Abstract
As a graduate student with Professor Richard Setlow at Yale in the late 1950s, I studied the effects of ultraviolet and visible light on the syntheses of DNA, RNA, and protein in bacteria. I reflect upon my research in the Yale Biophysics Department, my subsequent postdoctoral experiences, and the eventual analyses in the laboratories of Setlow, Paul Howard-Flanders, and myself that constituted the discovery of the ubiquitous pathway of DNA excision repair in the early 1960s. I then offer a brief perspective on a few more recent developments in the burgeoning DNA repair field and their relationships to human disease.
Keywords: nucleotide excision repair, DNA repair history, repair replication, transcription-coupled repair, ultraviolet light, xeroderma pigmentosum, Cockayne syndrome, UV sensitive syndrome
Introduction
The experiments that led to the discovery of DNA excision repair were initiated in the Yale Biophysics Department just a few years after James Watson and Francis Crick proposed the double-helical structure for DNA, based upon Rosalind Franklin’s X-ray crystallography and Erwin Chargaff’s base-pairing rules. The complementary strands of that duplex structure had immediately suggested a mechanism by which DNA might replicate, but the possibility had been overlooked that an intact strand might serve as a template for the precise reconstruction of a damaged region in the other strand.
I entered Yale as a biophysics graduate student in autumn 1954, having completed an undergraduate physics major at Oberlin College. I had come to the right place at the right time, and as it turned out, I was also going to have the opportunity to work with the right mentor on the right dissertation project [1]. Biophysics at Yale began as a program within the physics department. The charismatic program chair was Ernest C. Pollard, a nuclear physicist who had worked with Rutherford and Chadwick in the 1930s and had then constructed Yale’s first cyclotron before turning his interest to the biological effects of radiation in the mid-1950s. I completed an MS in physics during my first year and carried out directed readings with Harold Morowitz, an assistant professor in biophysics. He assigned the classic monograph, “What is Life?,” in which the theoretical physicist, Erwin Schröedinger, had elaborated on the prevailing view that DNA must be a remarkably stable crystal-like molecule in order to account for the low frequencies of spontaneous mutagenesis [2]. The predominant emphasis in the biophysics group was on the effects of ionizing radiation on living systems, and the weekly seminar discussions were rapidly converging upon DNA as a prime target, since it was becoming abundantly clear that DNA was the primary repository of the genetic blueprint. However, there was little biochemical understanding of the processing of the genetic material in living cells. The multiple functions of RNA and its role in information transfer were obscure, and the mechanism of protein synthesis was a total mystery. It had long been established, however, that short wavelength ultraviolet light (UV†) was mutagenic and could kill bacteria. In 1949, the phenomenon of photoreactivation was discovered as a process by which subsequent exposure to visible light somehow improved the survival of bacteria that had been irradiated with UV. The transient inhibition of DNA synthesis in UV-irradiated bacteria had also been shown, and the time was ripe for speculation about a recovery process that operated in the dark.
Graduate Study and Research at Yale
My favorite course in the first year was Modern Physical Measurements, for which the section on spectroscopy was taught by Professor Richard “Dick” Setlow in the sub-basement (two floors underground) of the Sloan Physics Building, in a labyrinth of rooms with flat-black walls, containing light sources, prisms, diffraction gratings, lenses, and film cassettes mounted precisely on optical benches. It was an exciting exercise to confirm basic physical principles by appropriately arranging these basic components within this clandestine maze. Meanwhile, the nascent biophysics program shared the attic “shell-space” of that same building with the ventilation equipment, which included several enormous exhaust fans that fortunately had been disconnected. Within a few years, Biophysics became established as a separate department and was relocated to half of the fourth floor in the new Gibbs Laboratory, while the other half accommodated the research group of a classic cytogeneticist, Norman Giles. Professor Giles taught another of my favorite courses, a practical laboratory in cytology, in which I was introduced to the fascinating world of chromosomes. We prepared chromosome spreads from different stages in the cell cycle from several types of plant and animal cells, we analyzed X-ray induced chromosomal aberrations, and we were introduced to the concept of aneuploidy as we examined the karyotype in HeLa cells. This was my first exciting view of cells from a human tumor.
