Abstract
Signal transducer and activator of transcription (STAT)3 is widely expressed in the CNS during development and adulthood. STAT3 has been implicated in the control of neuron/glial differentiation and leptin-mediated energy homeostasis, but the physiological role and degree of involvement of STAT3 in these processes is not defined and controversial because of the lack of a direct genetic model. To address this, we created mice with a neural-specific disruption of STAT3 (STAT3N-/-). Surprisingly, homozygous mutants were born at the expected Mendelian ratio without apparent developmental abnormalities but susceptible to neonatal lethality. Mutants that survived the neonatal period were hyperphagic, obese, diabetic, and infertile. Administering a melanocortin-3/4 receptor agonist abrogated the hyperphagia and hypothalamic immunohistochemistry showed a marked reduction in proopiomelanocortin with an increase in neuropeptide Y and agouti-related protein. Mutants had reduced energy expenditure and became hypothermic after fasting or cold stress. STAT3N-/- mice are hyperleptinemic, suggesting a leptin-resistant condition. Concomitant with neuroendocrine defects such as decreased linear growth and infertility with accompanying increased corticosterone levels, this CNS knockout recapitulates the unique phenotype of db/db and ob/ob obese models and distinguishes them from other genetic models of obesity. Thus, STAT3 in the CNS plays essential roles in the regulation of energy homeostasis and reproduction.
Signal transducer and activator of transcription (STAT) proteins are a group of cytokine-activated signaling molecules that can directly bind to DNA and activate or repress transcription of target genes. A myriad of cytokines activate STAT proteins through receptor-associated kinases. Activation of STATs occurs through tyrosine phosphorylation that is required for SH2 domain-mediated dimerization and DNA binding. Most STAT proteins have specific effects, but STAT3, the most ancient STAT, is broadly expressed and activated by a diverse array of cytokines and stresses (1-3). It was initially described as an acute phase protein involved in various biological and pathological processes. STAT3 deletion causes embryonic lethality before gastrulation via an unknown mechanism (4). STAT3 has unusually pleiotropic effects regulating murine embryonic stem cell maintenance (5), macrophage function (6, 7), immune regulation (8), and peripheral neuron survival after axotomy (9, 10) among others.
In the CNS, STAT3 is expressed during embryonic development, mostly at ventricular areas, where neuronal proliferation and differentiation take place. Consistently, STAT3 is strongly suggested by in vitro studies to play an instructive role in glial and neuron differentiation (11-14). In adults, STAT3 has long been implicated in the regulation of energy homeostasis through the fat-derived cytokine leptin. Although leptin can activate several STAT proteins, including STAT3, 5, and 6, in vitro (15), only STAT3 is activated in the hypothalamus in vivo upon leptin administration (16, 17). These data strongly suggest that STAT3 plays a role in leptin mediated energy balance.
Leptin regulates energy homeostasis and physiology by affecting diverse functions such as food intake, energy expenditure, and reproduction (18) through distinct proopiomelanocortin (POMC)- and neuropeptide Y (NPY)-related neuronal circuitry in hypothalamic nuclei (19, 20). Activation of STAT3 in response to leptin within the hypothalamus can be detected in both networks that mediate anorexogenic (POMC and CART) and orexogenic [NYP and agouti-related protein (AgRP)] physiology. Leptin signaling down-regulates NPY/AgRP and up-regulates POMC/CART neurons, thus limiting food intake and signaling peripheral energy stores. The biological evidence supporting a major role for STAT3 in energy homeostasis comes from the weight reducing effects of other STAT3 activating cytokines such as IL-6 (21) and ciliary neurotrophic factor (22), and leptin receptor point mutations (23), However, no direct test of STAT3 function in energy homeostasis has been conducted.
Multiple signaling pathways are activated by ligand activation of the leptin receptor including mitogen-activated protein kinase (24) and phosphatidylinositol 3-kinase (25, 26), which has been shown to control feeding. In addition, immediate electrical changes can be detected in hypothalamic neurons after leptin administration in a manner not amendable to transcriptional modification (27, 28). Other signals, such as insulin (29, 30) and long chain fatty acid (31, 32), can also affect energy metabolism through hypothalamic neuronal circuitry. This raises questions on the role and degree of involvement of STAT3 in leptin function. To address (i) the participation of STAT3 in the CNS and (ii) the degree to which STAT3 is involved in energy homeostasis, we generated a neural-specific deletion of STAT3 by using gene targeting to create a conditional STAT3 allele. We found that STAT3 can account for all of the essential effects of leptin action on energy balance and neuroendocrinology because neural STAT3 mutant mice recapitulate leptin receptor (db/db) and ligand deficiency (ob/ob).
Methods
Animals. Generation of mice with a conditional knockout (KO) of STAT3 has been described (7). Exons 18-20, which contain the SH2 domain of STAT3, were flanked by two lox P sites. Nestin-Cre transgenic mice [The Jackson Laboratory mice database: B6.Cg(SJL)-TgN(Nes-cre)1Kln] expressing Cre under the control of a rat nestin promoter/enhancer were described (34). The Nestin-Cre;STAT3f/f mutant mice, designated as STAT3N-/-, were generated through Nestin-Cre;STAT3f/+ mice crossed to STAT3f/f mice. The genotype was determined by PCR as described (7). All experiments were done in a C57BL/6 background. All procedures were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and under the approval of the Yale Medical School Animal Care and Use Committee.
