Abstract
Control of chromatin structure is crucial for multicellular development and regulation of cell differentiation. The CHD (chromodomain-helicase-DNA binding) protein family is one of the major ATP-dependent, chromatin remodeling factors that regulate nucleosome positioning and access of transcription factors and RNA polymerase to the eukaryotic genome. There are three mammalian CHD subfamilies and their impaired functions are associated with several human diseases. Here, we identify three CHD orthologs (ChdA, ChdB and ChdC) in Dictyostelium discoideum. These CHDs are expressed throughout development, but with unique patterns. Null mutants lacking each CHD have distinct phenotypes that reflect their expression patterns and suggest functional specificity. Accordingly, using genome-wide (RNA-seq) transcriptome profiling for each null strain, we show that the different CHDs regulate distinct gene sets during both growth and development. ChdC is an apparent ortholog of the mammalian Class III CHD group that is associated with the human CHARGE syndrome, and GO analyses of aberrant gene expression in chdC nulls suggest defects in both cell-autonomous and non-autonomous signaling, which have been confirmed through analyses of chdC nulls developed in pure populations or with low levels of wild-type cells. This study provides novel insight into the broad function of CHDs in the regulation development and disease, through chromatin-mediated changes in directed gene expression.
Keywords: SNF2, Transcriptome profiling, RNA-seq, Chemotaxis, Growth, Differentiation
INTRODUCTION
The packaging of DNA into chromatin physically limits access of transcription factors and other elements of the transcription machinery to their target genes. Throughout development, changes in chromatin organization are exploited to control gene expression. In particular, alterations in nucleosome positioning along genomic DNA, termed chromatin remodeling, impact transcriptional activity, both positively and negatively (Ho and Crabtree, 2010; Euskirchen et al., 2012). Importantly, as nucleosome positions are preserved during DNA replication, inherited differences in positioning can alter multicellular development or cellular properties during disease states.
ATP-dependent chromatin remodeling proteins use the energy of ATP hydrolysis to physically change the interaction between DNA and nucleosomes, which can promote or limit DNA access to a variety of regulators (Hopfner et al., 2012). The archetypal ATP-dependent chromatin remodeler was discovered in yeast where mutations in a single yeast gene, designated SWI2/SNF2, lead to aberrant mating-type switching (swi2) (Breeden and Nasmyth, 1987) and glucose repression (snf2) (Abrams et al., 1986). SWI2/SNF2 has a DNA-dependent (helicase-like) ATPase that is part of a large multiprotein complex. The Drosophila ortholog of SWI2/SNF2, Brahma (Brm), regulates cell fate by acting as a transcriptional co-activator of homeotic gene expression in the Antennapedia and Bithorax complexes (Tamkun et al., 1992), and the mammalian ortholog BRG1 (SMARCA4 - Mouse Genome Informatics), Brahma-related gene 1, is implicated in T-cell development, ES cell differentiation and tumor suppression (Hargreaves and Crabtree, 2011).
Other chromatin remodeling families have been identified that share the ATPase motif, but differ in other protein domains (Clapier and Cairns, 2009; Eisen et al., 1995; Ryan and Owen-Hughes, 2011). These include ISWI (imitation switch), INO80 (inositol requiring 80) and CHD (chromodomain, helicase, DNA binding) proteins (Ho and Crabtree, 2010). The first member of the CHD family was identified in mice (Delmas et al., 1993), but CHD family members were subsequently described in a variety of eukaryotic organisms, including humans, Drosophila, Saccharomyces cerevisiae, C. elegans and Arabidopsis (Flanagan et al., 2007). The human CHD family comprises nine members, CHD1-9, divided into three subfamilies - CHD1-2 (class I), CHD3-5 (class II) and CHD6-9 (class III) - based upon their chromodomain types and additional motif features (Yap and Zhou, 2011).
Functionally, the CHD proteins play important roles in the development of multicellular organisms. CHD1 is essential for maintaining pluripopotency of mouse ES cells (Gaspar-Maia et al., 2009), and Drosophila Chd1 nulls are both male and female sterile (McDaniel et al., 2008). Loss of Chd2 in mice causes embryonic lethality (Marfella et al., 2006a) and conditional inactivation of CHD4 in hematopoietic cells impairs their differentiation and development (Williams et al., 2004). Haploinsufficient mutations of CHD7 are observed in 60-80% of cases of CHARGE syndrome (Janssen et al., 2012; Lalani et al., 2006; Layman et al., 2010; Zentner et al., 2010), a severe developmental disorder in humans characterized by coloboma of the eye, heart defects, atresia of the choanae, retardation of growth, and genital and ear abnormalities (Pagon et al., 1981). Chd7+/- mice recapitulate some of the human symptoms, and Chd7-/- mice are embryonic lethal (E10.5) (Bosman et al., 2005; Hurd et al., 2007). Wild-type CHD7 can direct nucleosome sliding in vitro, and CHD7 proteins that recapitulate CHARGE mutations are significantly impaired in this activity (Bouazoune and Kingston, 2012). CHD7 is suggested to function on tissue-specific genes that regulate development and differentiation (Schnetz et al., 2009; Schnetz et al., 2010). CHD8 also has an important role in development, is highly expressed during early embryogenesis and appears to modulate WNT/β-catenin signaling through interaction and downregulation of response genes (Nishiyama et al., 2012; Thompson et al., 2008). Other CHD functions are associated with human diseases, including some neuroblastomas (CHD5), Hodgkin’s lymphoma (CHD3) (Lemos et al., 2003) and small-cell lung cancers (CHD7) (Pleasance et al., 2010).
It is not yet clear how the different CHDs may cause these multiple, diverse and distinct patterns of developmental deficiencies. Broad investigations of the developmental targets of the individual CHD proteins and their effect on cell behavior are required to establish the connectivity of CHD function with gene expression, developmental phenotypes and disease states. We have now identified the CHD orthologs of Dictyostelium and associate loss-of-function mutations with distinct developmental phenotypes and altered gene expression profiles.
Dictyostelium grow as single-celled organisms, in which cellular growth and division are dependent on an adequate food supply (McMains et al., 2008). Nutrient depletion triggers a developmental program to ultimately form a terminally differentiated, multicellular structure: the fruiting body. Development occurs in distinct stages as cells first sense starvation and high population density, and then aggregate via a chemotactic response to secreted cAMP signals to form the initial multicellular organism. These multicellular mounds then differentiate into patterned prespore and prestalk cell populations, before terminal differentiation into structures comprising mature spore and stalk cells (Kessin, 2001).
Using genome-wide transcript profiling (RNA-seq) during growth and development to assay gene expression in Dictyostelium (Loomis and Shaulsky, 2011; Parikh et al., 2010) that individually lack the different CHD family members, we correlate altered transcription profiles with the developmental phenotypes of the different chd nulls. We conclude that the different CHDs act to control diverse aspects of Dictyostelium development, perhaps by targeting distinct sets of genetic loci.
ChdC in Dictyostelium is an apparent ortholog of the mammalian Class III CHD group that is associated with the human CHARGE syndrome. Similar to CHD7 mutations in humans (Zentner et al., 2010), chdC nulls have the strongest phenotype in Dictyostelium, with impaired growth, chemotaxis, cell differentiation and developmental arrest. Analyses following transcriptome profiling indicate defects in both cell-autonomous and non-autonomous functions, phenotypes confirmed by chimeric development of chdC nulls with wild-type cells. These data yield novel insight into the broad function of CHDs, with particular relevance to the Class III CHD group and their association with highly severe genetic syndromes in humans.
