Abstract
Epicuticular wax forms a layer of hydrophobic material on plant aerial organs, which constitutes a protective barrier between the plant and its environment. We report here the identification of WIN1, an Arabidopsis thaliana ethylene response factor-type transcription factor, which can activate wax deposition in overexpressing plants. We constitutively expressed WIN1 in transgenic Arabidopsis plants, and found that leaf epidermal wax accumulation was up to 4.5-fold higher in these plants than in control plants. A significant increase was also found in stems. Interestingly, ≈50% of the additional wax could only be released by complete lipid extractions, suggesting that not all of the wax is superficial. Gene expression analysis indicated that a number of genes, such as CER1, KCS1, and CER2, which are known to be involved in wax biosynthesis, were induced in WIN1 overexpressors. This observation indicates that induction of wax accumulation in transgenic plants is probably mediated through an increase in the expression of genes encoding enzymes of the wax biosynthesis pathway.
Plant epidermal wax forms a hydrophobic layer covering aerial plant organs, which is deposited either outside of the cuticle (epicuticular wax), or within the cuticular matrix (intracuticular wax). In addition to repelling atmospheric water, epidermal wax plays an important role in water retention by the plant (1, 2). It has also been shown to be part of the plant's defense against biotic stresses. In particular, its chemical makeup and abundance are known to affect resistance to insects (3-5). Wax production is under developmental as well as environmental control: developmental stage, tissue type, and humidity changes, for example, are known to affect its accumulation (6-8).
The composition of epidermal wax varies from species to species. In Arabidopsis, wax components mostly consist of straight aliphatic chains 20-30 carbons in length, as well as primary and secondary alcohols, aldehydes, and ketones (8). The 18-carbon fatty acids from which wax lipids are derived go through cycles of elongation dependent on the action of elongation complexes thought to be associated with the endoplasmic reticulum (9-11). Two metabolic routes then contribute to the diversification of the long-chain products: the decarbonylation pathway, which leads predominantly to the production of alkanes from long-chain fatty acids and aldehydes, and the acyl reduction pathway, which leads to primary alcohols. Secondary alcohols and ketones result from the action of modifying enzymes on alkanes (12).
The identification of a number of Arabidopsis mutants with altered levels of stem glaucousness, eceriferum or cer (13, 14), or leaf glaucousness (15), has greatly helped to identify components of the pathway and its regulation. To date, 11 genes have been identified that are involved in wax biosynthesis or its regulation in plants. Seven of these genes, CER1, CUT1/CER6, KCS1, WAX2, and FIDDLEHEAD (FDH; all from Arabidopsis), GLOSSY1 (GL1), and GLOSSY8 (GL8; from maize), are predicted to encode enzymes or components of the secretory pathway (10, 11, 16-22). Another four encode regulatory proteins or proteins thought to play a regulatory role: GL2, GL15, CER2, and CER3 (23-27). However, even though some biosynthesis genes have been shown to be transcriptionally regulated (28, 29), genetic approaches have not yet led to the transcription factors that directly control their expression.
We report here the isolation, through a systematic functional genomics approach, of a transcriptional activator that can up-regulate wax production in Arabidopsis. We show that overexpression of this transcription factor is sufficient to increase wax load on aerial organs in transgenic plants, and that the wax accumulation phenotype is accompanied by an induction of several wax biosynthesis pathway genes.
Materials and Methods
Plant Materials and Genetic Transformation. All Arabidopsis plants were in the Col-0 genetic background.
Cloning of WIN1 and Construction of WIN1 Overexpressors. A fragment of the WIN1 ORF was initially detected in a Bacterial Artificial Chromosome end sequence (GenBank accession no. B30104). WIN1 contains one intron, and the full-length cDNA sequence of the gene was determined by 3′ and 5′ RACE. The complete genomic sequence of WIN1 was released by the Arabidopsis Genome Initiative, as part of bacterial artificial chromosome F9L1 (GenBank accession no. AC007591). For cloning, the WIN1 coding region was PCR-amplified from cDNA by using primer sequences GCACGCGTCGACATTACTCATCATCAAGTTCCTACTT and GGCTCTAGATAGGTACATATATATAAGCAAATAA. The resulting PCR product was cut with XbaI and SalI and was cloned into the binary vector pMen020 downstream of a double 35S enhancer, and upstream of the pea RBCS E9 terminator sequence (30). The resulting plasmid was introduced into Agrobacterium strain ABI, which was in turn used to transform Arabidopsis plants, by using the vacuum infiltration method (31). Transgenic plants were then selected on Murashige and Skoog medium containing 50 mg/liter kanamycin.
Scanning Electron Microscopy (SEM) Analysis. Fragments of fresh leaves were mounted on stubs and were coated with 15- to 20-Å grain-size gold particles by using a Bio-Rad E5400 sputter coater. The samples were then transferred to an Hitachi S-5000 scanning electron microscope for examination.