Immunoblotting. Immunoblotting was done as described with an anti-STAT3 or anti-STAT1 Ab (Santa Cruz Biotechnology) (33). Next, a horseradish peroxidase-conjugated secondary anti-rabbit Ab was used and visualized by using SuperSignal chemiilluminescent substrate (Pierce).
Histological Analysis. Tissue was collected and fixed in 4% paraformaldehyde for 4 h, then snap frozen in OCT, and sectioned or paraffin embedded. Frozen sections (10 μm) were cut, washed in PBS, and stained with hematoxylin and eosin by using standard techniques. Immunohistochemistry was performed by using anti-β-endorphin for POMC neurons (Chemicon) 1:5,000, anti-AgRP (Calbiochem) 1:5,000, and anti-NPY (Peninsula Laboratories) 1:10,000 by using standard techniques.
Plasma Parameters. Blood samples were taken from cut tailtips of conscious mice either in the fasting (≈14 h) or fed state at 1000 hours. Plasma was collected for measurement of glucose (glucose oxidase method; Analyzer 2, Beckman Coulter), insulin (double Ab RIA, Linco Research Immunoassay, St. Charles, MO), leptin (RIA, Linco Research Immunoassay), corticosterone (double Ab RIA, ICN), triglycerides (enzymatic method, Sigma), and nonesterified fatty acids (enzymatic method, NEFA-C, Wako Pure Chemicals, Osaka).
Glucose Tolerance Test. The i.p. glucose tolerance tests were performed. After 14 h of fasting, mice received a single i.p. injection of 20% glucose solution (1 g/kg), time 0. Plasma samples for glucose and insulin measurement were taken at -30, 15, 30, 60, and 120 min from cut tailtips.
Results
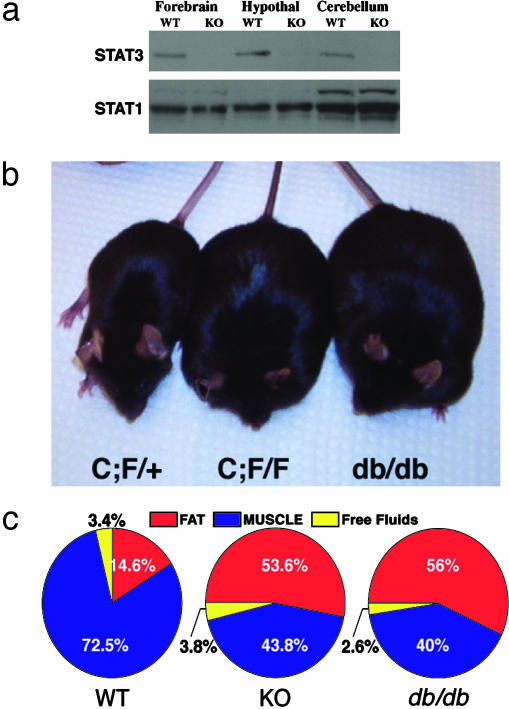
Neural-Specific STAT3 Deletion Results in Severe Obesity. To study the physiological role of STAT3 in the brain, we generated a neural-specific deletion of STAT3 by using gene targeting to create a conditional STAT3 allele. We initially generated a floxed STAT3 allele (7) that was mated to a rat nestin-Cre transgenic mouse (34) to generate a neural-specific STAT3 deletion. The rat nestin-Cre transgene expresses cre-recombinase in neuroepithelium as early as embryonic day 10.5 (29, 35) and yields a broad neural deletion of STAT3 in the brain as measured by immunoblotting (Fig. 1a), but not in other organs, such as liver and pancreas (data not shown). Embryonic day 14.5 fibroblast growth factor 2-responsive neurospheres derived from STAT3N-/- forebrain exhibited no detectible STAT3, supporting a complete deletion of STAT3 in early neuronal precursors (data not shown). To our surprise, STAT3N-/- mice were born alive at the expected Mendelian ratio with no apparent developmental abnormalities although STAT3 has been implicated in the control of gliogenesis (11, 12). They appeared to locomote and suckle normally but were susceptible to neonatal lethality. Upon necropsy, these pups were found to contain dramatically reduced milk in their stomachs. Reducing pup numbers to lower the competition for milk and to minimize neonatal stress was able to rescue a substantial proportion of the mutants.
Fig. 1.
Neural deletion of STAT3 results in severe obesity. (a) Western blot analysis of STAT3 in forebrain, hypothalamus, and cerebellum in WT and KO animals. The blot was stripped and reprobed with a STAT1 Ab as a control. (b) Heterozygous and obese homozygous Nestin-Cre; STAT3f/f (STAT3N-/-) animals compared to a db/db mutant. (c) NMR imaging by using a minispec MQ10 analyzer (Bruker, Woodlands, TX) of WT, KO, and db/db animals (n = 4) showing total fat and lean mass as well as free fluid. Data are given as percentage of total body weight.
The STAT3N-/- animals that survived the neonatal period started to develop an obese phenotype ≈6-8 weeks of age (Fig. 1b) and weighed twice as much as their littermates at adulthood (Fig. 2a). The accumulated mass was nearly exclusively adipose tissue revealed by a quantitative analysis of body composition done on live adults by using NMR imaging (minispec MQ10 analyzer). Total fat content of STAT3N-/- mice increased 5-fold compared to littermate controls to comprise >50% of their body weight, which is equal to the body composition of db/db mutants (Fig. 1c). In contrast, lean mass expressed as absolute weight was unchanged.