RESULTS
Dictyostelium possess three CHD proteins
We initiated separate insertional (REMI) mutagenic screens (Kuspa, 2006; Artemenko et al., 2011) to identify Dictyostelium strains that exhibited defects in chemotaxis to cAMP or that arrested development. An identical CHD-encoding gene was identified in these distinct screens as a novel genetic modifier of both chemotaxis and development. Extended searches for related sequences in the Dictyostelium genome identified two additional CHD proteins (supplementary material Fig. S1). We have termed the Dictyostelium CHD genes ChdA (DDB_G0284171), ChdB (DDB_G0280705) and ChdC (DDB_G0293012); ChdC was the gene found in the original mutagenic screens. Our sequence analyses also identified related SWI2/SNF2-type genes (Ryan and Owen-Hughes, 2011) in Dictyostelium, encoding members of the bona fide SWI2/SNF2 (snf2; DDB_G0271052 and DDB_G0285205), ISWI (isw; DDB_G0292948), SWR (swr1; DDB_G0267638) and INO80 (ino80; DDB_G0292358) chromatin remodeling families (see supplementary material Figs S1, S2). All of the encoded proteins possess the DNA-dependent ATPase domain and a DNA-binding motif, but differ in overall organization (supplementary material Fig. S2); the tandem chromodomains are a defining moiety for the CHDs (Blus et al., 2011). More distantly related SWI2/SNF2 members in Dictyostelium were also noted, including the Mot1 (modifier of transcription) and Rad54 families (supplementary material Fig. S1).
Dictyostelium ChdA shows the strongest similarity to human CHD subfamily I, clustering with human CHD1/2 and other species Chd1-subtypes through the entire protein sequence (Ryan et al., 2011), including the C-terminal DUF4208 (supplementary material Figs S1, S2). Dictyostelium ChdB possesses weak C-terminal CHDCT2 domains, which are generally characteristic of CHD subfamily II (Ryan and Owen-Hughes, 2011); however, ChdB lacks the N-terminal PHD finger present in most subfamily II members and globally aligns better with human CHD6-9 proteins (supplementary material Fig. S1). ChdC (DDB_G0293012) is the largest Dictyostelium CHD, and clusters most strongly with the CHD6-9, subfamily III group (supplementary material Figs S1, S2). Although ChdC has C-terminal SANT and SLIDE DBD motifs characteristic of other CHD III members (Ryan and Owen-Hughes, 2011), it lacks the conserved BRK site. Motif prediction in ChdC is complicated by the large number of homopolymeric residues embedded in its long C-terminal region. In addition, although BRK domains are of unknown function, they appear to be unique to the metazoa; thus, although the yeast SWI2/SNF2 and metazoan Brm/BRG are considered to be orthologs and functionally equivalent, only Brm/BRG proteins possess BRK domains (Ryan and Owen-Hughes, 2011). ChdC may uniquely possess a SUMO-like amino acid sequence of unknown function. Collectively, the individual CHDs of Dictyostelium would seem to embody different CHD types.
Developmental expression of ChdA, ChdB and ChdC
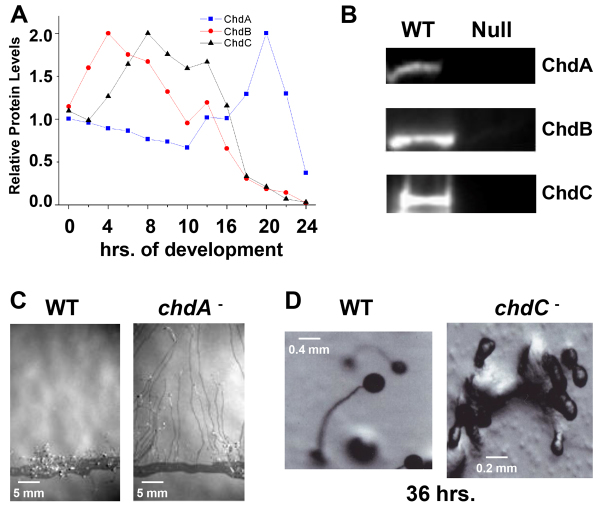
We examined developmental protein expression patterns of each CHD by immunoblotting, using protein-specific antibodies (Fig. 1A). Each CHD is detected during growth and the early stages of development, but they exhibit large expression differences following multicellular aggregation. ChdA has low relative expression during aggregation and early stages of multicellular development, but has increased abundance during terminal differentiation, after 16 hours. By contrast, ChdB shows peak abundance at 4 hours of development, the mid-stage of cellular aggregation (Fig. 1A). ChdB abundance then declines and is undetectable during late stages of development. Finally, ChdC shows peak abundance between 8 and 10 hours of development, as cells complete aggregate formation and initiate multicellular development. Again, as seen for ChdB, ChdC is undetected during late stages of development; thus, ChdA predominates at terminal differentiation.
Fig. 1.
The three CHD proteins of Dictyostelium have distinct developmental roles. (A) Whole-cell extracts were prepared from developing Dictyostelium at 2-hour intervals and ChdA, ChdB and ChdC proteins levels measured by immunoblot assay. Quantified data are expressed as normalized protein levels relative to expression at 0 hour. (B) Whole-cell extracts were prepared from growing wild-type, chdA-null, chdB-null and chdC-null cells, and ChdA, ChdB and ChdC proteins expression measured by immunoblot assay. (C) Wild-type and chdA-null cells were plated for development at the edge of nitrocellulose membranes and pseudoplasmodia migration assayed by exposure to a directional light source. (D) Wild-type and chdC-null cells were terminally developed. chdC nulls arrest development at multi-cell aggregation.
Major phenotyopic developmental defects in cells lacking chdA, chdB or chdC
To investigate CHD functions in Dictyostelium, we made deletion strains for each of the Chd genes; immunoblotting confirmed the absence of the targeted protein in each mutant strain (Fig. 1B). Mutants lacking each gene were viable, but possessed different developmental phenotypes that largely correlated with their peak expression patterns. Disruption of chdA causes a slight (∼1 hour) delay in streaming maxima, but no apparent effect on aggregate, tip or slug formation during early Dictyostelium development. However, the chdA nulls exhibited a significant delay at the onset of terminal differentiation (i.e. culmination), ∼16 hours post-starvation. This delay is best illustrated when Dictyostelium are developed on nitrocellulose filters where wild-type cells rarely develop migratory pseudoplasmodia, but instead rapidly progress to terminal differentiation. Under the same developmental conditions, chdA-null mutants form migratory pseudoplasmodia at high frequency (Fig. 1C) and persist at this stage for extended times, when compared with wild type.
ChdB is maximally expressed during early development and chdB-null mutants take ∼2-4 hours longer to form multicellular mounds than do wild type. The timing defect does not appear to be stage specific; initiation of streaming, loose mound formation and tight aggregation are proportionally delayed. It is currently unclear how this phenotype originates. Beyond this developmental delay, no prominent phenotypic effects were apparent.
The strongest developmentally defective chd-null phenotype was observed with cells lacking chdC. chdC-null cells exhibited a delay during aggregation of ∼2 hours and a developmental arrest at the multicellular mound stage, at ∼12 hours; development failed to progress to the tipped mound stage, but instead formed bulbous extensions when development terminated (Fig. 1D). The developmental arrest in chdC-null mutant cells was completely penetrant, with no migratory pseudoplasmodia or mature terminal wild-type structures formed.
Differential growth effects of the CHDs
During cell culture, we observed a slower growth rate for chdC nulls, than for the other strains in axenic suspension. To quantify growth rates, we identically diluted wild type, and chdA, chdB and chdC nulls from log phase growth (∼2×106 cells/ml) into fresh growth media, and monitored growth over several days (Fig. 2A). The growth rate for chdC nulls was consistently halved compared with all other strains. The growth defect of chdC nulls is not due to deficiencies in cytokinesis, and chdC nulls are not multi-nucleate.
Fig. 2.