Transmission Electron Microscopy (TEM) Analysis. Small leaf pieces were fixed with 5% glutaraldehyde in 0.05 M sodium phosphate buffer for 4 h under vacuum, followed by 1 h at room temperature. Samples were postfixed in 1% buffered osmium tetroxide overnight at 4°C, and were then dehydrated in an ethanol series, beginning with 30% and ending with two rinses in 100%. They were then infiltrated and were polymerized in Spurr's epoxy resin. Resin blocks were sectioned by using a Leica Ultracut S microtome. Silver-gold sections (60-90 nm thick) were mounted on copper grids and were stained with 2% uranyl acetate and 0.5% lead citrate. Micrographs were made by using a Philips 410 transmission electron microscope.
Analysis of Wax Composition. Sample preparation: Epicuticular wax analysis. A total of 100-200 mg of fresh leaf or stem tissue from multiple T2 overexpressors was pooled and epicuticular wax was extracted into 5 ml of chloroform containing 100 μg of triacontane (Sigma) for 5 min at room temperature. The extracts were dried under a stream of nitrogen and were dissolved into 100 μl of N,O-bis(trimethylsilyl)trifluoroacetamide):trimethylchlorosilane(99:1, Sigma). The samples were then derivatized at 80°C for 1 h.
Total leaf wax analysis. Leaf lipids from 100-200 mg of fresh tissue were transmethylated for 1 h at 80°C in 1 M methanolic HCl as described (32). Fatty acid methyl esters and wax components were extracted into hexane, were dried under a stream of nitrogen, and were derivatized as above.
Gas chromatography. One microliter of each sample was injected into an HP5-MS column (HP 19091J-433) fitted to an HP6890 gas chromatograph connected to an HP5972 mass spectrometer (split ratio 1:1). The temperature profile was 150-250°C (12°C per min), followed by 250-300°C (4°C per min), back to 150°C (25°C per min).
Northern Analysis. RNA was extracted from leaves of T2-transgenic plants by using TRI reagent according to the manufacturer's protocol (Molecular Research Center, Cincinnati). Samples were run on formaldehyde gels, and 5 μg of total RNA was loaded per lane. The NorthernMax kit (Ambion) was used for Northern analysis. Radiolabeled probes were generated by random hexamer labeling of PCR products amplified by using gene-specific primers (see Table 3, which is published as supporting information on the PNAS web site).
RT-PCR. Total RNA was extracted from stems, roots, flowers, leaves, siliques, and whole-plate-grown seedlings. Stems, flowers, leaves, and siliques were of mixed stages from soil-grown plants. Roots were obtained from plants grown in tissue culture. Template for PCR was generated by reverse transcription from poly(A)+ RNA by using SuperScript reverse transcriptase (Invitrogen). cDNAs that were used for semiquantitative RT-PCR were normalized based on the intensity of PCR-amplified actin2 fragments obtained by using primers AGAGATTCAGATGCCCAGAAGTCTTGTTCC and AACGATTCCTGGACCTGCCTCATCATACTC. WIN1 mRNA was amplified by using primers GTCGCTGAGATTCGTCATCCTCTCTTGA and TGCAAAGCAACCTTTTCTTCCTCATCCA.
Microarray Analysis. RNA samples from lines 13 and 22, and wild-type controls generated as described above, were used for microarray analysis. Preparation of fragmented biotin-labeled cRNA, hybridizations of Arabidopsis genome arrays (Affymetrix, 510429), washes and scanning of the slides were performed according to the manufacturer's instructions.
Analysis of microarray data were performed by using the Rosetta Resolver gene expression data analysis system (Rosetta Biosoftware, Kirkland, WA), comparing the hybridizations from lines 13 and 22 with the common wild-type reference. Resolver uses an empirical intensity-based error model, specific for the Affymetrix microarray platform, to obtain a conservative estimate of signal variability (33, 34). Intensity profiles and ratiometric data are provided as Tables 4-8, which are published as supporting information on the PNAS web site. Affymetrix data files (CEL, DAT, and EXP) have been deposited at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/geo) under accession nos. GSM17238-GSM17240.
Results
WIN1 Is an Ethylene Response Factor (ERF)-Type Transcription Factor Causing a Glossy Leaf Phenotype in Arabidopsis Overexpressors. In an effort to functionally characterize the Arabidopsis complement of transcription factors, a genomic approach was undertaken. All gene sequences predicted to encode proteins sharing conserved domains with cognate transcription factors (35) were submitted to a functional screen by using transgenic plants constitutively expressing the genes (under the control of the CaMV 35S promoter), or knockout plants in which gene function was abolished. Knockout and overexpressor plants were analyzed through a battery of tests to detect possible changes in their morphology, physiology, and biochemistry. In the course of the screen, a characteristic and consistent phenotype was identified in transgenic lines overexpressing the full-length sequence of an ERF-type transcription factor, At1g15360 [the ERF family has also been referred to as EREBP (36)]. Leaves of these plants were strikingly glossier than those of control plants (Fig. 1). Because this phenotype is suggestive of a possible change in leaf epicuticular wax (and biochemical analyses indicated that the change corresponded to an increase in wax accumulation, see below), the transcription factor was termed WIN1, for wax inducer 1. Comparison of the predicted protein sequence with those of all other Arabidopsis ERF factors showed that WIN1 is most related to two other proteins, encoded by genes At5g11190 and At5g25390. At the amino acid sequence level, At5g11190 and At5g25390 are predicted to be 54% and 55% identical to WIN1 (Fig. 6, which is published as supporting information on the PNAS web site), respectively, with significant conservation beyond the conserved AP2 DNA-binding domain (43% and 44% identity, respectively). This level of sequence identity suggests that At5g11190 and At5g25390 might be related in function to WIN1 (see below).