Fig. 2.
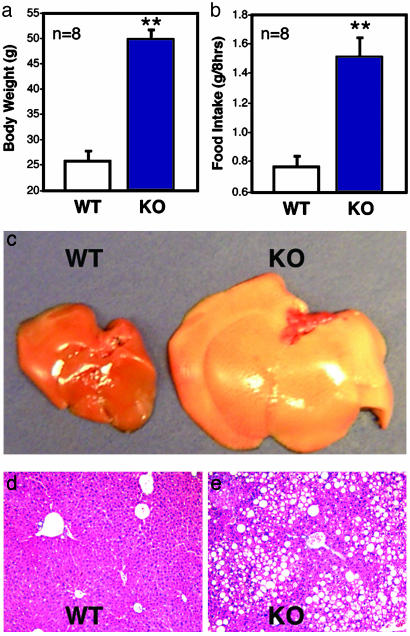
Neural STAT3 deletion results in hyperphagia and fatty livers. STAT3N-/- animals are twice as heavy as WT animals (a) and consume twice as much food (b). Livers of KO animals are severely enlarged and fatty (c) and accumulated a large amount of fat (fatty liver) (d and e). (**, P < 0.001.)
The livers of STAT3N-/- mice were severely enlarged with greatly increased fat deposits (Fig. 2 c-e) despite no local genetic defect of STAT3. In contrast, a hepatocyte-specific STAT3 KO under the same diet did not result in increased fat deposits in the liver (data not shown) consistent with selective KO of the leptin receptor (36). The restricted phenotypes resemble the dysfunction of hypothalamic leptin signaling, demonstrating STAT3 as a central and not peripheral mediator of energy homeostasis. No anatomical defects were detected in hypothalamic nuclei by histochemical analysis of serial brain sections in STAT3N-/- mice (data not shown).
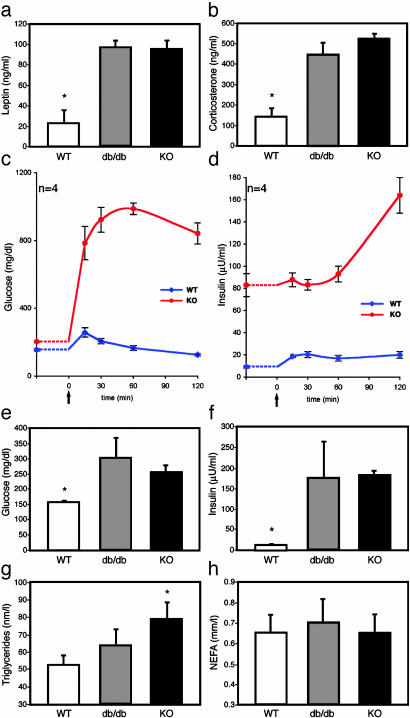
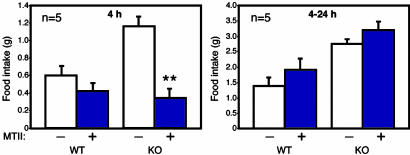
CNS STAT3 Is Necessary for Satiety Signals. As with leptin receptor-deficient (db/db) mice, STAT3N-/- mice are hyperphagic (Fig. 2b) in a hyperleptinemic background (see Fig. 4a) reflecting the increased fat mass and block of leptin signaling. The hyperphagia is a consequence of an imbalance in anorexogenic and orexogenic neuropeptides due to the disruption of leptin signaling. Physiologically, leptin signaling up-regulates anorexogenic neuropeptides, including α-melanocyte-stimulating hormone, a cleavage product of POMC, which functions to inhibit food intake mainly through the G protein-coupled melanocortin-4 receptor (MC4R) (37). POMC was suggested to be a direct STAT3 downstream target gene (38). In both WT and leptin-signaling mutants, one i.p. injection of the MC3/4R agonist, MTII (10 mg/kg), lowers food intake over a 4-h period (25, 39) and long-term application of MTII reverses obesity in ob/ob and db/db mice. MTII administration to STAT3N-/- mice completely reversed their hyperphagia over a 4-h period (Fig. 3). The food intake reduced to levels equal that of treated WT controls. This result suggests that the STAT3N-/- hyperphagia resulted from the down-regulation of anorexogenic peptides, consistent with studies of leptin-deficient mutants (40) as well as MC4R KO (39) and POMC KO (41). Moreover, plasma corticosterone levels were significantly elevated in STAT3N-/- mice (Fig. 4b), which is unique to ob/ob and db/db mutants but not to the other obese models (42). This further suggests a close genetic link between STAT3 and leptin signaling.
Fig. 4.
STAT3N-/- animals are leptin-resistant and have increased corticosterone and glucose dysregulation. Leptin was increased significantly in STAT3N-/- and db/db animals (a), and corticosterone concentration was significantly increased as well (b). (c and d) Glucose tolerance tests were performed by measuring baseline glucose and insulin. Then an i.p. bolus of glucose (20%, 1 g/kg) was administered (arrow), and plasma was collected at the times indicated. (e and f) Fasting glucose and insulin were increased in STAT3N-/- and db/db animals. (g) Plasma triglycerides. (h) Nonesterified fatty acids. (*, P < 0.005.)
Fig. 3.
