Abstract
In infections of Gram-negative bacteria, lysis is a three step process, with a choice of two effectors for each step. At a precise, allele-specific time, the inner membrane (IM) is fatally permeabilized by either a holin or a pinholin. This allows a muralytic enzyme, either a canonical endolysin, escaping from the cytoplasm, or a SAR endolysin, activated in the periplasm, to degrade the peptidoglycan. Surprisingly, a third class of lysis protein, the spanin, is required for disruption of the outer membrane (OM). Key steps are regulated by membrane protein dynamics, both in terms of bilayer topology and subcellular distribution, by the energization of the membrane, and by holin-specific inhibitors called antiholins.
Introduction
In the infection cycle of double-stranded DNA phages, the lysis pathway involves at least two proteins, a small membrane protein called the holin and a muralytic enzyme called the endolysin[1]. The holin accumulates harmlessly in the membrane until, at an allele-specific time, “triggering”, a term that stands for fatal permeabilization of the bilayer that allows the endolysin to escape from the cytoplasm and attack the peptidoglycan. Triggering occurs prematurely if the infected cells are subjected to conditions that cause a sudden decrease in the proton motive force (PMF); i.e., energy poisons, membrane damage by external agents, or sudden anaerobiosis. Historically, the fatal membrane lesions formed by holin triggering have been called “holes”, as opposed to channels, transporters, or porins.
The main goal here is to highlight progress in the last few years that, at least for phages of Gram-negative hosts, has advanced our understanding of this event, which is, numerically, the most common fate of earthly cells. At the outset, it must be noted that this overview devotes no space to consideration of lysis in Gram-positive bacteria, where most attention has been focused on the biotechnological utility of phage endolysins[2]. Also omitted is consideration of the “single gene lysis” systems of small ssDNA and ssRNA phages, in which a single phage protein channels the host into autolysis[3].
The lambda lysis paradigm
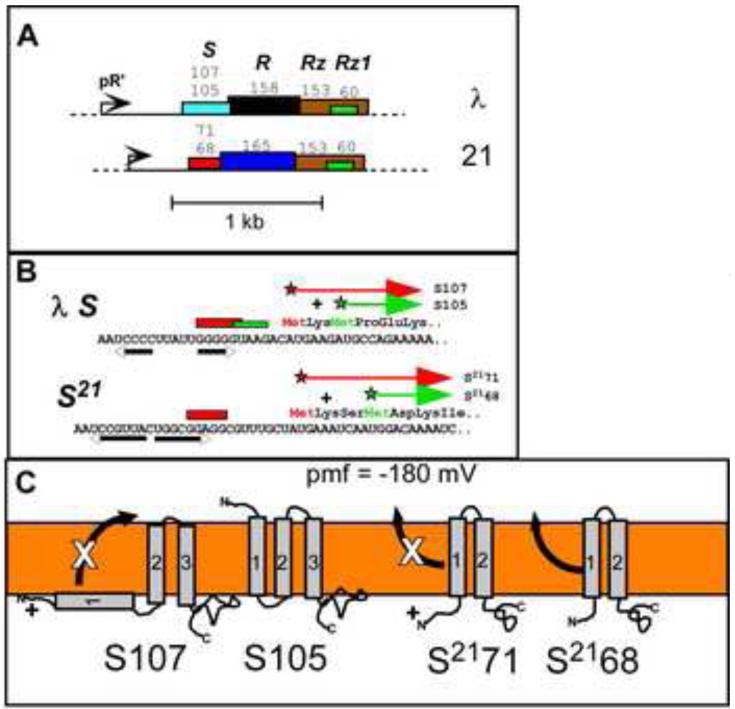
Lysis genes are often found in “cassettes” (Fig. 1), and, in the case of the best-studied lysis systems, phage lambda and the lambdoid phage 21, four genes producing five protein products are clustered immediately downstream of the single late gene promoter. For both, the S gene encodes the lysis effector, the lambda holin S105 and the phage 21 pinholin, S2168, that triggers for lethal hole formation; the protein names reflect the length in AA residues[4,5]. In both cases, an upstream translational start is used for production of an antiholin (S107; S2171). The negative-dominant character of each antiholin form is due to an N-terminal positively charged residue that confers a PMF-sensitive block on a topological change of the first transmembrane domain (TMD1) required for lytic function. The last genes in both cassettes encode the spanin subunits (See below)[6].
Figure 1. Lysis cassettes and holin topology.
A. The lysis cassettes of phages λ and lambdoid phage 21 are shown. In both lambdoid phages, the lysis genes are proximal to the late promoter, pR’. Lengths of primary translation products are shown above each gene; upper and lower numbers for S and S21 are for antiholin and holin. Gene color codes: turquoise = canonical holin, red = pinholin; black = canonical endolysin (R transglycosylase); dark blue = SAR endolysin; brown = i-spanin; green = o-spanin; white/black = u-spanin. Same color in two genes indicates sequence similarity.
B. Dual start motifs for λ holin and 21 pinholin genes. Start codons for each product indicated by star; red = antiholin, green = holin (S105) or pinholin (S2168). Red and green rectangles indicate Shine-Dalgarno sequences. Inverted arrows indicate RNA stem-loops that control choice of start codons.
C. Topological dynamics of holins and pinholins. In the energized membrane (PMF = ~180 mV), TMD1 of antiholin forms (S107 and S2171) are inhibited from entering (S107) or exiting (S2171) the bilayer, whereas TMD1 of the pinholin (S2168) exits spontaneously during pinhole pathway.