Fig. 1.
Phenotype of WIN1 overexpressors. (a) Arabidopsis wild-type plant (Ecotype Columbia). (b) Class B 35S:WIN1 plant (line 6). (c) Class C plant (line 22). Arabidopsis plants overexpressing WIN1 show glossier leaves. Pictures were taken at the same magnification.
To gain insight into the biochemical nature of the glossy leaf phenotype caused by WIN1 overexpression, and the molecular mechanisms underlying it, we undertook a detailed study of 35S:WIN1 overexpressors. To facilitate the analysis, we defined three arbitrary classes of overexpressors: A, B, and C, with low, medium, and high levels of leaf glossiness, respectively, and focused our efforts on five transgenic lines representing the different classes (Table 1).
Table 1. Classification of 35S:WIN 1-transgenic lines.
| Phenotypic class | No. of independent transgenic lines | Lines described in this report | WIN 1 overexpression level (leaves) | Leaf glossiness | Overall morphology |
|---|---|---|---|---|---|
| A | 6 | 3 and 5 | Low | Low | Wild-type |
| B | 17 | 6 and 13 | Medium | Medium | Smaller than wild-type |
| C | 16 | 22 | High | High | Stunted |
| Silenced lines | 1 | 22-1* | Half wild-type levels | None | Wild-type |
Derived from line 22, identified among progenies with a wild-type morphological phenotype.
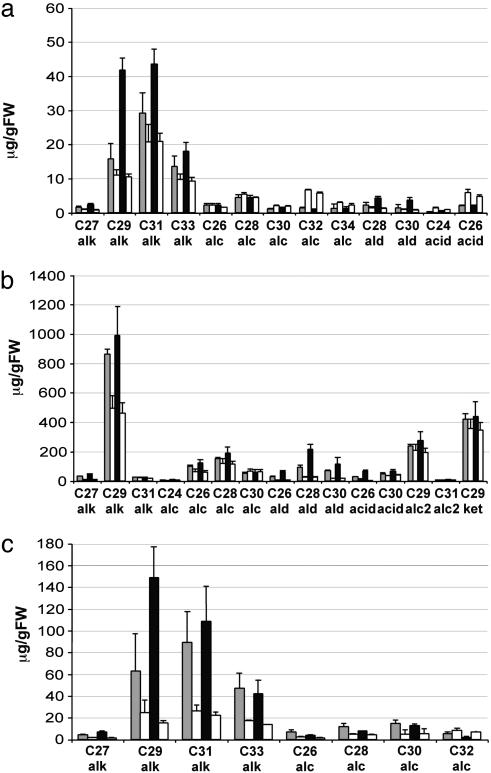
Leaves and Stems of Transgenic 35S:WIN1 Plants Show a Substantial Increase in Epicuticular Wax Accumulation. To confirm that the glossy leaf phenotype was caused by an alteration in epicuticular wax production, WIN1 overexpressors (line 22, class C) were examined by SEM. Whereas control Arabidopsis rosette leaves do not produce wax crystals, leaves of 35S:WIN1 plants display characteristic plate-like wax crystals that are typically 1-2 μm in length (Fig. 2). In an effort to further determine whether crystal production was the result of qualitative or quantitative changes in wax accumulation, rosette leaf wax of lines 6 (class B) and 22 (class C) were subjected to GC-MS analysis. As shown on Fig. 3a, there were major differences both in total wax content and wax composition between leaves of WIN1 overexpressors and of control plants. Average wax content, on a fresh weight basis, ranged between 1.1- and 2-fold wild-type values [line 6/control: 77.3/72.0 μg·grams of fresh weight (gfw)-1; line 22/control: 126.9/65.5 μg·gfw-1]. Increases in the most abundant wax species in wild-type leaves, such as hentriacontane (C31 alkane) and nonacosane (C29 alkane) accounted for most of the change in wax content (Fig. 3a).