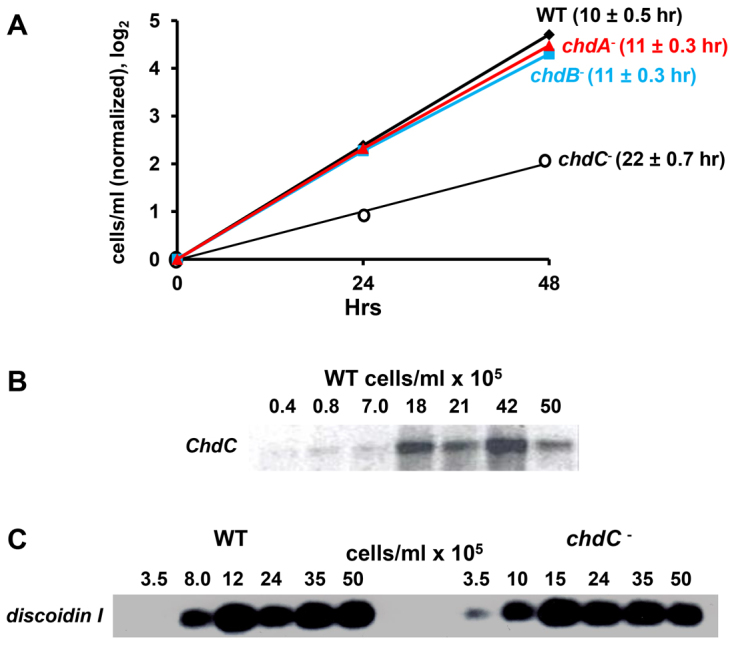
ChdC and cell growth. (A) Wild-type, chdA-, chdB- and chdC-null cells were grown to log phase (<2×106 cells/ml) and diluted ∼20× into fresh media and cell aliquots taken at various times to evaluate relative growth rate. Growth increase for each strain was normalized to the initial cell density and plotted as a log2 scale; doubling times for each strain are indicated. (B) RNA was prepared from growing wild-type cells at the indicated cell densities and hybridized by RNA gel blot with a ChdC probe. (C) RNA was prepared from growing wild-type and chdC- cells at the indicated cell densities and hybridized by RNA gel blot to a probe specific for the coding region of discoidin I.
We considered whether chdC nulls might be aberrantly regulated during the transition from growth to development in response to nutrient depletion. Prior to the onset of starvation, Dictyostelium monitor the accumulation of various secreted factors to sense an increase in cell density (Burdine and Clarke, 1995). Inappropriate cell density sensing by chdC nulls could lead to growth defects or delayed entry into development and subsequent asynchrony. Using RNA and protein samples taken from growing wild-type cultures at ranges of ∼5×104 to 5×106 cells/ml, we show that the relative expression of both ChdC mRNA (Fig. 2B) and ChdC protein increases as cell density rises from ∼5×105 to 1×106 cells/ml. Thus, ChdC regulation appears linked to a pre-starvation response.
To determine whether ChdC function might additionally participate in the pre-starvation response, we examined cell density-dependent expression of the prototypic pre-starvation response marker Discoidin I (Burdine and Clarke, 1995) in wild-type and chdC-null cells. Discoidin I mRNA expression was similarly enhanced in wild-type and chdC-null cells at a cell density transition from ∼3×105 to 1×106 cells/ml (Fig. 2C). Although the data indicate that ChdC expression is induced by the pre-starvation response, as with several other early developmental markers, they do not support a requirement for ChdC to mediate this process.
CHD effects on chemotaxis
To further assess CHD function during early development, we examined wild-type and chd-null cells that had been induced to differentiate in shaking culture in the presence of exogenous pulses of cAMP. These cAMP conditions mimic early stage developmental signaling and bypass the potential for defects in endogenous cAMP synthesis (McMains et al., 2008). Cells were then assayed for their ability to move by chemotaxis towards an exogenous cAMP source. Chemotactic behavior of chdB nulls is highly similar to wild type (Fig. 3). By contrast, both chdA- and chdC-null cells had significant chemotaxis defects, including reduced cell speed, increased cell turning and lower chemotactic indices. Nonetheless, chemotaxis to cAMP was not absent in either chdA or chdC nulls, consistent with their ability to form multicellular aggregates during development. The discrete phenotypes of each strain suggest that the individual CHDs have distinct actions, with only limited functional redundancy.
Fig. 3.
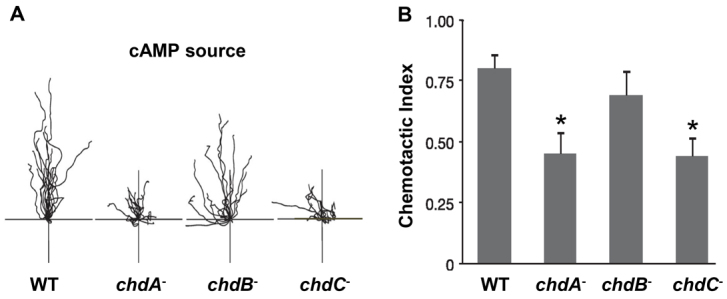
Differential CHD functions during chemotaxis. (A) Individual wild-type, chdA-null, chdB-null and chdC-null cells were imaged for chemotaxis toward a directed cAMP point source during a 15-minute period. Tracings reflect the migration paths for different cells of each strain. (B) The chemotactic index was plotted for each strain as mean±s.e.m. The Mann-Whitney test was used to evaluate significance. *P<0.001.
Aberrant and unique transcriptional profiles of chd nulls during growth and early development
To examine directly differential effects on gene expression upon loss of ChdA, ChdB or ChdC, we applied high-throughput transcriptome sequencing (RNA-seq) for the three mutant strains, and compared relative mRNA transcript levels of each strain with that of wild type and with each other. Transcriptome profiles were determined for cells during growth and following 5 hours of stimulation with exogenous 100 nM pulses of cAMP. These latter conditions promote early differentiation and bypass the requirement for endogenous extracellular cAMP signaling (McMains et al., 2008).
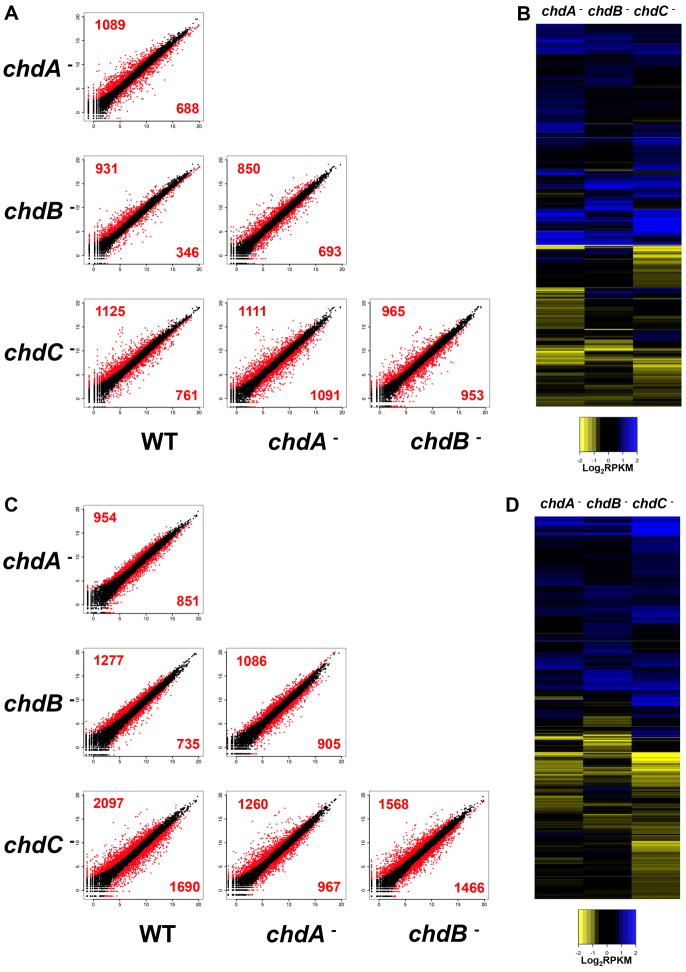
During growth, ∼10-15% of all genes are mis-expressed by over twofold (P<0.05) in each strain compared with wild type (Fig. 4A). Yet most wild-type gene expression is not altered upon loss of ChdA, ChdB or ChdC. When expression profile differences are compared between any two strains (Fig. 4A) or if all mis-expressed genes are collectively analyzed by comparative heat maps (Fig. 4B), they cluster as distinct gene sets. Although the gene sets for each strain are not fully unique, their observed intersections are no greater than would occur by the random distribution of independent variables. We conclude that ChdA, ChdB and ChdC predominantly regulate distinct gene sets during growth. We did not detect compensatory expression profile changes of chdA, chdB or chdC mRNAs in the various mutant strains.