A and C adapted from Wang et al. [50]. B adapted from Young [51].
Lambda lysis has been conveniently characterized by thermal induction of a lambda prophage, after which, under strictly-defined conditions, lysis of nearly all the cells of the culture occurs abruptly at 50 min, or some 42 min after the turn-on of late gene expression[7]. Other lysis systems have been studied in this convenient system by substituting genes for heterologous holins, endolysins, and spanins or even substituting whole lysis cassettes[5,8-10].
A unified model for phage lysis in Gram-negative hosts
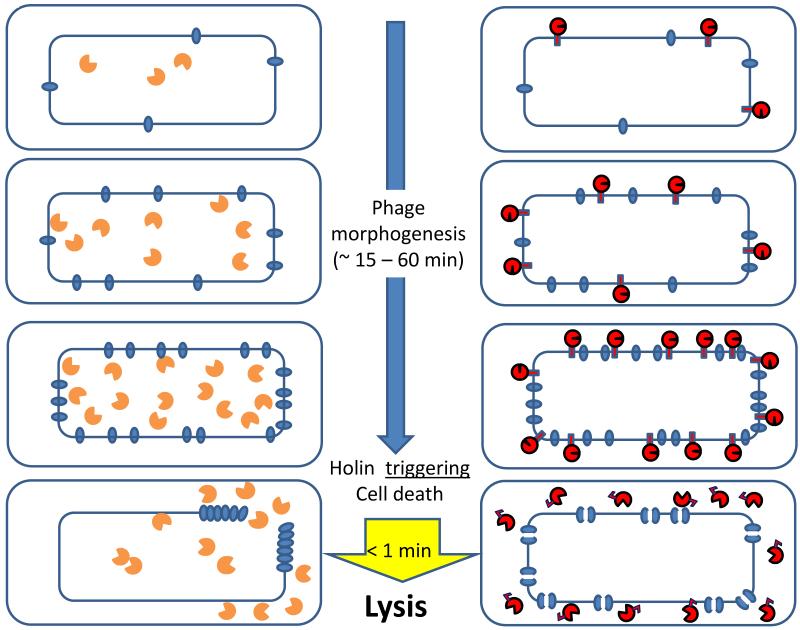
In the last few years there have been multiple studies using fluorescence, transmission, and cryoelectron microscopy that have profoundly affected our view of the lysis pathway[11-13]. The cartoon in Fig. 2 attempts to summarize in outline a coherent model of the two different molecular strategies by which the temporal scheduling of lysis is effected. In the left and right panels are depicted canonical holin-endolysin lysis, as exemplified by lambda, and the pinholin-SAR endolysin pathway, exemplified by phage 21, respectively. When late gene expression commences, the holin S105 of phage lambda and the pinholin S2168 begin to accumulate in the membrane, each as homodimers or as heterodimers with their cognate antiholins. These proteins continue to accumulate harmlessly, freely mobile in the cytoplasmic membrane, while virion assembly is proceeding in the cytoplasm. In both cases, when the holins and pinholins reach an allele-specific critical concentration, suddenly nucleation and rapid formation of large two-dimensional aggregates, or rafts, occurs, based on studies with holin-GFP and pinholin-GFP fusions (Supp. Fig. 1). Rafts are very large and number only a few per cell for the canonical lambda holin. In contrast, the rafts in the pinholin situation are smaller and more numerous. Raft formation is tightly coupled with triggering and hole formation.
Figure 2. The two pathways for phage lysis in Gram-negative hosts.
Schematic views of the holin-endolysin (left) and pinholin-SAR endolysin (right) pathways to lysis, beginning at the onset of late gene expression (phage morphogenesis) period. The inner (IM) and outer membranes (OM)of cells are shown, with holin or pinholin (blue ovals) accumulating in the IM. In the bottom figures, upon reaching a critical concentration, the holin triggers to form a large micron-scale hole (left). In contrast, the pinholin triggers to from many heptameric pinholes (represented by double ovals with a channel).
The orange symbols with the open “active site” represent the enzymatically active canonical endolysin accumulating in the cytosol. The red symbols with the closed and open “active sites” represent the inactive SAR endolysin accumulating in the IM and the activated SAR endolysin released into the periplasm, respectively.
S-holes
It has long been known that the S105 lesions were large and non-specific, as attested to by the ability of S105 (and other holins[9,10,14-16]) to support lysis with heterologous endolysins and, more dramatically, with hybrid endolysin-βgalactosidase fusions of ~0.5 MDa native mass[17]. However, it was not until recently that cryo-EM studies and tomography revealed the “S-holes” to be micron-scale interruptions in the IM, averaging >340 nm in diameter and ranging to ~1 μm, but small in number, 1-3 per cell (Supp. Fig. 2; upper panel)[11]. These massive lesions allow the endolysin R to escape from the cytoplasm and attack the peptidoglycan. The model is supported by the lysis morphology, as observed in phase-contrast videomicroscopy (Supp. Fig. 2; lower panels)[18]. Induced lysogens are seen to elongate until suddenly undergoing a “blow-out” at a localized region; the cell violently empties of refractile material, including the progeny virions, while at least momentarily maintaining rod shape. It is thought that the localized disruption reflects the huge holin lesion and the resultant massive degradation of the peptidoglycan opposite the hole (Fig. 2, left).