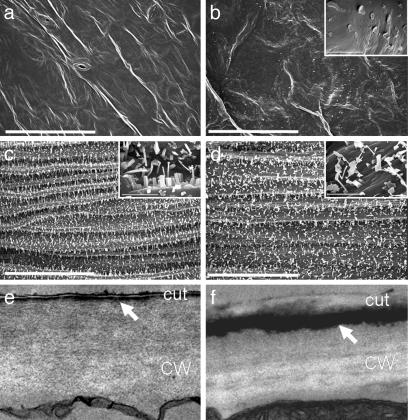
Fig. 2.
Electron microscopy analysis of 35S:WIN1 plants (line 22). (a and b) SEM images of leaves of wild-type (a) and 35S:WIN1 (b) plants. (b Inset) Plate-like wax crystals are observed in the 35S:WIN1 line. (c and d) SEM images of stems of control (c) and 35S:WIN1 (d) plants. (White bars in a-d, main image, 50 μm; white bars in Insets, 5 μm.) (c and d) TEM images of wild-type (e) and 35S:WIN1 (f) leaf epidermal cell sections. A thick layer of osmium dense material is observable in 35S:WIN1 epidermal cells underneath the epicuticular and peripheral cuticular layer (arrow). CW, cell wall; cut, cuticle. Images in e and f were taken at ×21,000 magnification.
Fig. 3.
Wax profile of selected tissues from transgenic 35S:WIN1 plants. (a) Leaf epicuticular wax composition. (b) Stem epicuticular wax composition. Leaf and stem wax constituents were extracted into chloroform, were TMS-derivatized, and were analyzed by GC-MS. (c) Quantitative analysis of total leaf wax. Wax components from methanolized leaf lipid extracts were analyzed by GC-MS after TMS derivatization. Each value for a wax constituent from a 35S:WIN1 line is followed by the corresponding value from control plants grown under the same conditions. All values are relative to fresh weight and represent the average of three independent experiments. Gray bars, line 6; black bars, line 22; and white bars, control line.
To evaluate the effect of WIN1 overexpression on a more major site of wax production in Arabidopsis, we also measured accumulation on stems. In contrast to leaves, abundance or shape of stem wax crystals did not appear to be significantly altered in the overexpressors (Fig. 2). However GC-MS analysis revealed that WIN1 also caused large increases in stem wax accumulation: on average, stems from transgenic plants produced between 1.5 (line 6/control: 2,178/1,459 μg·gfw-1) and 2 times (line 22/control: 2,699/1,385 μg·gfw-1) more wax per fresh weight than control extracts (Fig. 3b). Unlike the situation in leaves, however, only one of the major wax constituents contributed most to the increase (Fig. 3b).
WIN1 Overexpression Causes a Preferential Increase in Products of the Decarbonylation Pathway in Leaves but Not in Stems. In leaves of line 22, where the wax increase was highest, alkanes accounted for nearly all of the overall increase (line 22, Fig. 3a). As a result, alkanes, which make up ≈65% of the leaf wax in wild-type plants, constituted almost 85% of the wax in transgenic plants. In contrast, primary alcohol content was ≈35% lower than in wild-type leaves, with alcohols representing only 8% of the wax amount in line 22, as compared with ≈25% in control plants. Dotriacontanol (C32) was most affected, with a drop of almost 50%. Acids were also down in WIN1-overexpressing leaves, from 9% to 2% of the total, whereas aldehydes increased by ≈2.7-fold, from 3% to 6% (Fig. 3a). In stems, however, primary alcohols and alkanes both increased, by ≈2- and 1.5-fold, respectively. In fact, the relative contribution of the different compound classes to stem wax changed little, with the exception of aldehydes, which increased by ≈7-fold in strong overexpressors. However, as in leaves, alkanes, which more than doubled, strongly contributed to the increase in wax levels (Fig. 3b).
Cell Ultrastructure and Chemical Composition Suggest the Presence of an Internal Wax Layer in WIN1 Overexpressors. To characterize the pattern of epidermal wax deposition, leaf sections of transgenic and control plants were examined by TEM. Differences in the deposition of lipophilic compounds were apparent, starting at early stages of rosette leaf development. In particular, a thick layer of osmium-dense material was visible in the outer cell wall in transgenic but not in control plants. Because osmium staining does not discriminate between lipophilic compounds, we could not establish with certainty whether the dark layer was intra- or subcuticular (Fig. 2). In an effort to elucidate the chemical basis of this observation, we changed our extraction procedure to increase wax recovery. Increasing extraction time or solvent temperature did not increase the recovery of wax from transgenic plants (not shown). However, when leaf lipids were transmethylated by using boiling acidic methanol, more than twice as much wax was recovered from WIN1 overexpressors. In contrast, similar amounts were recovered from control plants (Fig. 3c). By using this method, leaf extracts from overexpressing lines contained between 2.7- (line 6/control: 243.5/91.8 μg·gfw-1) and 4.5-fold more wax than control extracts (line 22/control: 334/72.3 μg·gfw-1). Some of the minor wax components could not be measured by using this method. However, it was clear that although most compounds were more abundant in the methanolized extract, recovery of some appeared to be nearly complete in chloroform extracts. In line 22, nonacosane and triacontanol contents were more than triple that of chloroform extracts (3.6- and 9.3-fold respectively), whereas tritriacontane levels were equivalent (Fig. 3c). This finding may reflect compositional differences between internal and external wax layers in the epidermis of transgenic plants.