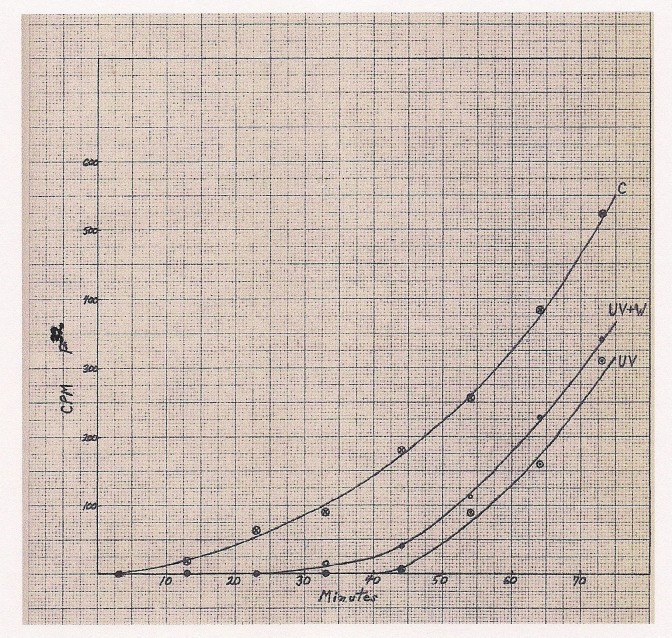
I started a research project on synchronous growth in bacterial cultures with Harold Morowitz, who was the most popular biophysics advisor, but after a few months, Pollard summoned me to his office to tell me that Morowitz was taking on too many students and that I would have to either switch to his lab or join Setlow’s group. Having enjoyed the spectroscopy course with Dick Setlow, the decision was a “no-brainer.” Setlow had been studying UV effects on enzyme activities and was becoming interested in extending his research to living cells. Reginald Deering, one year ahead of me in Setlow’s group, was measuring effects of UV on cell division in the bacterium, Escherichia coli [3]. Still a physicist at heart, I wanted to use UV to probe the inner workings of that cellular “black box” without breaking it open. Since the absorption spectra for the cellular macromolecules were well known, I wanted to obtain action spectra for cellular functions and to also determine how UV affected the synthesis of informational macromolecules such as DNA, RNA, and protein. Arthur Giese at Stanford University had used colorimetric assays to follow these syntheses in UV-irradiated bacteria over an 8-hour period [4]. I wanted to look more closely at what happened within the critical first hour after irradiation, so I developed an approach to resolve incorporated 32PO4 into DNA, RNA, and phospholipids to improve sensitivity of the analysis [5]. I confirmed that low dose UV, with an action spectrum implicating nucleic acids, caused a dose-dependent delay in the recovery of DNA synthesis and reduced rates of RNA and protein synthesis [6]. While completing my thesis in the summer of 1958, I mentored a high school student, John Buehler, in experiments revealing that visible light exposure, after the UV irradiation, shortened the lag in recovery of DNA synthesis (Figure 1). As an aside, I apologize for the poor quality of that hand-drawn figure from my PhD thesis and explain that in those years we typed our own theses and then prepared the required copies by the now-obsolete process of Thermofax. In my thesis, I stated: “The fact that DNA synthesis generally resumes, at a near normal rate, at some later time after irradiation would indicate that the process can be repaired. Visible light may just speed up this repair process. Since colony forming ability can also be photoreactivated after UV inactivation, there is evidence that the repair process can allow the cell to produce functional DNA again.” We documented our findings in a short paper [7]. Quoting from that paper: “It would appear that photoreactivation basically involves a repair of DNA integrity rather than a restoration of DNA synthesis,” since the recovery of protein synthesis and RNA synthesis, thought to be dependent upon DNA integrity, was similarly enhanced by photoreactivating light. However, it was really premature to talk about repair of DNA before we knew what kind of damage needed to be repaired.
Figure 1.
Photoreactivation of DNA synthesis. (From Philip Hanawalt’s PhD Thesis, Yale University, 1958). Time course of incorporation of 32P from inorganic phosphate into DNA in control culture of E.coli B (C); UV irradiated culture, after 150 ergs/mm2 at 265nm (UV); and culture exposed to visible light for 10 minutes beginning 10 minutes after UV irradiation (UV + W). Visible light was administered by G.E. Hg AH-4 lamp with lead glass and 2.5 percent CuCl2 filter to exclude wavelengths below 310nm and above 750nm, respectively, at intensity of 2.5 x 106 ergs/cm2/min. CPM: Counts per minute.