Fig. 4.
Large transcriptome differences among the chd nulls during growth or differentiation. Scatter plots of normalized (RPKM, log2) expression values for genes in wild-type, chdA-, chdB- and chdC-null cells during growth (A) or after 5 hours of cAMP-pulsing (C). Data points with more than twofold (P<0.05) expression differences between indicated strains are in red, and less than twofold (P<0.05) expression differences are black. Heat maps of log2-fold changes in RPKM values in all genes with expression differences [≥2-fold (P<0.05)] in chdA, chdB or chdC relative to wild type, during growth (B) or after 5 hours of cAMP pulsing (D).
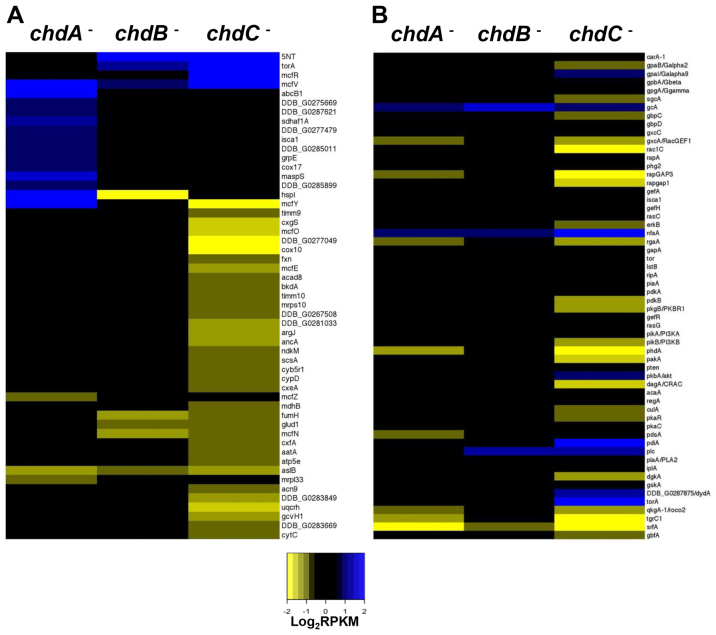
We next surveyed for functional gene classes that might be mis-expressed during growth of the various chd nulls, using Gene Ontology (GO) annotation. Despite the large number of genes that are downregulated in chdA and chdB nulls, little group biases could be discerned during growth. This is consistent with a lack of a significant growth phenotype for these strains. By contrast, chdC nulls displayed a poor growth phenotype and a large number of genes involved in mitochondrial and metabolic processes were poorly expressed exclusively in chdC nulls (Fig. 5A and supplementary material Fig. S3A).
Fig. 5.
Expression changes in GO clustered genes for growth or development. (A) Heat maps show log2-fold changes in RPKM values for 53 genes with GO annotations associated with mitochondrial functions in the chdA, chdB and chdC nulls relative to wild type during growth. (B) Fifty-nine gene functions have been mapped to a cAMP chemotaxis network that acts downstream of CAR1 (carA). Heat maps show log2-fold change in RPKM values for these genes in the chdA, chdB and chdC nulls relative to wild type, during differentiation in the presence of cAMP pulses for 5 hours. Genes are sorted approximately into separate signaling pathways downstream of the cAMP receptor.
Many regulators of aggregation also showed altered expression in growing chdC nulls. ctnA, cf45-1 and cf60, three gene members of the secreted countin complex that potentiate inter-cell signaling and chemotactic movement (Brock and Gomer, 1999; Brock et al., 2003; Brock et al., 2006), were downregulated in chdC nulls. Expression of lmcA, lmcB and srsA, which function similarly to countin, was also suppressed, while expression of other modulators of aggregate size (e.g. cnrJ, cnrG and cnrK) was upregulated.
We also mapped gene expression patterns in chdA, chdB and chdC nulls that had been induced to synchronously differentiate by cAMP pulsing, to a time point akin to ∼8 hours of development, when cells are chemotactically competent (Fig. 4C). As wild-type cells initiate development, there are large changes in the Dictyostelium transcriptome (Van Driessche et al., 2002; Loomis and Shaulsky, 2011). These changes are further augmented in the chd nulls, where individually 15-30% of all genes are mis-expressed by more than twofold (P<0.05). All of the mis-expressed genes were clustered in order to compare expression differences among all the cell lines, and displayed as heat maps (Fig. 4D). Although there is a trend for each strain to similarly exhibit more gene de-repression (i.e. overexpression) than repression (i.e. under-expression), the expression patterns for the chdA, chdB and chdC nulls display distinct gene expression profiles, underscoring their unique developmental phenotypes.
A complex series of interacting circuitries are shown to direct chemotactic movement and signal response to extracellular cAMP in Dictyostelium (Kortholt and van Haastert, 2008; McMains et al., 2008; Swaney et al., 2010). Although they primarily function downstream of the CAR1/Gα2βγ (cAMP receptor-G protein) complex, they diverge into several broad paths, including the cGMP-dependent and -independent actions of guanylyl cyclases sGC and GCA, the PIP3-dependent and -independent functions mediated via mTORC2, inter- and intracellular cAMP accumulation and signaling, and the interactions of Ca+2 and phospholipase A2 pathways (Kortholt and van Haastert, 2008). These networks have been defined biochemically and genetically and include ∼50 genes. Of these, 50% are mis-expressed in chdC nulls (Fig. 5B), suggesting defects in chemotactic response, but also in extracellular cAMP signal relay (e.g. Gα2, PKBR1, CRAC, ERK2). Other gene classes that influence chemotaxis via cAMP wave propagation and cytoskeletal organization (supplementary material Fig. S3B,C) are also preferentially mis-expressed in chdC nulls, and to a lesser degree in chdA nulls. Thus, although the ChdC and ChdA gene sets are distinct, there are some shared elements. Yet as many more chemotactic genes are mis-expressed in chdC-nulls than in chdA-nulls, the data also explain the more minor streaming defects of chdA nulls during development. In addition, as several independent signaling pathways regulate chemotaxis during Dictyostelium development, their collective and compensatory activities can dampen effects caused by deficiencies in selective elements (Van Haastert and Veltman, 2007; Kortholt and van Haastert, 2008). Accordingly, chemotaxis is compromised, but is not eliminated, in chdA and chdC nulls.