Constitutive WIN1 Overexpression Affects Plant Growth and Development. In addition to their wax phenotype, transgenic WIN1 overexpressors had characteristic alterations in growth and development. Typically, strong expressors were smaller in stature and slower growing than their wild-type counterparts (Fig. 1). They also tended to flower later, and often produced infertile flowers. As a result, they also produced less seeds than control plants. Strength of the morphological phenotype correlated with the level of WIN1 expression in the transgenic plants, as well as with the level of wax accumulation on aerial organs (Table 1 and Fig. 1).
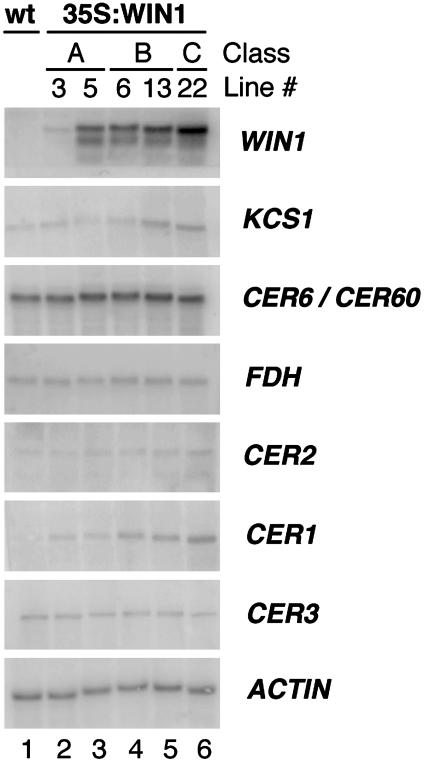
Genes Encoding Cognate Wax Biosynthetic Enzymes Are Induced in WIN1 Overexpressors. To determine the effects of WIN1 on gene expression, Northern analysis was performed by using total RNA isolated from leaves of overexpressing lines 3, 5, 6, 13, and 22. As expected, the level of WIN1 expression correlated with the severity of the morphological phenotype, line 22 (a class C overexpressor) presenting the highest levels of WIN1 expression (Fig. 4). Probes were generated from a set of genes known to be implicated in epidermal wax biosynthesis: CUT1/CER6 (11, 17), KCS1 (10), CER1 (16), CER2 (24, 26), CER3 (25), and FDH (20, 21). Among these genes, CER1 presented the most striking change in gene expression: its transcript level was clearly correlated with the strength of WIN1 overexpression. In addition to CER1, KCS1, which is predicted to encode a ketoacyl-CoA synthase involved in wax biosynthesis, and CER2, a presumed regulatory protein, also appeared to be up-regulated in overexpressing plants, although to a lesser extent. CER2 expression, in particular, changed only slightly, although we found that the change was significant (see below). In contrast, we could not detect any effect of WIN1 overexpression on CER6/CUT1 (and/or likely crosshybridizing CER60), CER3, and FDH transcript accumulation (Fig. 4).
Fig. 4.
Expression of genes encoding wax biosynthetic enzymes in 35S:WIN1-transgenic lines. Northern analysis of 35S:WIN1 lines 3 and 5 (both class A), 6 and 13 (class B), and 22 (class C). Probes detected WIN1, KCS1, CER6/CUT1, and CER60 (two genes highly related in sequence, which could not be distinguished), FDH, CER2, CER1, and CER3; actin was used as control. Higher levels of CER1 and KCS1 transcripts are detected in the transgenic lines.
To evaluate the effect of WIN1 overexpression on the expression of other genes, the transcript profiles of lines 13 and 22 were compared with that of wild-type plants by using a high-density Affymetrix oligonucleotide array that represents ≈8,200 genes. Samples for hybridization were derived from leaf RNA. The microarray data were analyzed by using the Rosetta Resolver system, using as filtering criteria for differential gene expression calls a P value cutoff of 0.001 and a fold-change cutoff of 2. With these criteria, 18 genes were found to be up-regulated in both lines, and 11 genes to be down-regulated. Among those, we identified genes with a known or likely involvement in wax biosynthesis. In particular, we found that CER1, CER2, and KCS1 were up-regulated in the overexpressors, in agreement with our Northern analysis, a finding that provided independent validation of the array results. Interestingly, a putative fatty acid elongase was significantly down-regulated in both 35S:WIN1 lines, suggesting that some of the components of the wax biosynthesis pathway may be differentially regulated in WIN1 overexpressors. Consistent with a change in lipid flux, we also identified a number of induced genes likely associated with lipid biosynthesis or catabolism (Table 2). The analysis also uncovered a number of genes with a likely involvement in cell wall metabolism or plant defense (see Table 9, which is published as supporting information on the PNAS web site).