Postdoctoral Studies in Copenhagen and at Caltech
Upon completion of my thesis in late 1958, I headed off to the University of Copenhagen for a 2-year postdoctoral apprenticeship with Professor Ole Maaløe, an expert in synchronizing division cycles in bacterial cultures, with the intent to resume that earlier research interest from the Morowitz lab and to carry out an analysis of UV effects at different stages of the cell cycle. However, I temporarily discontinued my UV studies and focused upon the phenomenon of thymineless death, extending experiments I had carried out at Yale on unbalanced bacterial growth in the absence of thymidine nucleotides. I introduced the subject of thymineless death to Maaløe’s group and my research led unexpectedly to a procedure for synchronizing the DNA replication cycle in E. coli [8]. I also learned from another postdoc, Don Cummings, how to study DNA replication by density-labeling with the thymine analogue, 5-bromouracil, an approach that was soon to figure prominently in my studies on DNA repair [9].
It was another timely piece of good fortune that I decided to take a third postdoctoral year at the California Institute of Technology with Robert Sinsheimer. Shortly after my arrival in Pasadena in September 1960, a major breakthrough in nucleic acid photochemistry was reported: UV irradiation of thymine in frozen solution created cyclobutane dimers [10]. Immediately realizing the potential significance of this discovery, Professor Max Delbrück offered an exciting course in UV photobiology that reinvigorated my interest in the effects of UV on DNA replication. It was soon reported that UV generated dimers between adjacent pyrimidines in cellular DNA and furthermore that the mechanism of photoreactivation involved direct enzymatic reversal of the dimer linkage without breaking the DNA backbone. At Caltech, I learned more about density labeling to study DNA replication from Sinsheimer and from Matt Meselson, who was just completing his PhD thesis before taking an assistant professorship at Harvard [11].
In the meantime, Setlow was recruited from Yale to Oak Ridge National Laboratories by the Director of the Biology Division, Alexander Hollaender, to lead a research program on the photochemistry of DNA and its cellular processing. On the basis of the enhanced survival and reduced mutagenesis when UV irradiated cells were held in buffer for an hour before plating on nutrient agar, Evelyn Witkin and others had surmised that there must be a DNA repair process that operated in the dark. A mutant deficient in the putative process was needed, and Setlow’s Yale colleague, Ruth Hill, soon obtained the first UV-sensitive mutant of E. coli B. Setlow and his coworkers detected little recovery of DNA synthesis when Ruth Hill’s mutant was irradiated, but survival was still enhanced by photoreactivation after very low UV doses, so the mechanisms of photoreactivation and dark recovery were evidently different [12].
Discovery of Repair Replication at Stanford
Upon joining the faculty at Stanford University in late 1961 as Research Biophysicist and Lecturer, I returned to the problem of what UV did to DNA replication, now that we knew the principal photoproducts. I wanted to understand the behavior of replication forks upon encountering pyrimidine dimers, and I was hoping to catch a blocked replication fork at a dimer. Using density labeling with 5-bromouracil and radioactive labeling of newly-synthesized DNA, we were able to observe partially replicated DNA fragments in E. coli [13]. However, in samples from UV irradiated bacterial cultures, the density patterns of nascent DNA indicated that much of the observed synthesis was in very short stretches, too short to appreciably shift the density of the DNA fragments containing them [14]. I communicated these results to Setlow by phone and learned that he had just discovered that pyrimidine dimers in wild type cells, but not in Ruth Hill’s UV sensitive mutant, were released from the DNA into an acid soluble fraction. We speculated in discussion that my student, David Pettijohn, and I were detecting a patching step by which a process of repair replication might use the complementary DNA strand as template to fill the single-strand gaps remaining after the pyrimidine dimers had been removed. At about the same time, Paul Howard-Flanders in the Department of Therapeutic Radiology at Yale had isolated a number of UV-sensitive mutants from E. coli K12 strains, and he was able to show that these mutants were also deficient in removing pyrimidine dimers from their DNA. The seminal discovery of dimer excision was published by the Setlow and Howard-Flanders groups, as the first indication of an excision repair pathway [15,16]. Of course, the excision per se is not a repair event but only the first step, since it generates another lesion, the gap in one strand of the DNA. We carried out more controls, to then claim that we had discovered a non-conservative mode of repair replication, constituting the presumed patching step in the postulated excision-repair pathway [17]. I later showed that DNA containing the repair patches could undergo semiconservative replication with no remaining blockage [18].