Behavioral defects in STAT3N-/- animals are linked to a deficiency in the melanocortin system. Over a 4-h period, one i.p. administration of MTII (10 mg/kg) inhibits food intake in KO animals (n = 5) significantly over saline-injected controls. After 4 h, food intake returns to normal. (**, P < 0.001.)
STAT3N-/- Animals Are Diabetic. Glucose dysregulation and development of type 2 diabetes were evident in STAT3N-/- mice. Plasma fasting glucose concentrations in STAT3N-/- and db/db mice were equivalent and increased compared to littermates (Fig. 4e). The hyperglycemia was accompanied by severe hyper-insulinemia (Fig. 4f) in the STAT3N-/- animals. Plasma insulin concentrations were as much as 14- and 10-fold higher in the fed and fasted state, respectively, suggesting severe insulin resistance. To characterize the dynamics of glucose metabolism with respect to insulin, we carried out a glucose tolerance test on fasted (14 h) animals. Basal glucose and insulin levels were measured before i.p. administration of a 20% glucose solution (1 g/kg). In STAT3N-/- mice, plasma glucose and insulin concentrations as measured by area under the curve (mg/dl/120 min) increased 5-fold after glucose challenge (glucose, P < 0.0002; insulin, P < 0.003) with a prolonged peak time. Basal insulin concentrations, which were 9- to 10-fold higher in mutants were not significantly induced by a dramatic increase of plasma glucose levels for the first hour but doubled during the second hour. In contrast, WT controls exhibited an immediate insulin induction (15 min, Fig. 4d) and a quick reduction of plasma glucose concentration. These observations revealed a severe alteration of the glucose/insulin dynamics in response to glucose challenge in STAT3N-/- animals. Plasma triglyceride but not nonesterified fatty acids were changed between groups probably indicating a peripheral effect of leptin (Fig. 4 e and f).
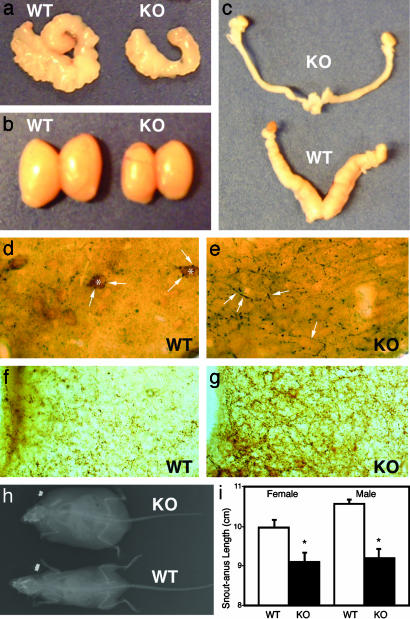
STAT3N-/- Animals Are Infertile with Reduced Linear Growth. Like db/db mutants, STAT3N-/- animals are infertile with accompanying hypogonadism in both male and female animals (Fig. 5 a-c). Heterozygous animals are fertile and both sexes were used for breeding resulting in normal litters. Fifteen STAT3N-/- animals (seven males and eight females) were housed with WT animals of the opposite sex with known fertility for 6-8 mo. No pregnancy was detected from any male or female STAT3N-/- mouse. Male STAT3N-/- animals showed a marked reduction in both testicular (Fig. 5b) and seminal vesicle size (Fig. 5a), whereas female STAT3N-/- mice showed a reduction in the size of the uterine horns (Fig. 5c). Normal gonadotropin-independent follicular development was observed in STAT3N-/- females, but corpa lutea were never observed indicating ovulation did not occur. This is consistent with the hypogonadotropic hypogonadism seen in leptin-deficient mice (43).
Fig. 5.
STAT3N-/- animals are infertile, hypogonadal, and short. Seminal vesicles (a) as well as testes (b) are smaller in STAT3N-/- males. (c) Uterine horns of female KO mice are markedly decreased in size as well. (d and e) Immunocytochemistry of the lateral arcuate nucleus in WT and KO with anti-β-endorphin for POMC neurons diaminobenzidine (brown) and anti-NPY nickel diaminobenzidine (dark) (magnification, ×40). POMC staining (*) was not seen in KO animals. NPY neurons (arrows) were seen in direct contact with POMC neurons and overexpressed in KO animals. (f and g) KO animals showed overexpression of AgRP in the medial arcuate nucleus. WT and KO medial arcuate nuclei were stained by using anti-AgRP (Calbiochem) at 1:5,000. (h)An x-ray image of WT and KO animals showing decreased linear size and increased body mass. (i) Snout-anus measurements.
Moreover, the linear growth of STAT3N-/- mice as measured by snout-anus length was reduced (Fig. 5 h and i). Mutants are ≈8% shorter than their littermate controls, a level of reduction that is comparable with the linear growth reduction in db/db mice (≈5%). STAT3N-/- mice showed a marked reduction in POMC and an increase in NPY (Fig. 5 d and e) and AgRP (Fig. 5 f and g) by immunohistochemistry. Taken together, the STAT3N-/- model strongly argues a major role for STAT3-dependent leptin signaling in hypothalamic neuroendocrine function in reproduction and linear growth.