Table 2. Relative expression of genes involved in lipid metabolism in 35S:WIN1 and control lines.
| Fold change
|
|||
|---|---|---|---|
| Locus | Annotation | Line 13 | Line 22 |
| At1G02205 | CER1 protein | 3.63 | 10 |
| At2G41540 | Glycerol-3-phosphate dehydrogenase | 7.33 | 8.22 |
| At2G04570 | GDSL-motif lipase/hydrolase protein | 13.67 | 7.78 |
| At2G38110 | Phospholipid/glycerol acyltransferase family | 3.25 | 6.19 |
| At4G24510 | CER2 | 2.09 | 4.23 |
| At1G01120 | Fatty acid elongase 3-ketoacyl-CoA synthase 1 | 3.71 | 3.62 |
| At4G14440 | Enoyl-CoA hydratase/isomerase family | 3.44 | 3.28 |
| At2G15090 | Fatty acid elongase 1, putative | −3.75 | −20.89 |
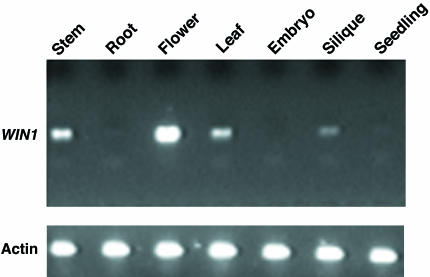
WIN1 Is Preferentially Expressed in Floral Tissues. To determine the site of expression of WIN1, we used semiquantitative RT-PCR to measure its expression levels in a variety of tissues. As shown in Fig. 5, WIN1 is expressed at higher levels in flowers than in any other tissues. Expression was also detectable, although at lower levels, in stems, siliques, and rosette leaves. WIN1 transcript was not detectable in roots or embryos by using our approach, and appeared to be very weakly expressed in germinating seedlings.
Fig. 5.
Analysis of WIN1 tissue expression. WIN1 expression was determined by semiquantitative RT-PCR in a variety of tissues from wild-type Columbia plants (Upper). RT-PCR was also performed with actin primers as control (Lower). WIN1 RT-PCR products are shown after 36 PCR cycles, and actin products are shown after 28 PCR cycles.
Discussion
During the course of a large-scale functional genomics approach targeting Arabidopsis transcription factors, we identified a gene encoding an ERF-type protein, WIN1, which caused a significant increase in wax accumulation when overexpressed in transgenic Arabidopsis plants. This phenotype was accompanied by an increase in the expression of genes implicated in wax biosynthesis.
The large increase in total amount of wax in WIN1 overexpressors is consistent with an increase in flux through the wax biosynthetic pathway. Furthermore, the disproportionate buildup, in leaves, of long-chain alkanes, in contrast with the reduction in primary alcohol levels, suggests a preferential increase in flux through the decarbonylation pathway, to the detriment of the acyl reduction pathway. Stems, in contrast with leaves, did not show a measurable reduction in primary alcohol accumulation. However, the relative increase in alkane content was the largest seen for any stem wax constituent, which also suggests the preferential production of alkanes (Fig. 7, which is published as supporting information on the PNAS web site). It was surprising to discover that leaf glossiness was the result of increased, rather than decreased wax accumulation, as in the wax mutants (15). However, the change in leaf aspect may be attributable to changes in light reflection/refraction due to the presence of wax crystals and/or to wax compositional modifications in the overexpressor. In stems, the effect of increased accumulation was not obvious on the wax crystal shape or abundance. It is possible that such an increase translates into higher crystal volumes and abundance, which may not have been easily detected by SEM analysis.
TEM and chemical analyses suggested the presence of an internal layer of wax in the epidermis of WIN1 overexpressors, possibly in the outer cell wall. Although wax accumulation in cell layers internal to the epidermis is a possibility, we could not detect it in our TEM analysis or by using lipophilic dyes (not shown). The epidermal wax buildup suggested by our observations may occur as a result of wax compounds being synthesized more rapidly than they can transit through the cuticular matrix. We cannot rule out, however, that wax is not the only cause of observed ultrastructural changes, and that buildup of cuticular material also occurs.
The wax phenotype of WIN1 overexpressors was consistent with observed differences in gene expression as compared with control plants. In particular, there was a significant increase in the expression of CER1, KCS1, and CER2. The fact that CER1 was one of the most highly induced genes in WIN1 overexpressors, together with the massive increase in alkane accumulation in these plants, supports the hypothesis that CER1 encodes a fatty aldehyde decarbonylase or, possibly, a protein involved in the selective secretion of alkanes from epidermal cells. The increase in expression of KCS1 is also consistent with an increase in pathway flux. Interestingly, microarray analysis revealed the induction, in overexpressing lines, of a number of genes involved in lipid metabolism. This finding suggests that increasing wax biosynthesis may necessitate redirecting intracellular lipid fluxes.