Pinholes and SAR endolysins
In the pinholin/SAR endolysin pathway, where after triggering the rafts were found to be smaller and more numerous, cross-linking, cysteine-accessibility and modeling studies have indicated that the entire pinholin population of ~7000 molecules participate in the formation of ~103 heptameric “pinholes”, estimated to have a ~ 2 nm diameter (Supp. Fig. 3)[13,19]. Lesions of this size are clearly too small to allow transit of mature endolysins. Instead, pinholins require a SAR endolysin secreted by the host sec system[20]. SAR stands for signal-anchor-release; SAR domains are N-terminal TMDs (or signal anchors) that serve to engage the sec pathway and then anchor the exported endolysin to the bilayer in a membrane potential-dependent fashion (Fig. 3). Premature degradation of the murein is avoided because the tethered enzyme is inactive. Depolarization of the membrane by the pinholin causes quantitative release of the tethered SAR endolysin molecules, which then refold into an enzymatically active form, leading to generalized murein degradation. In consequence, the pinholin-endolysin system has a distinct morphological signature observable by phase-contrast videomicroscopy: after pinholin triggering, the cell begins to shorten and round up before suddenly exploding (Supp. Fig. 2)[18]. Presumably this reflects the uniform dispersal of the SAR endolysin within the periplasm, such that murein degradation is not localized to the region opposite a massive holin lesion but is instead distributed throughout the periplasm after release and activation (Fig. 2, right).
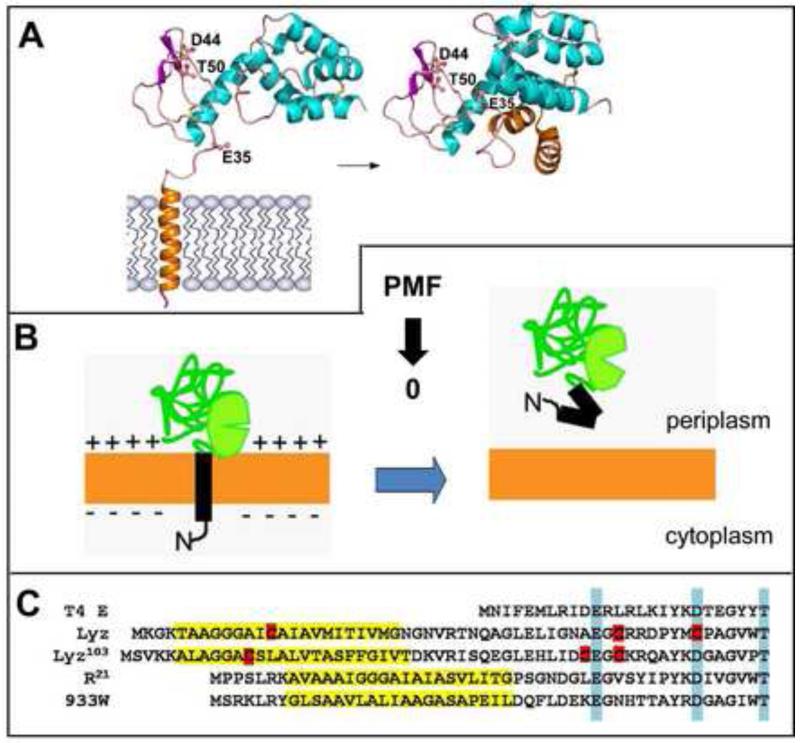
Figure 3. SAR endolysins.
A. Structures of inactive, membrane-tethered (left) and active, released R21 (right). SAR domain is modeled as TMD in structure of inactive form. In this activation event, the catalytic triad Glu35-Asp44-Thr50 is assembled when SAR domain folds up against the body of the protein. Adapted from Sun [22].
B. General regulation of SAR endolysins. These enzymes are catalytically inactive when tethered to the membrane. They will release spontaneously at a slow rate but quantitatively if the PMF collapses. Release of the SAR domain causes refolding to the active state.
C. SAR domains (yellow highlights) cause different mechanisms of activation. SAR endolysins are homologs of the canonical lysozyme T4 E, with the Ex8D/Cx5T catalytic triad (highlighted in blue). SAR domains have a high proportion of Gly/Ala/Ser residues, at the expense of Leu, compared to normal TMDs [20]. SAR domains that include a Cys residue (red) cause disulfide bond isomerization after release, resulting in liberation of a catalytic Cys (Lyz of phage P1)[21] or uncaging of the catalytic Glu (Lyz of phage ERA103)[23]. R21 activation is non-covalent (panel A). The activation of R933W, the enzyme that causes release of Shiga toxin from E. coli O157:H7 cells[52], is under investigation (Q. Sun, G. Kuty, T. Pang, J. Sacchettini and R. Young, unpublished).
Regulation of lysis
The operational cartoon in Fig. 2 shows the common strategy but different tactics by which holins and pinholins control the timing of lysis. Underlying this simplified operational scheme is a remarkable pattern of membrane protein dynamics at the level of bilayer topology and subcellular distribution.
Diverse gymnastics of SAR endolysins
The molecular basis of SAR endolysin regulation has been studied for three different phages, with crystal structures determined for the inactive and active forms of two of these (Fig. 3)[20-23]. All three systems have different regulatory schemes, including dramatic conformational rearrangements and two different pathways of disulfide bond isomerization, all consequent to the escape of the SAR domain from the bilayer. SAR domains are always N-terminal TMDs with a relatively hydrophilic character (reduced content of Leu residues compensated by enriched Gly/Ala/Ser). Moreover, SAR domains exit the bilayer spontaneously at a specific low rate but are instantly released when the PMF is collapsed.