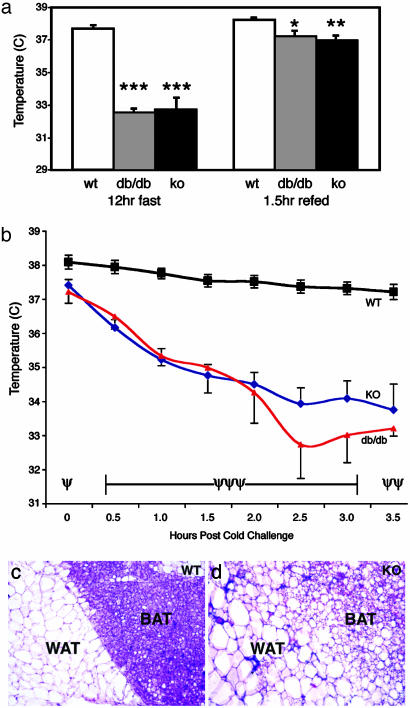
STAT3N-/- Animals Have Reduced Energy Expenditure. In leptin signaling mutant animals, both hyperphagia and lower energy expenditure contribute to their obese phenotype. To address this, we next asked whether STAT3N-/- mice had disrupted energy expenditure as measured by body temperature. In the fed state, STAT3N-/- animals showed a slightly but significantly lower body temperature (≈0.5°C) when compared with littermate controls suggesting a lower level of basal metabolism. Nevertheless, these animals were able to stabilize their body temperatures at a normal range by consuming twice as much food. After a 12-h fast, while littermate control mice maintained a normal body temperature, STAT3N-/- and db/db animals became significantly hypothermic as their body temperature dropped >4°C below normal (Fig. 6a), an effect that was reversed after 1.5 h of refeeding. Because body energy supply as measured by plasma glucose and fatty acid levels are not reduced during fast, these observations suggest that a constant feeding stimulation is essential for maintaining body temperature in the mutants.
Fig. 6.
Thermogensis and brown adipose tissue are altered in STAT3N-/- animals. (a) WT, db/db, and STAT3N-/- animals were fasted overnight, and the body temperatures were taken rectally. A 4°C drop in body temperature was seen after the fast in both db/db and KO animals, but body temperature was restored and maintained at 1.5 h of ad libitum refeeding. Note that, even at refeeding and fed status, temperature is significantly lower in both mutants (WT and KO, n = 8; db/db, n = 4). (b) Animals were subjected to a 4°C cold stress for 3.5 h. Body temperature dropped to ≈3°C below baseline temperature whereas WT animals maintained their body temperature. One db/db mutant died at the 3-h time point (WT and KO, n = 6; db/db, n = 4). (c and d) Hematoxylin and eosin staining of 10-μm frozen sections of brown and white adipose tissue. KO animals have larger white adipose cells and brown adipose tissue with more diffuse morphology resembling white adipose tissue in structure. (***, P < 0.00001; **, P < 0.005; *, P < 0.012) (ψψψ, P < 0.00001; ψψ, P < 0.0003; ψ, P < 0.02.)
The induction of body thermogenesis is controlled by the sympathetic nervous system in response to food or cold in brown adipose tissue and white adipose tissue (44). In leptin mutants, such as db/db mice, sympathetic activity is reduced. To examine whether neuronal STAT3 disruption causes similar defects, we measured cold-induced thermogenesis. Animals were housed at 4°C for 3.5 h and body temperature was measured at 30-min intervals. Although control littermates were able to maintain their body temperature throughout the cold challenge, STAT3N-/- and db/db mice dropped ≈1°C in body temperature every 30 min until they reached 34°C and were able to stabilize this temperature throughout the rest of the experiment (Fig. 6b). These data are consistent with the decreased sympathetic activity seen in leptin signaling mutants but are different from the result obtained from the “β-less” triple β-adrenergic receptor KO mice, which fully disrupted β-adrenergic receptor function and animals fail to have any thermogenic response to cold challenge (45). Histological examination of STAT3N-/- animals revealed that the brown adipose tissue became diffuse and resembled white adipose tissue in structure (Fig. 6c). The delineation between white and brown adipose tissue was less evident. This morphological change is less severe than that of β-less mice but is consistent with the impaired levels of thermogenesis in the β-less and STAT3N-/- animals.
Discussion
The acute phase response signaling/transcription factor STAT3 is broadly expressed in the CNS and is activated by a number of extracellular stimuli. We have shown that, during normal embryonic development and adulthood, the major function of neuronal STAT3 is for homeostatic regulation. There is a broad data set describing the effect of STAT3 on glial differentiation late in embryonic development (11-13). Our early deletion of STAT3 by using the nestin promoter surprisingly did not result in an absence of GFAP+ astrocytes as expected (Q.G. and M.J.W., unpublished data). Therefore, in vivo, STAT3 may have a limited function during neural differentiation. The phenotype of the STAT3N-/- mouse closely resembles that of leptin-deficient mice. The restricted phenotype was surprising given the number of STAT3-activating cytokines produced in the brain. Although other STAT3-activating cytokines have weight reducing characteristics under artificial or pathological conditions (21, 22), the only candidate cytokine that can signal body energy stores efficiently in normal physiology is the fat-derived cytokine leptin. STAT3N-/- animals are hyperleptinemic but obese, and therefore leptin does not act to lower body weight in these animals.