The culmination defect of chdA nulls is a post-slug formation, late developmental event. The regulatory pathways for this stage are complex (Harwood et al., 1992; Guo et al., 1999) and we do not observe gene expression defects at early stages that are predictive of the phenotype. Although chdB nulls show large numbers of mis-expressed genes, the cells do not exhibit a strong early phenotype and the mis-expressed genes do not group into simple GO collections.
cAMP signaling also directs developmentally regulated gene expression changes. Cells respond differentially to low (nM) level pulses of cAMP during early development and to non-varying, higher (μM) cAMP levels at the multicellular stage. Although only 13 genes are currently classified as exclusively pulse-dependent (Iranfar et al., 2003), 11 of these are under-expressed in chdC-nulls and 10 are under-expressed chdA nulls (supplementary material Fig. S3C); only 1 gene is affected in chdB nulls. In contrast to pulse-regulated gene expression, aggregation and cell-type specific gene expression is mediated, in part, by response to a non-varying cAMP stimulus, involving regulatory genes such as tgrC1/lagC, gbfA and srfA (Fig. 5B). Only the chdC nulls show developmental arrest during aggregation and only chdC nulls show impaired expression of all these essential regulatory factors.
ChdC-mediated regulation of multicellular development and pattern formation
The observed chemotaxis (see Fig. 3) and morphology (see Fig. 1) defects of the chdC nulls fit precisely with those identified during our initial genetic screens for aberrant chemotaxis and multicellular development. The RNA-seq data from the 5-hour pulsed set indicate poor expression of genes (e.g. tgrC1/lagC, gbfA, srfA) that are required for multi-cell differentiation. Indeed, the strongest phenotype of chdC-null mutant cells arises as cells begin multicellular development, where wild-type cells in the aggregation mound differentiate into the precursors of the terminally spore and stalk cells. Prespore cells comprise ∼80% of mound cells, with the remaining prestalk cells that give rise to the stalk, basal disc and supporting cup structures (Williams, 2006). We, thus, examined multi-cellular differentiation of the chdC nulls.
Endogenous prespore pspA and cotB and prestalk ecmA and ecmB mRNA (Williams, 2006) levels are diminished in chdC nulls compared with wild type (Fig. 6A). In vivo reporter expression using pspA promoter-lacZ fusions shows decreased reporter activity and restricted expression patterning compared with wild type (Fig. 6B). In addition, the expression of 53% of genes annotated as prespore-specific (including pspA and cotB) is halved in chdC-nulls as assayed by RNA-seq at the early mound stage, fully consistent with the defects described for prespore/spore differentiation (see Fig. 6).
Fig. 6.
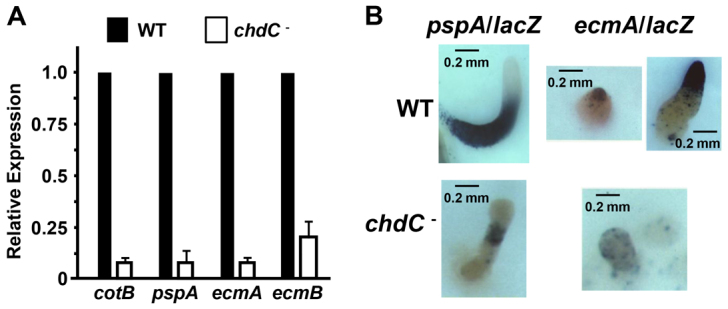
Regulation of prespore/spore and prestalk/stalk differentiation by ChdC. (A) RNA was prepared from 12-hour developed cells, and relative expression levels of the prespore genes pspA and cotB and the prestalk genes ecmA and ecmB were quantified by RT-qPCR. (B) Wild-type and chdC-null cells were transformed with either the prespore-specific expression construct pspA/lacZ or the prestalk-specific expression construct ecmA/lacZ, plated for development on nitrocellulose filters and stained for β-galactosidase activity.
Prestalk cell differentiation is more complex, marked by multiple sub-compartments of defined expression termed the pstA, pstO and pstB regions, which are primarily defined by subpromoter regions of prestalk genes ecmA and ecmB (Jermyn et al., 1987). In chdC-null mutants, ecmA/lacZ is expressed with a weaker and scattered pattern, without defined sorting to the tip of the mound (Fig. 6B). Global transcriptional profiling also gave a clear view of the observed broad prestalk defects, where the expression of 56% of genes annotated as prestalk specific is halved in chdC nulls.
Other GO terms that were enriched in the downregulated population of chdC nulls at the loose mound stage broadly segregate as regulators of cell division or metabolism. The former group includes cell cycle genes (cdk1, cdc45, cycB), DNA replication genes (polA1, polA3, polD2, pcna, rcf3, rcf4, rcf5) and mitotic genes (nek2, kif2, kif12). Although controversy surrounds the requirement of cell cycle progression during Dictyostelium development (Chen et al., 2004; Muramoto and Chubb, 2008), this gene set is significantly upregulated during wild-type development (Strasser et al., 2012).
Autonomous and non-autonomous defects in chdC-nulls
The RNA-seq data indicate that chdC nulls have defects in both cell-cell signaling (e.g. cAMP production) and cell-autonomous (e.g. the TgrC1/LagC-GbfA) regulatory pathways. These data suggest that although certain developmental aspects of chdC-null development might be rescued by exogenous wild-type signals, they would be insufficient to bypass defects in cell-autonomous functions.
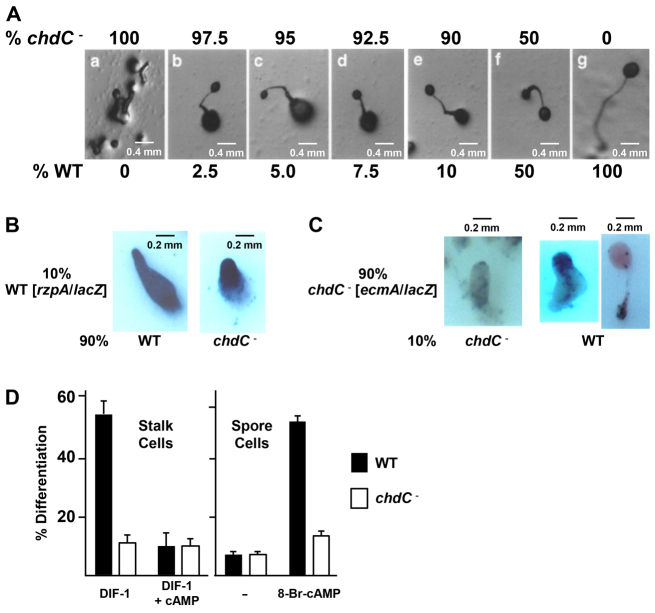
To examine possible defects in both cell-autonomous and non-autonomous pathways in chdC nulls, we investigated cell pattern specification in a series of chimeric, mixed cell developments. chdC nulls were mixed at various cell ratios with the parental wild type. Remarkably, although 100% chdC nulls are unable to form terminal fruiting body structures, chimeric organisms with as few as 2.5% wild-type cells will progress to fruiting bodies, albeit with a reduced spore mass and a significantly larger basal disk (Fig. 7A).
Fig. 7.
Developmental defects of chdC nulls are partially rescued by chimeric development with wild-type cells. (A) The indicated percentages of wild-type and chdC- cells were mixed prior to plating for development. Images were obtained after 36 hours of development. (B) Wild-type cells were transformed with the ubiquitously expressed rzpA/lacZ reporter construct, mixed and developed as a 10% ratio with 90% unmarked wild-type cells or with 90% unmarked chdC- cells, and stained for β-galactosidase activity. (C) chdC- cells were transformed with the prestalk-specific reporter construct ecmA/lacZ, mixed and developed as a 90% ratio with 10% unmarked wild-type cells or with 10% unmarked chdC- cells, and stained for β-galactosidase activity. (D) Stalk cell assays: wild-type and chdC-null cells were plated at low cell density in monolayer with DIF-1 or with DIF-1 + cAMP. The percentage of cells that were fully vacuolated (mature stalk cells) was determined using phase-contrast microscopy. Spore cell assays: wild-type and chdC-null cells were plated at low cell density in monolayer without or with 8Br-cAMP to promote sporulation. Assays were scored by phase-contrast microscopy for the percentage of phase-bright fully mature spore cells.