Topological changes in the pinhole pathway
The topological behavior of the S21 pinholin is even stranger, as revealed by recent genetic, biochemical, and high resolution real-time fluorescence microscopy studies[13,19,24-27]. The pinholin accumulates as a dimer in which both TMD1, which is a SAR domain, and TMD2 of each monomer are stabilized in the membrane by intramolecular and intermolecular helix-helix interactions (Supp. Fig. 2). It is not until both TMD1s spontaneously exit the membrane, and then interact as aqueous-phase helices, that the pinholin dimer is activated for the raft formation pathway. Thus TMD1 is an intrinsic intramolecular inhibitor of lysis while in the bilayer but a positive effector once relocalized to the periplasm, whereas TMD2 is the actual pinhole-former. The antiholin product, S2171, has an extra positively charged residue at the N-terminus that inhibits escape of TMD1 from the bilayer (Fig. 1). Thus, S2171, if dimerized with the pinholin S2168 form, creates an inactive heterodimer and slows the “lysis clock”.
The lambda antiholin: penetration however slight
Opposite topological dynamics underlie the inhibitory role of S107, the lambda antiholin. In this case, the extra positive charge at the N-terminus of the antiholin prevents TMD1 from entering the membrane, giving rise to inactive S105:S107 heterodimers, again slowing the lysis clock (Fig. 1)[28-33]. In both cases, the depolarization that occurs at the instant of triggering removes a barrier for the topological change of TMD1 (entry for S107, exit for S2171) and thus converts the inactive holin-antiholin heterodimers into fully active molecules. Since in normal conditions, the holin-antiholin ratio is ~2:1, and the antiholin preferentially heterodimerizes with the holin, the number of active holin or pinholin dimers would triple at the instant of triggering.
It must be emphasized that the dynamics of these antiholin systems, however elegant, are not the key to lysis timing in either the holin or pinholin pathways. Precisely-timed triggering still occurs in mutants in which the antiholin forms are not produced, albeit a few minutes earlier. RNA secondary structures (Fig.1) control the ratio of holins (or pinholins) to antiholins, and mutations that alter these structures such that the antiholin form predominates are not only non-lytic alleles but also have dominant character (which is how they were discovered.)[4,5,7,34,35] There is some evidence that host factors may regulate the efficiency of expression of the lambda holin gene or the holin-antiholin ratio by affecting the formation of these structures (K. Nam, “Translational regulation of the S gene of bacteriophage lambda”, Ph.D. thesis, Texas A&M University, 1991;)[34].
Triggering and the critical concentration
A lynchpin of the current model is that the holins and pinholins exhibit critical concentration behavior for raft formation, based on the precedent of the well-studied bacteriorhodopsin system (Fig. 2; Supp. Fig. 4)[36,37]. After induction, mobile bacteriorhodopsin monomers accumulate in the membrane until, at a critical concentration, beginning the assembly of the “purple membrane” array. Missense mutations at intermolecular interfaces can raise or lower the critical concentration significantly. Presumably, similar effects explain the phenotypic variance seen in genetic studies of lambda S, which, although the wt allele triggers at 50 min, can give rise to missense mutants triggering as early as 11 min or as late as 90 min [7,38](K. To, unpublished).
Also implicit in this scheme is the notion that the holin or pinholin rafts spontaneously convert to holes (or pinholes). Possible clues to this behavior come from studies on the pinhole pathway[13,27]. Genetics and chemical probing studies indicated that in the pinhole, TMD2 of the pinholin has a more hydrophilic surface facing the lumen, whereas in the pre-triggering dimer, the same hydrophilic faces are sequestered against each other and the still-integrated TMD1. It is proposed that raft formation involves intimate helical packing, resulting in a lipid-depleted two-dimensional aggregate that leaks ions along the interfaces of the hydrophilic surfaces (Supp. Fig. 3). Local reduction in the membrane potential could stimulate reorientation of these surfaces to allow hydration, more ion leakiness, and thus a concerted shift to the pinhole arrangement. A few local pinholes could then lead to more depolarization and, ultimately, total collapse of the PMF.
Although short on specifics, this scheme accounts for the universal holin susceptibility to premature triggering by anything that reduces membrane energization. S105 has two TMDs, 1 and 3, with relatively hydrophilic faces; experiments are in progress to test if these faces line the lumen of the S-hole (K. To, unpublished).
Spanins
One might expect that once the murein has been degraded, lysis and dispersal of the viral progeny would be automatic. It was perhaps not too surprising to discover that this was not true for phages of the mycolata, which encode, in addition to the holin and endolysin, an enzyme,LysB, to degrade the mycolate esters of the waxy outer membrane-equivalent[39-41]. However, it was definitely unexpected to find that, even after destruction of the peptidoglycan, lysis of Gram-negative hosts requires yet another functional class of protein, a spanin, for disruption of the outer membrane (OM)[18]. Moreover, two completely different types of spanins, two-component spanins and unimolecular spanins, have been identified[6].