The fact that the STAT3 deletion, which disrupts one distinct signaling output from the leptin receptor, can so closely resemble leptin deficiency is surprising and intriguing because recent studies have shown that in the hypothalamus, leptin activates multiple signaling events (24, 25). Specific inhibitors of some of these pathways can cause inhibitory actions to the effects of leptin. In addition, immediate electrical changes (27, 28) were also observed in hypothalamic neurons after leptin administration, which presumably is not the direct function of STAT3. Even recently, the role of STAT3 in hypothalamic signaling has come into question. Indeed, the conclusion from this study is contradictory to a recent report that concluded that STAT3 is not involved in neuroendocrinology including reproduction, and linear growth, and only moderately involved in glucose regulation (23). These conclusions were based on a receptor knock-in that mutated the putative STAT3 docking site (Y to S change at position 1,138 on the leptin receptor, LRbs1138). We have found that the reproductive and growth characteristics of leptin receptor mutant mice are controlled by STAT3. The inconsistency between STAT3N-/- mice and LRbs1138 mice is probably due to the nature of the mutations, with STAT3N-/- mice ablating STAT3 directly and LRbs1138 having only diminished STAT3 activation by leptin (24). This is supported by in vitro data (24) and by other genetic models of leptin signaling such as lipodystrophic mice that have very low leptin levels and show the metabolic characteristics of leptin deficiency (46). However, these animals are fertile. STAT3N-/- animals also show the same neuropeptide levels as db/db animals indicating they are infertile via the same mechanism, lending further support for our model.
The infertility, decreased linear growth and high plasma corticosterone levels show that STAT3N-/- mice recapitulate the unique phenotypes of db/db mutants, which delineate STAT3N-/- and db/db mutants from other obese genetic models that often exhibit only a subset of the db/db phenotype. The fact that STAT3N-/- mutants exhibit all four characteristic defects (obesity, decreased linear growth, infertility, and high corticosterone) of db/db mutants provide convincing evidence that STAT3 mediates most, if not all, hypothalamic leptin function. This notion is further supported by comparative studies of energy expenditure between STAT3N-/- and db/db mutants measured by body temperature in response to fasting or cold stress. Therefore, the overlapping phenotypes between db/db mutants and STAT3N-/- mice unlikely resulted from a leptin-independent STAT3 function.
Leptin is not the only hormone that controls body weight and diabetes. There are multiple peripheral signals that can modify body weight independent of leptin. For example, insulin (29, 30) and long chain fatty acids (31, 32) regulate energy homeostasis within the hypothalamus. Insulin can act through insulin receptor substrates (47, 48) and downstream pathways such as Akt/protein kinase B (49) to modulate energy homeostasis. How are these divergent pathways orchestrated? Are they organized in parallel or form convergent pathways to regulate energy balance? Because disruption of STAT3 causes all of the major phenotypes of leptin signaling, it is possible that different pathways activated by leptin or independent of leptin are convergent and function through STAT3. For example, both insulin and leptin activate phosphatidylinositol 3-kinase pathway in the hypothalamus. At least in some systems, the activation of this pathway requires STAT3 (50). It is also possible that leptin-STAT3 plays a dominant role in energy balance, whereas other pathways play modifying roles, which may be essential for age related and/or diet induced obesity and diabetes. Leptin is functional in humans, and leptin resistance in humans is rarely caused by direct defects on leptin or its receptor (51, 52). Therefore, the integration of these divergent pathways in energy homeostasis may be essential to garner a clear understanding, and better direct clinically relevant therapies. Further study of this model will address the interaction between Janus kinase/STAT signaling and other signaling pathways and elucidate the transcriptional control of neuropeptides and other down-stream target genes as well as the action of leptin within the CNS.
Acknowledgments
We thank Y. Xi for help with x-ray images. X.-Y.F. was a recipient of a Career Development Award from the National Institutes of Health. G.I.S. is an Investigator of Howard Hughes Medical Institute. This work was supported by National Institutes of Health Grants AI34522 and EY13607 (to X.-Y.F.), RO1DK-40936 and P30DK-45735 (to G.I.S.), and RR-014451 and DK-060711 (to T.L.H.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: STAT, signal transducer and activator of transcription; POMC, proopiomelanocortin; KO, knockout; NPY, neuropeptide Y; AgRP, agouti-related protein.