In chimeras with unmarked 90% chdC nulls, the 10% ubiquitously marked rZIP/lacZ (Balint-Kurti et al., 1997) wild-type cells preferentially, but not exclusively, populate the anterior prestalk region (Fig. 7B). In a reverse experiment, where the 90% chdC nulls are marked with ecmA/lacZ and mixed with 10% unmarked wild-type cells, stained chdC nulls can localize to the prestalk/stalk regions (Fig. 7C). These data suggest that the failure of tip formation in chdC nulls may result from defects in signal propagation and not simply from a loss of an autonomous intracellular function.
To investigate non-autonomous parameters further, we examined terminal cell formation under monolayer induction conditions (Kay, 1987). Stalk cell differentiation is induced by the sequential exposure of cells to first cAMP and then to DIF-1. Under these conditions ∼55% of wild-type cells were induced to form vacuolated stalk cells in the presence of DIF-1, a process that was highly suppressed by simultaneous exposure to cAMP (Fig. 7D). By contrast, only ∼10-12% of chdC nulls could form vacuolated cells in the presence of exogenous DIF-1, regardless of the presence of cAMP (Fig. 7D).
In low cell density monolayers, spore formation can be directly induced with the membrane-permeant cAMP analog 8Br-cAMP, which directly activates cAMP receptors and the cAMP-dependent protein kinase (PKA) and bypasses other signaling requirements (Kay, 1987). Under these conditions ∼60% of wild-type cells differentiated into spores, whereas only 15% of chdC null cells formed spores, efficiencies similar to that of uninduced wild-type cells (Fig. 7D), indicating an intrinsic loss in spore-forming ability of chdC nulls.
We also examined spore differentiation following chimeric development of 10% wild type with 90% chdC nulls. Spores were harvested and germinated in the absence and presence of blasticidin to differentiate and quantify wild-type and chdC nulls. Here, fewer than 10% of total spores from the mix were blasticidin-resistant chdC nulls. Although spore differentiation is not totally blocked in chdC nulls, it was substantially reduced to a frequency of ∼10% that of WT, confirming a cell-autonomous defect for spore differentiation in chdC nulls. Although exogenous wild-type cells are able to influence developmental patterning in chdC nulls, they are unable to rescue terminal cell differentiation of chdC nulls.
DISCUSSION
Of the multiple SWI2/SNF2, ATPase-type chromatin remodeling proteins in eukaryotes, the CHD family is among the largest and most diverse (Ryan and Owen-Hughes, 2011). Humans have nine distinct CHDs that group into three structural subfamilies, CHD I, CHD II and CHD III. The CHD I subfamily is ubiquitous, found in the yeasts, plants and metazoa, whereas CHD II and CHD III members are generally restricted to multicellular organisms, with the latter seemingly absent in plants (Ryan and Owen-Hughes, 2011). It is, thus, striking that Dictyostelium, a facultative metazoan, encodes two proteins that cluster with the CHD III subfamily. Accordingly, Dictyostelium provides a unique model system for analyzing multiple and diverse regulatory aspects of different CHD family members; Dictyostelium does not seem to encode CHD II members, which are defined by the presence of PHD fingers (Ryan and Owen-Hughes, 2011).
Although CHD I members are suggested to facilitate an open chromatin organization at active promoters and to regulate nucleosome spacing more distally (Hennig et al., 2012; Persson and Ekwall, 2010; Zentner et al., 2013), their interactions at these promoter sites do not seem to be functionally essential at a global level. S. cerevisiae are viable without CHD1 (Tsukiyama et al., 1999) and D. melanogaster that lack CHD1 develop to maturity, although both males and females are sterile (McDaniel et al., 2008). Nonetheless, mouse ES cells depleted of CHD1 exhibit defects in pluripotency (Gaspar-Maia et al., 2009). Although Chd1-/- mice have not been reported, Chd2-/- mice are embryonically lethal (Marfella et al., 2006b). Therefore, CHD I family members can have crucial functions in the control of developmentally essential gene sets.
chdA-null Dictyostelium are viable, without apparent impact upon growth on bacteria or in axenic culture. However, chdA nulls exhibit a highly defined early developmental defect, involving poor transcriptional response to the extracellular activating signal cAMP. Furthermore, the morphological transitions and cell fate determinations required for terminal differentiation, which occur co-temporaneously with maximal levels of ChdA expression in wild-type cells, are effectively arrested in cells lacking ChdA. To some extent, the phenotypic effects of ChdA (i.e. a CHD1/2 ortholog) loss in Dictyostelium parallels the essential developmental requirement for CHD I members in mouse.
Consistent with the observed phenotypes, we also show that loss of ChdA in Dictyostelium causes a restricted, rather than global, impact on gene expression. Fewer than 15% of the Dictyostelium genome shows altered patterns of gene expression during growth or early development when parental and chdA-null cells are compared. We observe a similar distribution of genes that exhibit up- or downregulation. The data indicate that ChdA in Dictyostelium, and potentially CHD1 in other regulatory networks, is not essential for global chromatin organization and gene expression induction. ChdA is likely to have more confined operational targets and integrate with other chromatin organizing components.
Studies of the CHD III class in Drosophila (e.g. Kismet) or in humans (e.g. CHD7) suggest unique developmental functions compared with other CHD members. Kismet regulates cell fate determination of Drosophila and can antagonize the repressive effects of Polycomb in homeotic patterning (Srinivasan et al., 2008). Similarly, loss-of-function mutations or haploinsufficiencies of CHD7 in mouse, humans and zebrafish (Layman et al., 2010; Patten et al., 2012) have complex developmental defects. In humans, the eye, heart, ear and genitalia are specifically affected, and many phenotypes are recapitulated in mouse and zebrafish. Although CHD7 is not essential for self renewal, pluripotency or somatic reprogramming of mouse ES cells, it has been suggested that a significant defect in human CHARGE syndrome may involve lineage effects in the proliferation, cell migration and specification of neural crest cells (Zentner et al., 2010).
As in the metazoa, Dictyostelium has multiple (ChdB and ChdC) CHD III subfamily members, with ChdC having the most essential and complex regulatory functions of all CHD members. At the phenotypic level, chdC-null cells are viable, but have reduced growth rates. They respond to nutrient depletion for induction of multi-cell development, but rapidly display abnormalities that temporally parallel the upregulation of ChdC in wild-type cells. Response to cAMP, chemotaxis and aggregate formation are impaired, as are cell-fate commitments.
Many of the defects in chdC nulls can be directly attributed to specific abnormalities in gene expression patterns compared with parentals. Although only a small population of genes are mis-regulated during growth of chdC nulls, they include a large number of cellular metabolic and energetic regulators. By contrast, the mis-regulated genes in chdB nulls do not simply cluster in a few functionally related groups, but are distributed broadly among GO classes, and, under the conditions studied, ChdB seems to be only minimally required.
During aggregation, Dictyostelium organize a complex inter-cell signaling pathway, which directs chemotaxis and multi-cell aggregation, but also cytodifferentiation and morphogenetics. Central to this pathway is the production and secretion (signal-relay) of the chemoattractant cAMP (Kortholt and van Haastert, 2008; McMains et al., 2008; Swaney et al., 2010). Receptor-mediated response to cAMP involves a highly integrated pathway that collectively promotes signal amplification, cytoskeletal reprogramming, cell polarization and non-terminal, cell-fate specification; many individual components of the cAMP signaling pathway are required for all cellular responses (Kortholt and van Haastert, 2008; McMains et al., 2008; Swaney et al., 2010). We show that ∼50% of the genes, which lie most immediate to receptor sensing, are mis-regulated in chdC nulls. Their mis-regulations are predicted to compromise cell-autonomous requirements of intracellular signaling that directs chemotaxis and differentiation, but also the non-autonomous, extracellular elements involved in cAMP signal relay. Thus, although certain non-autonomous defects of chdC nulls may be rescued by co-development with a small population of wild-type cells, the cell-autonomous pathways that are regulated by intracellular signaling remain compromised even in the presence of exogenous cAMP or wild-type factors.