Excision Repair Pathways in Bacteria and in Human Cells
Richard Boyce and Howard-Flanders at Yale also documented excision of lesions induced by mitomycin C in E. coli K12 strains, indicating some versatility of excision repair [19]. In a collaboration with Robert Haynes, I found a similar pattern of repair replication after nitrogen mustard exposure to that following UV, and we concluded that “it is not the precise nature of the base damage that is recognized, but rather some associated secondary structural alteration …” We speculated that “[s]uch a mechanism might even be able to detect accidental mispairing of bases after normal replication,” thus predicting the existence of a mismatch repair pathway [20]. Mismatch repair was reported by Wagner and Meselson a decade later [21] and yet another excision repair mode, termed base excision repair, was discovered by Tomas Lindahl [22].
At the same time that we were characterizing repair replication in bacteria, Robert Painter, at the Ames Research Laboratories near Stanford, was observing a peculiar phenomenon in which tritiated thymidine labeling revealed DNA synthesis in non S-phase human cells following UV irradiation, as detected in autoradiographs. Painter learned the density-labeling protocol from us and was then able to validate this “unscheduled DNA synthesis” as a measure of repair replication instead of adventitious events of genomic replication. James Cleaver, a postdoc in Painter’s lab then contributed a major discovery that propelled the infant DNA repair field into notoriety, when he discovered that cells from patients with the sun-sensitive, cancer prone hereditary disease xeroderma pigmentosum were deficient in carrying out repair replication [23]. Some years later, Richard Wood, upon completing a postdoc with Franklin Hutchinson at Yale, joined Tomas Lindahl at the Imperial Cancer Research Fund in England, where he developed the first mammalian cell free system for nucleotide excision repair, facilitating complementation analyses to learn the responsible enzymes and complexes [24].
The Ongoing History of DNA Repair
The rich history of the DNA repair field has been recounted by Errol Friedberg through interviews with many, if not most, of the early contributors [25]. He has also orchestrated a compendium that defines the rapidly progressing fields of DNA repair and mutagenesis up to 2006 [26]. I would conclude this essay with reflections on a few of the areas in which my laboratory has continued to contribute to the ongoing DNA repair story. Some of these are detailed in a recent autobiographical sketch that presents a more colorful and anecdotal treatment of my life in science [27]. Our discovery in the mid-1980s of transcription-coupled repair (TCR) led to the realization that a translocating RNA polymerase is a very sensitive detector of alterations in the template DNA strand that may be transparent to recognition by the global genomic repair pathway. It also led to the discovery that the sun-sensitive hereditary disease Cockayne syndrome (CS), characterized by severe developmental and neurological defects but without predisposition to cancer, is specifically deficient in TCR and then some years later that another rare disease with no cancers, UV sensitive syndrome (UVSS), is similarly deficient in TCR of UV induced damage, while lacking the serious developmental issues of CS. Although experiments with reporter genes had shown sensitivity to oxidative damage in CS, while UVSS behaved like wild type, recent results from comet-FISH studies reveal that CS and UVSS are equally deficient in TCR of the major oxidative lesion, 8-oxo-guanine [28-30]. Thus, oxidative damage sensitivity and developmental issues in CS remain to be understood, as does the remarkable lack of cancer predisposition in both of these diseases. There has been increased interest in TCR with reports that the mutation profiles in cells from tumors reveal a predictable strand bias, superimposed upon the mutagenic signature from the likely causal agent [31,32], and there is a strong inverse correlation between somatic mutation frequencies and gene expression levels (averaged over many cell lines) in cancers and in matched normal tissues [33]. A recent focus of our research interest is on the behavior of RNA polymerases encountering DNA sequences that are guanine rich and/or can form unusual secondary structures that can block transcription; the blockage could trigger apoptosis or may result in a gratuitous form of TCR that could be deleterious. A possible example relates to the behavior of transcription in repetitive DNA sequences, such as the CAG triplet repeats, which are amplified in Huntington’s disease [34,35]. We also hope to develop approaches in which we can make the very act of transcription lethal for particular classes of cells (e.g., in tumors) that are uniquely expressing particular genes for which we can target transcription blockage through generation of stable R-loops (RNA/DNA hybrids) [36,37]. Many exciting translational applications of our growing understanding of DNA repair are emerging since those pioneering discussions among Yale biophysicists half a century ago.