References
- 1.Akira, S., Nishio, Y., Inoue, M., Wang, X. J., Wei, S., Matsusaka, T., Yoshida, K., Sudo, T., Naruto, M. & Kishimoto, T. (1994) Cell 77, 63-71. [DOI] [PubMed] [Google Scholar]
- 2.Zhong, Z., Wen, Z. & Darnell, J. E., Jr. (1994) Science 264, 95-98. [DOI] [PubMed] [Google Scholar]
- 3.Wegenka, U. M., Lutticken, C., Buschmann, J., Yuan, J., Lottspeich, F., Muller-Esterl, W., Schindler, C., Roeb, E., Heinrich, P. C. & Horn, F. (1994) Mol. Cell. Biol. 14, 3186-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeda, K., Noguchi, K., Shi, W., Tanaka, T., Matsumoto, M., Yoshida, N., Kishimoto, T. & Akira, S. (1997) Proc. Natl. Acad. Sci. USA 94, 3801-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niwa, H., Burdon, T., Chambers, I. & Smith, A. (1998) Genes Dev. 12, 2048-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeda, K., Clausen, B. E., Kaisho, T., Tsujimura, T., Terada, N., Forster, I. & Akira, S. (1999) Immunity 10, 39-49. [DOI] [PubMed] [Google Scholar]
- 7.Welte, T., Zhang, S. S., Wang, T., Zhang, Z., Hesslein, D. G., Yin, Z., Kano, A., Iwamoto, Y., Li, E., Craft, J. E., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 1879-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoo, J. Y., Huso, D. L., Nathans, D. & Desiderio, S. (2002) Cell 108, 331-344. [DOI] [PubMed] [Google Scholar]
- 9.Alonzi, T., Middleton, G., Wyatt, S., Buchman, V., Betz, U. A., Muller, W., Musiani, P., Poli, V. & Davies, A. M. (2001) Mol. Cell. Neurosci. 18, 270-282. [DOI] [PubMed] [Google Scholar]
- 10.Schweizer, U., Gunnersen, J., Karch, C., Wiese, S., Holtmann, B., Takeda, K., Akira, S. & Sendtner, M. (2002) J. Cell Biol. 156, 287-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonni, A., Sun, Y., Nadal-Vicens, M., Bhatt, A., Frank, D. A., Rozovsky, I., Stahl, N., Yancopoulos, G. D. & Greenberg, M. E. (1997) Science 278, 477-483. [DOI] [PubMed] [Google Scholar]
- 12.Sun, Y., Nadal-Vicens, M., Misono, S., Lin, M. Z., Zubiaga, A., Hua, X., Fan, G. & Greenberg, M. E. (2001) Cell 104, 365-376. [DOI] [PubMed] [Google Scholar]
- 13.Takizawa, T., Nakashima, K., Namihira, M., Ochiai, W., Uemura, A., Yanagisawa, M., Fujita, N., Nakao, M. & Taga, T. (2001) Dev. Cell 1, 749-758. [DOI] [PubMed] [Google Scholar]
- 14.Moon, C., Yoo, J. Y., Matarazzo, V., Sung, Y. K., Kim, E. J. & Ronnett, G. V. (2002) Proc. Natl. Acad. Sci. USA 99, 9015-9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghilardi, N., Ziegler, S., Wiestner, A., Stoffel, R., Heim, M. H. & Skoda, R. C. (1996) Proc. Natl. Acad. Sci. USA 93, 6231-6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darnell, J. E., Jr. (1996) Proc. Natl. Acad. Sci. USA 93, 6221-6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaisse, C., Halaas, J. L., Horvath, C. M., Darnell, J. E., Jr., Stoffel, M. & Friedman, J. M. (1996) Nat. Genet. 14, 95-97. [DOI] [PubMed] [Google Scholar]
- 18.Friedman, J. M. & Halaas, J. L. (1998) Nature 395, 763-770. [DOI] [PubMed] [Google Scholar]
- 19.Elmquist, J. K., Ahima, R. S., Elias, C. F., Flier, J. S. & Saper, C. B. (1998) Proc. Natl. Acad. Sci. USA 95, 741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elmquist, J. K., Ahima, R. S., Maratos-Flier, E., Flier, J. S. & Saper, C. B. (1997) Endocrinology 138, 839-842. [DOI] [PubMed] [Google Scholar]
- 21.Wallenius, V., Wallenius, K., Ahren, B., Rudling, M., Carlsten, H., Dickson, S. L., Ohlsson, C. & Jansson, J. O. (2002) Nat. Med. 8, 75-79. [DOI] [PubMed] [Google Scholar]
- 22.Lambert, P. D., Anderson, K. D., Sleeman, M. W., Wong, V., Tan, J., Hijarunguru, A., Corcoran, T. L., Murray, J. D., Thabet, K. E., Yancopoulos, G. D., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 4652-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bates, S. H., Stearns, W. H., Dundon, T. A., Schubert, M., Tso, A. W., Wang, Y., Banks, A. S., Lavery, H. J., Haq, A. K., Maratos-Flier, E., et al. (2003) Nature 421, 856-859. [DOI] [PubMed] [Google Scholar]
- 24.Banks, A. S., Davis, S. M., Bates, S. H. & Myers, M. G., Jr. (2000) J. Biol. Chem. 275, 14563-14572. [DOI] [PubMed] [Google Scholar]
- 25.Niswender, K. D., Morton, G. J., Stearns, W. H., Rhodes, C. J., Myers, M. G., Jr. & Schwartz, M. W. (2001) Nature 413, 794-795. [DOI] [PubMed] [Google Scholar]
- 26.Zhao, A. Z., Huan, J. N., Gupta, S., Pal, R. & Sahu, A. (2002) Nat. Neurosci. 5, 727-728. [DOI] [PubMed] [Google Scholar]