Gene expression patterns have also been studied in Chd7+/+and Chd7-/- mouse ES cells (Schnetz et al., 2010). Data indicate an impact on only a restricted gene set, with significant enrichment of genes primarily expressed in the ES population (Schnetz et al., 2010). In addition, more CHD7-binding sites map to enhancers of these genes than to non-ES class genes. Interestingly, these ES-enriched and CHD7-bound gene classes are generally upregulated in the absence of CHD7, suggesting a modulating repressive function on this ES gene set. Nonetheless, CHD7 is not entirely repressive, as other gene sets are downregulated upon the loss of CHD7. Regardless of the impact of CHD7 on ES cell transcriptional patterns as both a positive and negative effector, processes of self renewal, pluripotency and somatic cell reprogramming are not impaired in CHD7-deficient ES cells (Schnetz et al., 2010).
The directed and interactive roles of the different CHDs in multicellular differentiation are complex and still largely undefined. Dictyostelium provides a unique system to separately dissect these functions. We have shown that phenotypes resulting from deficiencies of ChdA, ChdB or ChdC in Dictyostelium are not global and, thus, parallel those in the more complex metazoa. It is particularly striking that proliferative, migration and cell specification defects of chdC-null Dictyostelium are mirrored in Chd7-/- neural crest cells. We further speculate that defective inter- and intra-cell signaling, as seen in chdC-null Dictyostelium, may also contribute to the developmental abnormalities seen in CHARGE syndrome.
Our data extend conclusions on the role for CHDs in gene expression. Each of the CHDs in Dictyostelium regulates different developmental patterns and different gene sets. The CHDs do not appear to act globally and can have both repressive and activating functions. We suggest that the mechanistic functions of CHD in Dictyostelium are similarly shared in the metazoa and that more-complete analyses will contribute significantly to the understanding for the ultimate treatment of genetic diseases, such as CHARGE syndrome, caused by loss-of-function mutations in CHD loci.
MATERIALS AND METHODS
Bioinformatics of CHD and SWI2/SNF2-related proteins
Identification of genes in Dictyostelium was performed by BLAST search of the Dictyostelium genome (http://dictybase.org/tools/blast). Three CHD proteins were identified: ChdA is DDB_G0284171, ChdB is DDB_G0280705 and ChdC is DDB_G0293012 (Basu et al., 2013). Additional domains were identified using EMBL SMART software (Letunic et al., 2012) and NCBI conserved domain software (Marchler-Bauer et al., 2011). The phylogenic tree was created using http://phylogeny.lirmm.fr/phylo_cgi/index.cgi (Dereeper et al., 2008) and full-length proteins. Similar clusterings were generated using only the chromodomains or helicase motifs.
Dictyostelium strains
Dictyostelium Ax2 (wild-type) cells were grown axenically in HL5 medium at 20°C. Gene deletion constructs were produced using standard PCR-based methods. Gene regions were disrupted by the floxed blasticidin resistance cassette from pLPBLP (Faix et al., 2004) using TOPO-Blunt II (Invitrogen). Gene deletion strains were generated by transformation with 15 μg of linearized plasmid using electroporation and selection in HL5 media supplemented with 10 μg/ml Blasticidin S. Transformants were screened by PCR and gene disruptions confirmed by immunoblotting.
Dictyostelium development and phenotype analyses
To analyze development, growing cells in log phase (1-3×106 cells/ml) were washed twice in KK2 buffer (15 mM KH2PO4, 3 mM K2HPO4) and developed on 0.45 μm nitrocellulose filters. Developmental expression of the individual CHD proteins were analyzed every 2 hours by immunoblotting. Slug migration assays were carried out in a ‘slug can’. Washed cells were plated at one edge of a nitrocellulose filter, developed to the slug stage, and subjected to unidirectional light; images were taken after 48 hours. For chemotaxis, 5×107 cells/ml were shaken in KK2 buffer for 5 hours while being pulsed every 6 minutes with cAMP to a final concentration of 10-7 M. The cells were then placed in the Zigmond chamber (Z02; Neuro Probe, Gaithersburg, MD) in a cAMP gradient (source at 10-6 M cAMP, sink with no cAMP). Differential interference contrast (DIC) images of cells were captured at 6-second intervals for 15 minutes. Cell movement was analyzed using Dynamic Image Analysis System (DIAS) software (version 3.4.1) (Soll Technologies, Iowa City, IA; (Wessels et al., 2009). Stalk and spore cell differentiation in monoloayer assays were performed at a cell density of 1×104 cells/cm2 (Kay, 1987) and scored by phase-contrast microscopy.
Antibodies and immunoblotting
Antibodies specific to ChdA, ChdB and ChdC were raised in rabbits by immunizing with peptides (∼20 amino acids) specific for each protein (Openbiosystems, 70 day protocol). Immunoblotting was by standard methodologies, using whole-cell, SDS lysates, 3-8% Tris-acetate gels and blotting overnight at low voltage.
RNA-seq; sequencing and bioinformatics
RNA was isolated from all strains using TRIzol (Life Technologies). RNA quality was analyzed using the Agilent 2100 Bioanlyzer. 5 μg of total RNA was used for the Illumina libraries. RNA was polyA enriched and libraries were single-end, sequenced for 50 bases, using the Illumina HiSeq2000. Filtered reads from the manufacturers recommended pipeline (version 1.7) were aligned to the Dictyostelium genome with Tophat (version 1.3.0) (Trapnell et al., 2009) while masking the duplicated region of chromosome 2 (nucleotides 3,015,984 to 3,768,555) using the gene models from dictybase (http://dictybase.org/) (Basu et al., 2013). Reads were counted and normalized, and differential expression was calculated with HTSeq and DESeq (Anders and Huber, 2010). Heat maps were generated with the heatmap.2 function in the gplots package (Warnes, 2012) scatterplots were also produced in R (R Development Core Team, 2012). GO term enrichment used Orange (Curk et al., 2005). The RNA-seq data have been deposited in the GEO database (Accession Number, GSE47222).
Supplementary Material
Acknowledgments
We are indebted to Dr Harold Smith and the NIDDK genomics core.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
J.L.P., B.J.R., K.C.R., A.J.H. and A.R.K. developed the concepts and approaches. J.L.P., B.J.R., K.C.R. and A.R.K. performed experiments. J.L.P., B.J.R., K.C.R., A.J.H. and A.R.K. analyzed the data. J.L.P., A.J.H. and A.R.K. prepared the manuscript prior to submission.