Genes like Russian dolls
The last genes of the lambda lysis cassette are also the strangest: Rz, which encodes an integral IM protein, and Rz1, which is wholly embedded in the +1 reading frame of the Rz gene and encodes an OM lipoprotein (Fig. 1). Recently phase contrast videomicroscopy has revealed that both genes are required for lysis. In the absence of Rz or Rz1 function, the lysis process terminates in spherical cells lacking the cell wall and bounded by the outer membrane (OM) (Supp. Fig. 2)[18]. At the molecular level, the Rz and Rz1 proteins were shown to be the subunits of a complex that spans the entire periplasm, connecting the IM and OM, and thus designated as a spanin (Fig. 4)[42]. Rz is an 153 AA integral membrane protein, or i-spanin, with an N-terminal TMD and a periplasmic domain shown to be both highly helical and subject to coiled-coil oligomerization[6,43]. Mature Rz1 is a 40 AA OM lipoprotein, or o-spanin, with a proline content of 25%. Co-precipitation experiments have shown that the Rz-Rz1 spanin complex assembles in the envelope during the morphogenesis period, via C-terminal interactions[42]. To add to the litany of unique features, both Rz and Rz1 have recently been shown to accumulate as covalent homodimers, linked by three intermolecular disulfide bonds (two in the Rz dimer, one in the Rz1 dimer)[44]. Thus the spanin complex is actually an Rz2:Rz12 tetramer. Oddly, complementation studies have shown that spanin function requires that at least one of the two subunit homodimers is covalent linked, although it does not matter which one.
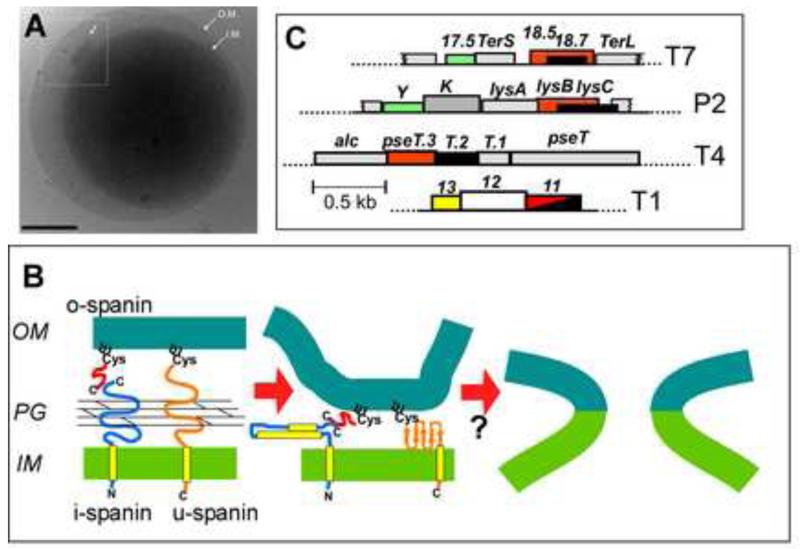
Figure 4. Spanin function and gene architecture.
A. Holin-endolysin function in the absence of spanin function results in spherical cells bounded by the OM. Cryo-EM image of E. coli cell in which SR genes have been expressed, after triggering. Note inset: shows efflux of cytosolic material from S-hole. Scale bar is 500 nm. From Dewey et al. [11].
B. Model for spanin structure and function. Two component spanin complexes consists of an integral IM subunit, the i-spanin (Rz) and an OM lipoprotein, the lipoprotein o-spanin (Rz1), that interact via a C-terminal/C-terminal interaction, spanning the periplasm through the meshwork of the peptidoglycan (PG). Alternatively, a unimolecular spanin, u-spanin (T1 gp11), which is an OM lipoprotein but also has a C-terminal TMD, spans the periplasm as a covalent chain. The Rz/Rz1 complex is shown as a heterodimer for simplicity; in vivo, each subunit is a disulfide-linked homodimer. The current model is that the spanin complex undergoes oligomerization when freed from the meshwork of the PG, providing free energy for a collapsing conformational change that brings the two membranes together. (Only the collapsing change is shown.) In the two-component system, the oligomerization is due to the high coiled-coil propensity of the highly α-helical periplasmic domain of the i-spanin. For the u-spanin, the collapsing change probably involves beta sheet formation. The resultant adjacency of the bilayers is thought to lead to membrane fusion, thus disrupting the OM.
C. Spanin gene architecture. Two component spanin genes (red = i-spanin; black = o-spanin) are highly diverse and are found in three architectures, each with multiple, dissimilar sequences: embedded (o-spanin gene entirely within i-spanin gene) (e.g., T7; also see Fig. 1); overlapped (o-spanin gene extends beyond end of i-spanin gene; e.g., P2); and separated (genes completely separated; e.g., T4). U-spanin (red-black hatch; T1) genes are also diverse.
Panels B and C adapted from Summer et al. [6].
Spanin function
The interesting questions are: (1) how does the Rz2:Rz12 spanin complex function to disrupt the OM? (2) how is this regulated, in view of the finding that the complexes accumulate throughout the morphogenesis period. Fig. 4 shows the current model, partly based on biochemical studies with the soluble periplasmic domains of the spanin subunits and partly conceived as shavings from Ockham’s Razor[18]. The regulation is thought to be inherent in the fact that since the complexes span the periplasm, they are trapped within the lacunae of the cross-linked peptidoglycan. After holin triggering and cell wall degradation by the endolysin, the spanin complexes are free to diffuse laterally and form aggregates, providing the free energy for a conformational change that disrupts the OM. The nature of the disruption is unknown but the simplest notion is that the oligomerized complexes bring the surfaces of the OM and IM together and cause membrane fusion.