- 27.Spanswick, D., Smith, M. A., Groppi, V. E., Logan, S. D. & Ashford, M. L. (1997) Nature 390, 521-525. [DOI] [PubMed] [Google Scholar]
- 28.Cowley, M. A., Smart, J. L., Rubinstein, M., Cerdan, M. G., Diano, S., Horvath, T. L., Cone, R. D. & Low, M. J. (2001) Nature 411, 480-484. [DOI] [PubMed] [Google Scholar]
- 29.Bruning, J. C., Gautam, D., Burks, D. J., Gillette, J., Schubert, M., Orban, P. C., Klein, R., Krone, W., Muller-Wieland, D. & Kahn, C. R. (2000) Science 289, 2122-2125. [DOI] [PubMed] [Google Scholar]
- 30.Obici, S., Feng, Z., Karkanias, G., Baskin, D. G. & Rossetti, L. (2002) Nat. Neurosci. 5, 566-572. [DOI] [PubMed] [Google Scholar]
- 31.Obici, S., Feng, Z., Arduini, A., Conti, R. & Rossetti, L. (2003) Nat. Med. 9, 756-761. [DOI] [PubMed] [Google Scholar]
- 32.Loftus, T. M., Jaworsky, D. E., Frehywot, G. L., Townsend, C. A., Ronnett, G. V., Lane, M. D. & Kuhajda, F. P. (2000) Science 288, 2379-2381. [DOI] [PubMed] [Google Scholar]
- 33.Kano, A., Wolfgang, M. J., Gao, Q., Jacoby, J., Chai, G. X., Hansen, W., Iwamoto, Y., Pober, J. S., Flavell, R. A. & Fu, X. Y. (2003) J. Exp. Med. 198, 1517-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tronche, F., Kellendonk, C., Kretz, O., Gass, P., Anlag, K., Orban, P. C., Bock, R., Klein, R. & Schutz, G. (1999) Nat. Genet. 23, 99-103. [DOI] [PubMed] [Google Scholar]
- 35.Graus-Porta, D., Blaess, S., Senften, M., Littlewood-Evans, A., Damsky, C., Huang, Z., Orban, P., Klein, R., Schittny, J. C. & Muller, U. (2001) Neuron 31, 367-379. [DOI] [PubMed] [Google Scholar]
- 36.Cohen, P., Zhao, C., Cai, X., Montez, J. M., Rohani, S. C., Feinstein, P., Mombaerts, P. & Friedman, J. M. (2001) J. Clin. Invest. 108, 1113-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huszar, D., Lynch, C. A., Fairchild-Huntress, V., Dunmore, J. H., Fang, Q., Berkemeier, L. R., Gu, W., Kesterson, R. A., Boston, B. A., Cone, R. D., et al. (1997) Cell 88, 131-141. [DOI] [PubMed] [Google Scholar]
- 38.Bousquet, C., Zatelli, M. C. & Melmed, S. (2000) J. Clin. Invest. 106, 1417-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marsh, D. J., Hollopeter, G., Huszar, D., Laufer, R., Yagaloff, K. A., Fisher, S. L., Burn, P. & Palmiter, R. D. (1999) Nat. Genet. 21, 119-122. [DOI] [PubMed] [Google Scholar]
- 40.Fan, W., Boston, B. A., Kesterson, R. A., Hruby, V. J. & Cone, R. D. (1997) Nature 385, 165-168. [DOI] [PubMed] [Google Scholar]
- 41.Yaswen, L., Diehl, N., Brennan, M. B. & Hochgeschwender, U. (1999) Nat. Med. 5, 1066-1070. [DOI] [PubMed] [Google Scholar]
- 42.Bray, G. A. & York, D. A. (1979) Physiol. Rev. 59, 719-809. [DOI] [PubMed] [Google Scholar]
- 43.Caprio, M., Fabbrini, E., Isidori, A. M., Aversa, A. & Fabbri, A. (2001) Trends Endocrinol. Metab. 12, 65-72. [DOI] [PubMed] [Google Scholar]
- 44.Lowell, B. B. & Spiegelman, B. M. (2000) Nature 404, 652-660. [DOI] [PubMed] [Google Scholar]
- 45.Bachman, E. S., Dhillon, H., Zhang, C. Y., Cinti, S., Bianco, A. C., Kobilka, B. K. & Lowell, B. B. (2002) Science 297, 843-845. [DOI] [PubMed] [Google Scholar]
- 46.Shimomura, I., Hammer, R. E., Richardson, J. A., Ikemoto, S., Bashmakov, Y., Goldstein, J. L. & Brown, M. S. (1998) Genes Dev. 12, 3182-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun, X. J., Wang, L. M., Zhang, Y., Yenush, L., Myers, M. G., Jr., Glasheen, E., Lane, W. S., Pierce, J. H. & White, M. F. (1995) Nature 377, 173-177. [DOI] [PubMed] [Google Scholar]
- 48.Withers, D. J., Gutierrez, J. S., Towery, H., Burks, D. J., Ren, J. M., Previs, S., Zhang, Y., Bernal, D., Pons, S., Shulman, G. I., et al. (1998) Nature 391, 900-904. [DOI] [PubMed] [Google Scholar]
- 49.Cho, H., Mu, J., Kim, J. K., Thorvaldsen, J. L., Chu, Q., Crenshaw, E. B., III, Kaestner, K. H., Bartolomei, M. S., Shulman, G. I. & Birnbaum, M. J. (2001) Science 292, 1728-1731. [DOI] [PubMed] [Google Scholar]
- 50.Pfeffer, L. M., Mullersman, J. E., Pfeffer, S. R., Murti, A., Shi, W. & Yang, C. H. (1997) Science 276, 1418-1420. [DOI] [PubMed] [Google Scholar]
- 51.Montague, C. T., Farooqi, I. S., Whitehead, J. P., Soos, M. A., Rau, H., Wareham, N. J., Sewter, C. P., Digby, J. E., Mohammed, S. N., Hurst, J. A., et al. (1997) Nature 387, 903-908. [DOI] [PubMed] [Google Scholar]
- 52.Clement, K., Vaisse, C., Lahlou, N., Cabrol, S., Pelloux, V., Cassuto, D., Gourmelen, M., Dina, C., Chambaz, J., Lacorte, J. M., et al. (1998) Nature 392, 398-401. [DOI] [PubMed] [Google Scholar]