Acknowledgments
I express my deep appreciation to Richard Setlow for initiating me into a lifetime career in the fields of UV photobiology and DNA repair, to the many talented students and postdocs whose ideas and efforts have enriched my academic experience, and for steadfast support from my wife and collaborator, Graciela Spivak.
Abbreviations
- UV
ultraviolet light
- NER
nucleotide excision repair
- XP
xeroderma pigmentosum
- CS
Cockayne syndrome
- UVSS
UV sensitive syndrome
Funding sources
Research in Hanawalt’s laboratory at Stanford was initially supported by a contract with the Atomic Energy Commission and a grant (GM09901) from the Institute of General Medical Sciences, NIH. Currently the work is supported by grants (CA077712) from the National Cancer Institute and (ES018834) from the National Institute of Environmental Health Sciences, NIH.
References
- Setlow RB. My early days in photobiology with Philip Hanawalt. Mutat Res. 2005;577(1-2):4–8. doi: 10.1016/j.mrfmmm.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Schrödinger E. What is life? Cambridge: Cambridge University Press; 1944. [Google Scholar]
- Deering RA, Setlow RB. Inhibition of cell division of Escherichia coli by low doses of ultraviolet light. Science. 1957;126:397–398. doi: 10.1126/science.126.3270.397. [DOI] [PubMed] [Google Scholar]
- Iverson RM, Giese AC. Synthesis of nucleic acids in ultraviolet-treated Escherichia coli. Biochim Biophys Acta. 1957;25:62–68. doi: 10.1016/0006-3002(57)90417-1. [DOI] [PubMed] [Google Scholar]
- Hanawalt PC. Use of phosphorus-32 in microassay for nucleic acid synthesis in Escherichia coli. Science. 1959;130:386–387. doi: 10.1126/science.130.3372.386. [DOI] [PubMed] [Google Scholar]
- Hanawalt PC, Setlow RB. Effect of monochromatic UV on macromolecular synthesis in Escherichia coli. Biochim Biophys Acta. 1960;41:283–294. doi: 10.1016/0006-3002(60)90011-1. [DOI] [PubMed] [Google Scholar]
- Hanawalt PC, Buehler J. Photoreactivation of macromolecular synthesis in Escherichia coli. Biochim Biophys Acta. 1960;37:141–143. doi: 10.1016/0006-3002(60)90088-3. [DOI] [PubMed] [Google Scholar]
- Maaløe O, Hanawalt PC. Thymine deficiency and the normal DNA replication cycle I. J Mol Biol. 1961;3:144–155. doi: 10.1016/s0022-2836(61)80041-7. [DOI] [PubMed] [Google Scholar]
- Hanawalt PC, Maaløe O, Cummings D, Schaecter M. The normal DNA replication cycle II. J Mol Biol. 1961;3:156–165. doi: 10.1016/s0022-2836(61)80042-9. [DOI] [PubMed] [Google Scholar]
- Beukers R, Berends W. Isolation and identification of the irradiation product of thymine. Biochim Biophys Acta. 1960;41:550–551. doi: 10.1016/0006-3002(60)90063-9. [DOI] [PubMed] [Google Scholar]
- Hanawalt PC. Density matters: the semiconservative replication of DNA. Proc Natl Acad Sci USA. 2004;101:17889–17894. doi: 10.1073/pnas.0407539101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow RB, Swenson PA, Carrier WL. Thymine dimers and inhibition of DNA synthesis by ultraviolet irradiation of cells. Science. 1963;142:1464–1466. doi: 10.1126/science.142.3598.1464. [DOI] [PubMed] [Google Scholar]
- Hanawalt PC, Ray DS. Isolation of the growing point in the bacterial chromosome. Proc Natl Acad Sci USA. 1964;52:125–132. doi: 10.1073/pnas.52.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettijohn D, Hanawalt PC. Deoxyribonucleic acid replication in bacteria following ultraviolet irradiation. Biochim Biophys Acta. 1963;72:127–129. [PubMed] [Google Scholar]