The membrane fusion model arose from the kernel ideas that there has to be some reason for the spanins to connect the IM and OM and that there is no evidence of enzymatic activity or catalytic domains. The idea was indirectly supported by the discovery that many phages, including the iconic coliphage T1 (Fig. 1), an entirely distinct type of spanin, the unimolecular or u-spanin (Fig. 4), in which features of the i-spanin and o-spanin subunits are combined and which complements an RzRz1 double defect[6]. The prototypic u-spanin, T1 gp11, is an OM lipoprotein, so that its N-terminal lipoylated Cys is exported by the Lol system to the inner leaflet of the OM[45], but it also has a C-terminal TMD. Besides the different topology, the periplasmic domain of gp11 is predicted to be mostly beta-sheet and has no Cys residues, so is clearly a completely unrelated solution to the same operational task. The notion here is that the gp11 protomers, once liberated from the meshwork of the peptidoglycan by endolysin action, undergo oligomerization that provides the free energy for a collapsing conformational change and membrane fusion.
Definitive evidence addressing the membrane fusion model should soon be available. In any case, whatever the mode by which these spanin proteins disrupt the OM, the genetic diversity involved turned out to be quite profound[6]. Not only are there are many unrelated families of embedded spanin genes arranged like Rz/Rz1 (Figs. 1, 4), but also there are multiple families in which the o-spanin gene extends beyond the end of the i-spanin gene (overlapped) and also where the two genes are entirely distinct (separated). Add to this mix the existence of multiple u-spanin genes like T1 11 and one is left with evolutionary puzzle that should prove intriguing to theorists.
Conclusions and Perspective
Considerable progress has been made in fleshing out the operational aspects of the lysis pathway in Gram-negative hosts. In overview, the pathway has three steps, each with two fundamentally different options: timed triggering by the holin or pinholin, cell wall degradation by the endolysin or SAR endolysin, and then OM disruption by the i-spanin/o-spanin complex or the u-spanin. There are thus eight possible combinations for these three steps, but only six are functionally compatible, considering that pinholins require SAR endolysins. Remarkably, all six have been found in nature or shown to function in the laboratory. One combination, comprising a pinholin, SAR endolysin, and i-spanin/o-spanin ensemble, has been shown to be responsible for the release of Shiga-toxin in EHEC infections[46], so there is even some clinical relevance to this story. More information is needed about the structure of the S-holes, specifically, and in the holes formed by canonical holins in general. Beyond this, technological innovation, possibly using planar lipid bilayer systems, will be required to get at the specifics of the current model, especially the interactions of the holin proteins with the energized membrane. Also needed are studies directly addressing the proposed membrane-fusion model for spanin function. If the general idea is validated, the spanin systems should be regarded as simple, genetically facile platforms for studying the membrane fusion process that underlies so many key biological processes. Finally, space limitations have precluded discussion of new developments in the oldest experimental system in molecular genetics, T4 lysis-inhibition, or LIN. For interested readers it is worth noting that, besides its historical status, LIN is the only case where it is known that environmental information is used for real-time, antiholin-mediated control of holin triggering [47-49].
Supplementary Material
Highlights.
Holins or pinholins form membrane holes upon reaching a critical concentration
Murein degradation is effected either by cytoplasmic or SAR endolysins
The outer membrane must be disrupted by two-component or unimolecular spanins
Lysis proteins are regulated by tertiary, quaternary and topological dynamics
Acknowledgements
This work was supported by National Institutes of Health grant NIGMS27099 to the author, as well as the Sadie Hatfield Professorship in Agriculture at Texas A&M University. The author is grateful to all of the students, post-doctoral scientists, and faculty collaborators for their many consequential insights into this fundamental biological phenomenon over more than 35 years.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Young R, Wang IN. Phage Lysis. In: Calendar R, editor. In The Bacteriophages. edn 2nd Vol. 104. Oxford University Press; 2006. p. 126. [Google Scholar]
- 2.Fischetti VA. Bacteriophage endolysins: a novel anti-infective to control Gram-positive pathogens. Int J Med Microbiol. 2010;300:357–362. doi: 10.1016/j.ijmm.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernhardt TG, Wang IN, Struck DK, Young R. Breaking free: “protein antibiotics” and phage lysis. Res. Microbiol. 2002;153:493–501. doi: 10.1016/s0923-2508(02)01330-x. [DOI] [PubMed] [Google Scholar]
- 4.Bläsi U, Nam K, Hartz D, Gold L, Young R. Dual translational initiation sites control function of the λ S gene. EMBO Journal. 1989;8:3501–3510. doi: 10.1002/j.1460-2075.1989.tb08515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonovich MT, Young R. Dual start motif in two lambdoid S genes unrelated to lambda S. J Bacteriol. 1991;173:2897–2905. doi: 10.1128/jb.173.9.2897-2905.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Summer EJ, Berry J, Tran TA, Niu L, Struck DK, Young R. Rz/Rz1 lysis gene equivalents in phages of Gram-negative hosts. J. Mol. Biol. 2007;373:1098–1112. doi: 10.1016/j.jmb.2007.08.045. [DOI] [PubMed] [Google Scholar]
- 7.Raab R, Neal G, Sohaskey C, Smith J, Young R. Dominance in lambda S mutations and evidence for translational control. J. Mol. Biol. 1988;199:95–105. doi: 10.1016/0022-2836(88)90381-6. [DOI] [PubMed] [Google Scholar]
- 8.Roof WD, Young R. èX174 E complements lambda S and R dysfunction for host cell lysis. J.Bacteriology. 1993;175:3909–3912. doi: 10.1128/jb.175.12.3909-3912.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramanculov ER, Young R. Functional analysis of the T4 t holin in a lambda context. Mol.Genet.Genomics. 2001;265:345–353. doi: 10.1007/s004380000422. [DOI] [PubMed] [Google Scholar]
- 10.To K, Dewey JS, Savva C, Park T, Young R. P2 Y: functional analysis of a class I holin. 2012 [Google Scholar]
- 11.Dewey JS, Savva CG, White RL, Vitha S, Holzenburg A, Young R. Micron-scale holes terminate the phage infection cycle. Proc Natl Acad Sci U S A. 2010;107:2219–2223. doi: 10.1073/pnas.0914030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12**.White R, Chiba S, Pang T, Dewey JS, Savva CG, Holzenburg A, Pogliano K, Young R. Holin triggering in real time. Proc Natl Acad Sci U S A. 2011;108:798–803. doi: 10.1073/pnas.1011921108. First real-time study of holin triggering using GFP fusions
- 13**.Pang T, Fleming TC, Pogliano K, Young R. Visualization of pinholin lesions in vivo. Proc Natl Acad Sci U S A. 2013;110:E2054–2063. doi: 10.1073/pnas.1222283110. Shows that pinholins also undergo the massive redistribution after triggering.