The fact that wax accumulation phenotypes were the result of WIN1 overexpression raises the question of the true biological role of the WIN1 protein. Unfortunately, we were unable to identify knockout lines in public collections, or in our own collection, despite screening >100,000 T-DNA lines. Also, we could not obtain fully silenced transgenic lines. However, several observations come in support of a native role of WIN1 in wax biosynthesis. First, the gene is expressed in tissues where wax production is most significant. Second, several wax biosynthesis pathway genes are very strongly activated by WIN1, and up-regulation occurs both in weak and in strong lines, suggesting that wax accumulation is not just a pleiotropic effect of strong WIN1 overexpression. Third, few metabolic mutants have been described to date, despite the large cumulative number of gain- and loss-of-function transcription factor mutants generated, which argues against metabolic effects such as wax accumulation being only a side effect of WIN1 overexpression (37-42). We have recently obtained preliminary biochemical and phenotypic evidence that overexpression of WIN1 paralogs At5g11190 and At5g25390 also causes an increase in leaf glossiness and wax accumulation on leaves and stems (data not shown). The fact that these three similar genes confer a striking wax phenotype that is unmatched by other lines in a screen of overexpressors for >1,500 transcription factors (including all ERF-type factors) strongly suggests, in our opinion, a biological role for WIN1 and its paralogs in the regulation of Arabidopsis wax biosynthesis.
It should be pointed out, however, that our microarray experiments also suggest that other pathways may also be affected in WIN1 overexpressors. Because WIN expression was constitutive in these plants, one cannot yet conclude as to whether these changes were the direct consequence of WIN1 overexpression. Combining inducible expression of WIN1 with genome-wide gene expression profiling and other analyses should be helpful to separate primary from secondary effects.
High levels of WIN1 overexpression had deleterious effects on plant growth and development. Although such effects could, in theory, be caused by altered levels and/or composition of wax, they may instead be a pleiotropic effect of high levels, in particular cell types, of a strong transcriptional activator. In fact, high expression of genes encoding other transcription factors, including ERF-type factors, has been reported to also cause stunting (43). If high constitutive WIN1 expression is indeed the cause of these phenotypic alterations, judicial temporal and/or tissue-specific expression of the transgene (for example under the control of an epidermal promoter) may help minimize these effects.
In conclusion, WIN1 represents, to our knowledge, the first example of a transcriptional activator affecting the expression of a lipid pathway in transgenic plants. We are optimistic that the manipulation of WIN1 levels will lead to a better understanding of wax biosynthesis and the associated secretory mechanisms, as well as provide a connecting thread between metabolism and differentiation in epidermal cells.
Supplementary Material
Acknowledgments
We thank Dr. R. Creelman for technical assistance with wax analysis, and Dr. Yan Liu, Mrs. Ning Sun, and Yuka Schwartz for experimental assistance. This work was supported by the Monsanto Company.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TEM, transmission electron microscopy; SEM, scanning electron microscopy; gfw, grams of fresh weight; ERF, ethylene response factor.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY378101).
References
- 1.Riederer, M. & Schreiber, L. (2001) J. Exp. Bot. 52, 2023-2032. [DOI] [PubMed] [Google Scholar]
- 2.Kerstiens, G. (1996) J. Exp. Bot. 47, 1813-1832. [Google Scholar]
- 3.Eigenbrode, S. D. & Espelie, K. E. (1995) Annu. Rev. Entomol. 40, 171-194. [Google Scholar]
- 4.Belding, R. D., Sutton, T. B., Blankenship, S. M. & Young, E. (2000) Plant Dis. 84, 767-772. [DOI] [PubMed] [Google Scholar]
- 5.Marcell, L. M. & Beattie, G. A. (2002) Mol. Plant-Microbe Interact. 15, 1236-1244. [DOI] [PubMed] [Google Scholar]
- 6.Jenks, M. A., Tuttle, H. A. & Feldmann, K. A. (1996) Phytochemistry 42, 29-34. [Google Scholar]
- 7.Jetter, R. & Schaffer, S. (2001) Plant Physiol. 126, 1725-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lemieux, B., Koornneef, M. & Feldmann, K. A. (1994) in Arabidopsis, eds. Meyerowitz, E. M. & Somerville, C. R. (Cold Spring Harbor Lab. Press, Plainview, NY), Vol. 27, pp. 1031-1047. [Google Scholar]
- 9.Evenson, K. J. & Postbeittenmiller, D. (1995) Plant Physiol. 109, 707-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Todd, J., Post-Beittenmiller, D. & Jaworski, J. G. (1999) Plant J. 17, 119-130. [DOI] [PubMed] [Google Scholar]
- 11.Millar, A. A., Clemens, S., Zachgo, S., Giblin, E. M., Taylor, D. C. & Kunst, L. (1999) Plant Cell 11, 825-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenks, M. A., Eigenbrode, S. D. & Lemieux, B. (2002) in The Arabidopsis Book, eds. Somerville, C. R. & Meyerowitz, E. M. (Am. Soc. Plant Biol., Rockville, MD).
