. Opin. Mol. Ther. 5, 302-309. [PubMed] [Google Scholar]
- 44.Richter-Landsberg, C. (2001) Microsc. Res. Tech. 52, 628-636. [DOI] [PubMed] [Google Scholar]
- 45.Mugnaini, E., Osen, K. K., Schnapp, B. & Friedrich, V. L., Jr. (1977) J. Neurocytol. 6, 647-668. [DOI] [PubMed] [Google Scholar]
- 46.Bhat, S. & Pfeiffer, S. E. (1985) J. Neurochem. 45, 1356-1362. [DOI] [PubMed] [Google Scholar]
- 47.Wagner, O. I., Lifshitz, J., Janmey, P. A., Linden, M., McIntosh, T. K. & Leterrier, J. F. (2003) J. Neurosci. 23, 9046-9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jellinger, K. A. (2003) J. Neural Transm. Suppl. 101-144. [DOI] [PubMed]
- 49.Huber, L. A., Madison, D. L., Simons, K. & Pfeiffer, S. E. (1994) FEBS Lett. 347, 273-278. [DOI] [PubMed] [Google Scholar]
- 50.Colello, R. J., Fuss, B., Fox, M. A. & Alberti, J. (2002) Electrophoresis 23, 144-151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.