- 14.Berkmen M, Benedik MJ, Bläsi U. The Serratia marcescens NucE protein functions as a holin in Escherichia coli. J.Bacteriology. 1997;179:6522–6524. doi: 10.1128/jb.179.20.6522-6524.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin AC, Lopez R, Garcia P. Functional analysis of the two-gene lysis system of the pneumococcal phage Cp-1 in homologous and heterologous host cells. J.Bacteriology. 1998;180:210–217. doi: 10.1128/jb.180.2.210-217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rydman PS, Bamford DH. Identification and mutational analysis of bacteriophage PRD1 holin protein P35. J. Bacteriol. 2003;185:3795–3803. doi: 10.1128/JB.185.13.3795-3803.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang IN, Deaton JF, Young R. Sizing the holin lesion with an endolysin-βgalactosidase fusion. J.Bacteriology. 2003;185:779–787. doi: 10.1128/JB.185.3.779-787.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18**.Berry JD, Rajaure M, Pang T, Young R. The spanin complex is essential for lambda lysis. J Bacteriol. 2012;194:5667–5674. doi: 10.1128/JB.01245-12. First systematic videomicroscopy of lysis morphology and evidence for spanin essentiality.
- 19**.Pang T, Savva CG, Fleming KG, Struck DK, Young R. Structure of the lethal phage pinhole. Proc Natl Acad Sci U S A. 2009;106:18966–18971. doi: 10.1073/pnas.0907941106. Discovery of SAR sequences, a signal conferring dynamic membrane topology.
- 20.Xu M, Struck DK, Deaton J, Wang IN, Young R. The signal arrest-release (SAR) sequence mediates export and control of the phage P1 endolysin. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6415–6420. doi: 10.1073/pnas.0400957101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu M, Arulandu A, Struck DK, Swanson S, Sacchettini JC, Young R. Disulfide isomerization after membrane release of its SAR domain activates P1 lysozyme. Science. 2005;307:113–117. doi: 10.1126/science.1105143. [DOI] [PubMed] [Google Scholar]
- 22.Sun Q, Kuty GF, Arockiasamy A, Xu M, Young R, Sacchettini JC. Regulation of a muralytic enzyme by dynamic membrane topology. Nat Struct Mol Biol. 2009;16:1192–1194. doi: 10.1038/nsmb.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuty GF, Xu M, Struck DK, Summer EJ, Young R. Regulation of a phage endolysin by disulfide caging. J Bacteriol. 2010;192:5682–5687. doi: 10.1128/JB.00674-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park T, Struck DK, Deaton JF, Young R. Topological dynamics of holins in programmed bacterial lysis. Proc. Natl. Acad. Sci. U.S.A. 2006;103:19713–19718. doi: 10.1073/pnas.0600943103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park T, Struck DK, Dankenbring CA, Young R. The pinholin of lambdoid phage 21: control of lysis by membrane depolarization. J Bacteriol. 2007;189:9135–9139. doi: 10.1128/JB.00847-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pang T, Park T, Young R. Mutational analysis of the S21 pinholin. Mol Microbiol. 2010;76:68–77. doi: 10.1111/j.1365-2958.2010.07080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pang T, Park T, Young R. Mapping the pinhole formation pathway of S21. Mol Microbiol. 2010;78:710–719. doi: 10.1111/j.1365-2958.2010.07362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang YF, Renshaw HW, Young R. Pneumonic pasteurellosis: examination of typable and untypable Pasteurella haemolytica strains for leukotoxin production, polasmid content, and antimicrobial susceptibiity. Am.J.Vet.Res. 1987;48:378–384. [PubMed] [Google Scholar]
- 29.Bläsi U, Chang CY, Zagotta MT, Nam K, Young R. The lethal λ S gene encodes its own inhibitor. EMBO Journal. 1990;9:981–989. doi: 10.1002/j.1460-2075.1990.tb08200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steiner M, Bläsi U. Charged amino-terminal amino acids affect the lethal capacity of lambda lysis proteins S107 and S105. Mol Microbiol. 1993;8:525–533. doi: 10.1111/j.1365-2958.1993.tb01597.x. [DOI] [PubMed] [Google Scholar]
- 31.Graschopf A, Bläsi U. Molecular function of the dual-start motif in the λ S holin. Mol.Microbiol. 1999;33:569–582. doi: 10.1046/j.1365-2958.1999.01501.x. [DOI] [PubMed] [Google Scholar]
- 32.Graschopf A, Bläsi U. Functional assembly of the lambda S holin requires periplasmic localization of its N-terminus. Arch.Microbiol. 1999;172:31–39. doi: 10.1007/s002030050736. [DOI] [PubMed] [Google Scholar]