- 13.Koornneef, M., Hanhart, C. J. & Thiel, F. (1989) J. Hered. 80, 118-122. [Google Scholar]
- 14.Mcnevin, J. P., Woodward, W., Hannoufa, A., Feldmann, K. A. & Lemieux, B. (1993) Genome 36, 610-618. [DOI] [PubMed] [Google Scholar]
- 15.Jenks, M. A., Rashotte, A. M., Tuttle, H. A. & Feldmann, K. A. (1996) Plant Physiol. 110, 377-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aarts, M. G. M., Keijzer, C. J., Stiekema, W. J. & Pereira, A. (1995) Plant Cell 7, 2115-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiebig, A., Mayfield, J. A., Miley, N. L., Chau, S., Fischer, R. L. & Preuss, D. (2000) Plant Cell 12, 2001-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, X., Goodwin, S. M., Boroff, V. L., Liu, X. & Jenks, M. A. (2003) Plant Cell 15, 1170-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu, X. J., Dietrich, C. R., Delledonne, M., Xia, Y. J., Wen, T. J., Robertson, D. S., Nikolau, B. J. & Schnable, P. S. (1997) Plant Physiol. 115, 501-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yephremov, A., Wisman, E., Huijser, P., Huijser, C., Wellesen, K. & Saedler, H. (1999) Plant Cell 11, 2187-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pruitt, R. E., Vielle-Calzada, J. P., Ploense, S. E., Grossniklaus, U. & Lolle, S. J. (2000) Proc. Natl. Acad. Sci. USA 97, 1311-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen, J. D., Pyee, J., Xia, Y., Wen, T. J., Robertson, D. S., Kolattukudy, P. E., Nikolau, B. J. & Schnable, P. S. (1997) Plant Physiol. 113, 1091-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tacke, E., Korfhage, C., Michel, D., Maddaloni, M., Motto, M., Lanzini, S., Salamini, F. & Doring, H. P. (1995) Plant J. 8, 907-917. [DOI] [PubMed] [Google Scholar]
- 24.Negruk, V., Yang, P., Subramanian, M., McNevin, J. P. & Lemieux, B. (1996) Plant J. 9, 137-145. [DOI] [PubMed] [Google Scholar]
- 25.Hannoufa, A., Negruk, V., Eisner, G. & Lemieux, B. (1996) Plant J. 10, 459-467. [DOI] [PubMed] [Google Scholar]
- 26.Xia, Y. J., Nicolau, B. J. & Schnable, P. S. (1996) Plant Cell 8, 1291-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moose, S. P. & Sisco, P. H. (1996) Genes Dev. 10, 3018-3027. [DOI] [PubMed] [Google Scholar]
- 28.Xia, Y. J., Nikolau, B. J. & Schnable, P. S. (1997) Plant Physiol. 115, 925-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunst, L., Clemens, S. & Hooker, T. (2000) Biochem. Soc. Trans. 28, 651-654. [PubMed] [Google Scholar]
- 30.Fluhr, R., Moses, P., Morelli, G., Coruzzi, G. & Chua, N. H. (1986) EMBO J. 5, 2063-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bechtold, N., Ellis, J. & Pelletier, G. (1993) C. R. Acad. Sci. Ser. III 316, 1194-1199. [Google Scholar]
- 32.Browse, J., Warwick, N., Somerville, C. R. & Slack, C. R. (1986) Biochem. J. 235, 25-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajagopalan, D. (2003) Bioinformatics 19, 1469-1476. [DOI] [PubMed] [Google Scholar]
- 34.Stoughton, R. S. & Dai, H. (2002) U.S. Patent 6,351,712.
- 35.Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., Adam, L., Pineda, O., Ratcliffe, O. J., Samaha, R. R., et al. (2000) Science 290, 2105-2110. [DOI] [PubMed] [Google Scholar]
- 36.Riechmann, J. L. & Meyerowitz, E. M. (1998) Biol. Chem. 379, 633-646. [DOI] [PubMed] [Google Scholar]
- 37.Bender, J. & Fink, G. R. (1998) Proc. Natl. Acad. Sci. USA 95, 5655-5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borevitz, J. O., Xia, Y., Blount, J., Dixon, R. A. & Lamb, C. (2000) Plant Cell 12, 2383-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grotewold, E., Drummond, B. J., Bowen, B. & Peterson, T. (1994) Cell 76, 543-553. [DOI] [PubMed] [Google Scholar]
- 40.Ludwig, S. R., Habera, L. F., Dellaporta, S. L. & Wessler, S. R. (1989) Proc. Natl. Acad. Sci. USA 86, 7092-7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nesi, N., Jond, C., Debeaujon, I., Caboche, M. & Lepiniec, L. (2001) Plant Cell 13, 2099-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Fits, L. & Memelink, J. (2000) Science 289, 295-297. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, J. Z. (2003) Curr. Opin. Plant Biol. 6, 430-440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.