- Setlow RB, Carrier WL. The disappearance of thymine dimers from DNA: An error-correcting mechanism. Proc Natl Acad Sci USA. 1964;51:226–231. doi: 10.1073/pnas.51.2.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce RP, Howard-Flanders P. Release of ultraviolet light-induced thymine dimers from DNA in E. coli K-12. Proc Natl Acad Sci USA. 1964;51:293–300. doi: 10.1073/pnas.51.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettijohn D, Hanawalt PC. Evidence for repair-replication of ultraviolet damaged DNA in Bacteria. J Mol Biol. 1964;9:395–410. doi: 10.1016/s0022-2836(64)80216-3. [DOI] [PubMed] [Google Scholar]
- Hanawalt PC. Normal replication of DNA after repair replication in bacteria. Nature. 1967;214:269–270. doi: 10.1038/214269a0. [DOI] [PubMed] [Google Scholar]
- Boyce RP, Howard-Flanders P. Genetic control of DNA breakdown and repair in E. coli K-12 treated with mitomycin C or ultraviolet light. Zeitschrift für Vererbungslehre. 1964;95:345–350. doi: 10.1007/BF01268667. [DOI] [PubMed] [Google Scholar]
- Hanawalt PC, Haynes R. Repair replication of DNA in bacteria: irrelevance of chemical nature of base defect. Biochem Biophys Res Commun. 1965;19:162–167. doi: 10.1016/0006-291x(65)90147-6. [DOI] [PubMed] [Google Scholar]
- Wagner R, Meselson M. Repair tracts in mismatched DNA heteroduplexes. Proc Natl Acad Sci USA. 1976;73(11):4135–4139. doi: 10.1073/pnas.73.11.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. New class of enzymes acting on damaged DNA. Nature. 1976;259:64–66. doi: 10.1038/259064a0. [DOI] [PubMed] [Google Scholar]
- Cleaver JE. Defective repair replication of DNA in xeroderma pigmentosum. Nature. 1968;218(5142):652–656. doi: 10.1038/218652a0. [DOI] [PubMed] [Google Scholar]
- Wood R, Robins P, Lindahl T. Complementation of the xeroderma pigmentosum DNA repair defect in cell-free extracts. Cell. 1988;53:97–106. doi: 10.1016/0092-8674(88)90491-6. [DOI] [PubMed] [Google Scholar]
- Friedberg EC. Correcting the Blueprint of Life: An Historical Account of the Discovery of DNA Repair Mechanisms. Cold Spring Harbor Press; 1997. [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA. DNA Repair and Mutagenesis (Second Edition) ASM Press; 2006. [Google Scholar]
- Hanawalt PC. Repairing DNA for 80 years: The timeline of my life. DNA Repair (Amst) 2012;11(5):452, e1-11. doi: 10.1016/j.dnarep.2011.05.007. [DOI] [PubMed] [Google Scholar]
- Hanawalt PC, Spivak G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- Vermeulen W, Fousteri M. Mammalian Transcription-Coupled Excision Repair. Cold Spring Harb Perspect Biol. 2013;5(8):a012625. doi: 10.1101/cshperspect.a012625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Hanawalt PC, Spivak G. Comet-FISH with strand-specific probes reveals transcription-coupled repair of 8-oxoGuanine in human cells. Nucleic Acids Res. 2013;41:7700–7712. doi: 10.1093/nar/gkt524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED. et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED. et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas-Rios V, Belotserkovskii BP, Hanawalt PC. DNA slip-outs cause RNA polymerase II arrest in vitro: potential implications for genetic instability. Nucleic Acids Res. 2011;39(17):7444–7454. doi: 10.1093/nar/gkr429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Wilson JH. Nucleotide excision repair, mismatch repair and R-loops modulate convergent transcription-induced cell death and repeat instability. PLoS One. 2012;7(10):e46807. doi: 10.1371/journal.pone.0046807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskii BP, Neil AJ, Saleh SS, Shin JHS, Mirkin SM, Hanawalt PC. Transcription blockage by homopurine DNA sequences: role of sequence composition and single-strand breaks. Nucleic Acids Res. 2013;41:1817–1828. doi: 10.1093/nar/gks1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskii BP, Mirkin SM, Hanawalt PC. DNA sequences that interfere with transcription: Implications for genome function and stability. Chem Rev. 2013 Aug 23; doi: 10.1021/cr400078y. Epub ahead of print. [DOI] [PubMed] [Google Scholar]