- 33.White R, Tran TA, Dankenbring CA, Deaton J, Young R. The N-terminal transmembrane domain of λ S is required for holin but not antiholin function. J Bacteriol. 2010;192:725–733. doi: 10.1128/JB.01263-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang CY, Nam K, Bläsi U, Young R. Synthesis of two bacteriophage λ S proteins in an in vivo system. Gene. 1993;133:9–16. doi: 10.1016/0378-1119(93)90218-r. [DOI] [PubMed] [Google Scholar]
- 35.Chang CY, Nam K, Young R. S gene expression and the timing of lysis by bacteriophage λ. J.Bacteriology. 1995;177:3283–3294. doi: 10.1128/jb.177.11.3283-3294.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isenbarger TA, Krebs MP. Role of helix-helix interactions in assembly of the bacteriorhodopsin lattice. Biochemistry. 1999;38:9023–9030. doi: 10.1021/bi9905563. [DOI] [PubMed] [Google Scholar]
- 37.Isenbarger TA, Krebs MP. Thermodynamic stability of the bacteriorhodopsin lattice as measured by lipid dilution. Biochemistry. 2001;40:11923–11931. doi: 10.1021/bi0106585. [DOI] [PubMed] [Google Scholar]
- 38.Zheng Y, Struck DK, Dankenbring CA, Young R. Evolutionary dominance of holin lysis systems derives from superior genetic malleability. Microbiology. 2008;154:1710–1718. doi: 10.1099/mic.0.2008/016956-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gil F, Catalao MJ, Moniz-Pereira J, Leandro P, McNeil M, Pimentel M. The lytic cassette of mycobacteriophage Ms6 encodes an enzyme with lipolytic activity. Microbiology. 2008;154:1364–1371. doi: 10.1099/mic.0.2007/014621-0. [DOI] [PubMed] [Google Scholar]
- 40.Payne K, Sun Q, Sacchettini J, Hatfull GF. Mycobacteriophage Lysin B is a novel mycolylarabinogalactan esterase. Mol Microbiol. 2009;73:367–381. doi: 10.1111/j.1365-2958.2009.06775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gil F, Grzegorzewicz AE, Catalao MJ, Vital J, McNeil MR, Pimentel M. Mycobacteriophage Ms6 LysB specifically targets the outer membrane of Mycobacterium smegmatis. Microbiology. 2010;156:1497–1504. doi: 10.1099/mic.0.032821-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berry J, Summer EJ, Struck DK, Young R. The final step in the phage infection cycle: the Rz and Rz1 lysis proteins link the inner and outer membranes. Mol Microbiol. 2008;70:341–351. doi: 10.1111/j.1365-2958.2008.06408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berry J, Savva C, Holzenburg A, Young R. The lambda spanin components Rz and Rz1 undergo tertiary and quaternary rearrangements upon complex formation. Protein Sci. 2010;19:1967–1977. doi: 10.1002/pro.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berry JD, Rajaure M, Young R. Spanin function requires subunit homodimerization through intermolecular disulfide bonds. Mol Microbiol. 2013;88:35–47. doi: 10.1111/mmi.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tokuda H. Biogenesis of outer membranes in Gram-negative bacteria. Biosci Biotechnol Biochem. 2009;73:465–473. doi: 10.1271/bbb.80778. [DOI] [PubMed] [Google Scholar]
- 46.Neely MN, Friedman DI. Functional and genetic analysis of regulatory regions of coliphage H-19B: location of shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol.Microbiol. 1998;28:1255–1267. doi: 10.1046/j.1365-2958.1998.00890.x. [DOI] [PubMed] [Google Scholar]
- 47.Tran TAT, Struck DK, Young R. Periplasmic domains define holin-antiholin interactions in T4 lysis inhibition. J. Bacteriol. 2005;187:6631–6640. doi: 10.1128/JB.187.19.6631-6640.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tran TA, Struck DK, Young R. The T4 RI antiholin has an N-terminal signal anchor release domain that targets it for degradation by DegP. J Bacteriol. 2007;189:7618–7625. doi: 10.1128/JB.00854-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moussa SH, Kuznetsov V, Tran TA, Sacchettini JC, Young R. Protein determinants of phage T4 lysis inhibition. Protein Sci. 2012;21:571–582. doi: 10.1002/pro.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang IN, Smith DL, Young R. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 2000;54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- 51.Young R. Bacteriophage holins: deadly diversity. J.Mol.Microbiol.Biotechnol. 2002;4:21–36. [PubMed] [Google Scholar]
- 52.Wagner PL, Livny J, Neely MN, Acheson DW, Friedman DI, Waldor MK. Bacteriophage control of Shiga toxin 1 production and release by Escherichia coli. Mol Microbiol. 2002;44:957–970. doi: 10.1046/j.1365-2958.2002.02950.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.