Funding
This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), the National Institute of Diabetes and Digestive and Kidney Diseases, and a Wellcome Trust/NIH Programme Studentship to J.L.P. Deposited in PMC for immediate release.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.099879/-/DC1
References
- Abrams E., Neigeborn L., Carlson M. (1986). Molecular analysis of SNF2 and SNF5, genes required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 6, 3643–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artemenko Y., Swaney K. F., Devreotes P. N. (2011). Assessment of development and chemotaxis in Dictyostelium discoideum mutants. Methods Mol. Biol. 769, 287–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balint-Kurti P., Ginsburg G., Rivero-Lezcano O., Kimmel A. R. (1997). rZIP, a RING-leucine zipper protein that regulates cell fate determination during Dictyostelium development. Development 124, 1203–1213 [DOI] [PubMed] [Google Scholar]
- Basu S., Fey P., Pandit Y., Dodson R. J., Kibbe W. A., Chisholm R. L. (2013). DictyBase 2013: integrating multiple Dictyostelid species. Nucleic Acids Res. 41 Database issue, D676–D683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blus B. J., Wiggins K., Khorasanizadeh S. (2011). Epigenetic virtues of chromodomains. Crit. Rev. Biochem. Mol. Biol. 46, 507–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosman E. A., Penn A. C., Ambrose J. C., Kettleborough R., Stemple D. L., Steel K. P. (2005). Multiple mutations in mouse Chd7 provide models for CHARGE syndrome. Hum. Mol. Genet. 14, 3463–3476 [DOI] [PubMed] [Google Scholar]
- Bouazoune K., Kingston R. E. (2012). Chromatin remodeling by the CHD7 protein is impaired by mutations that cause human developmental disorders. Proc. Natl. Acad. Sci. USA 149, 19238–19243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeden L., Nasmyth K. (1987). Cell cycle control of the yeast HO gene: cis- and trans-acting regulators. Cell 48, 389–397 [DOI] [PubMed] [Google Scholar]
- Brock D. A., Gomer R. H. (1999). A cell-counting factor regulating structure size in Dictyostelium. Genes Dev. 13, 1960–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock D. A., Hatton R. D., Giurgiutiu D. V., Scott B., Jang W., Ammann R., Gomer R. H. (2003). CF45-1, a secreted protein which participates in Dictyostelium group size regulation. Eukaryot Cell 2, 788–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock D. A., van Egmond W. N., Shamoo Y., Hatton R. D., Gomer R. H. (2006). A 60-kilodalton protein component of the counting factor complex regulates group size in Dictyostelium discoideum. Eukaryot Cell 5, 1532–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdine V., Clarke M. (1995). Genetic and physiologic modulation of the prestarvation response in Dictyostelium discoideum. Mol. Biol. Cell 6, 311–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. K., Shaulsky G., Kuspa A. (2004). Tissue-specific G1-phase cell-cycle arrest prior to terminal differentiation in Dictyostelium. Development 131, 2619–2630 [DOI] [PubMed] [Google Scholar]
- Clapier C. R., Cairns B. R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304 [DOI] [PubMed] [Google Scholar]
- Curk T., Demsar J., Xu Q., Leban G., Petrovic U., Bratko I., Shaulsky G., Zupan B. (2005). Microarray data mining with visual programming. Bioinformatics 21, 396–398 [DOI] [PubMed] [Google Scholar]
- Delmas V., Stokes D. G., Perry R. P. (1993). A mammalian DNA-binding protein that contains a chromodomain and an SNF2/SWI2-like helicase domain. Proc. Natl. Acad. Sci. USA 90, 2414–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.-F., Guindon S., Lefort V., Lescot M., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36 Web Server issue, W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J. A., Sweder K. S., Hanawalt P. C. (1995). Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 23, 2715–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen G., Auerbach R. K., Snyder M. (2012). SWI/SNF chromatin-remodeling factors: multiscale analyses and diverse functions. J. Biol. Chem. 287, 30897–30905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J., Kreppel L., Shaulsky G., Schleicher M., Kimmel A. R. (2004). A rapid and efficient method to generate multiple gene disruptions in Dictyostelium discoideum using a single selectable marker and the Cre-loxP system. Nucleic Acids Res. 32, e143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan J. F., Blus B. J., Kim D., Clines K. L., Rastinejad F., Khorasanizadeh S. (2007). Molecular implications of evolutionary differences in CHD double chromodomains. J. Mol. Biol. 369, 334–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A., Alajem A., Polesso F., Sridharan R., Mason M. J., Heidersbach A., Ramalho-Santos J., McManus M. T., Plath K., Meshorer E., et al. (2009). Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 460, 863–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo K., Anjard C., Harwood A., Kim H. J., Newell P. C., Gross J. D. (1999). A myb-related protein required for culmination in Dictyostelium. Development 126, 2813–2822 [DOI] [PubMed] [Google Scholar]
- Hargreaves D. C., Crabtree G. R. (2011). ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 21, 396–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood A. J., Hopper N. A., Simon M. N., Driscoll D. M., Veron M., Williams J. G. (1992). Culmination in Dictyostelium is regulated by the cAMP-dependent protein kinase. Cell 69, 615–624 [DOI] [PubMed] [Google Scholar]
- Hennig B. P., Bendrin K., Zhou Y., Fischer T. (2012). Chd1 chromatin remodelers maintain nucleosome organization and repress cryptic transcription. EMBO Rep. 13, 997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L., Crabtree G. R. (2010). Chromatin remodelling during development. Nature 463, 474–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner K. P., Gerhold C. B., Lakomek K., Wollmann P. (2012). Swi2/Snf2 remodelers: hybrid views on hybrid molecular machines. Curr. Opin. Struct. Biol. 22, 225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd E. A., Capers P. L., Blauwkamp M. N., Adams M. E., Raphael Y., Poucher H. K., Martin D. M. (2007). Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mamm. Genome 18, 94–104 [DOI] [PubMed] [Google Scholar]
- Iranfar N., Fuller D., Loomis W. F. (2003). Genome-wide expression analyses of gene regulation during early development of Dictyostelium discoideum. Eukaryot. Cell 2, 664–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen N., Bergman J. E., Swertz M. A., Tranebjaerg L., Lodahl M., Schoots J., Hofstra R. M., van Ravenswaaij-Arts C. M., Hoefsloot L. H. (2012). Mutation update on the CHD7 gene involved in CHARGE syndrome. Hum. Mutat. 33, 1149–1160 [DOI] [PubMed] [Google Scholar]
- Jermyn K. A., Berks M., Kay R. R., Williams J. G. (1987). Two distinct classes of prestalk-enriched mRNA sequences in Dictyostelium discoideum. Development 100, 745–755 [DOI] [PubMed] [Google Scholar]
- Kay R. R. (1987). Cell differentiation in monolayers and the investigation of slime mold morphogens. Methods Cell Biol. 28, 433–448 [DOI] [PubMed] [Google Scholar]
- Kessin R. H. (2001). Dictyostelium - Evolution, Cell Biology and the Development of Multicellularity. Cambridge, UK: Cambridge University Press; [Google Scholar]
- Kortholt A., van Haastert P. J. (2008). Highlighting the role of Ras and Rap during Dictyostelium chemotaxis. Cell. Signal. 20, 1415–1422 [DOI] [PubMed] [Google Scholar]
- Kuspa A. (2006). Restriction enzyme-mediated integration (REMI) mutagenesis. Methods Mol. Biol. 346, 201–209 [DOI] [PubMed] [Google Scholar]
- Lalani S. R., Safiullah A. M., Fernbach S. D., Harutyunyan K. G., Thaller C., Peterson L. E., McPherson J. D., Gibbs R. A., White L. D., Hefner M., et al. (2006). Spectrum of CHD7 mutations in 110 individuals with CHARGE syndrome and genotype-phenotype correlation. Am. J. Hum. Genet. 78, 303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layman W. S., Hurd E. A., Martin D. M. (2010). Chromodomain proteins in development: lessons from CHARGE syndrome. Clin. Genet. 78, 11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos T. Ã. A., Passos D. O., Nery F. C., Kobarg J. (2003). Characterization of a new family of proteins that interact with the C-terminal region of the chromatin-remodeling factor CHD-3. FEBS Lett. 533, 14–20 [DOI] [PubMed] [Google Scholar]
- Letunic I., Doerks T., Bork P. (2012). SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40 Database issue, D302–D305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis W. F., Shaulsky G. (2011). Developmental changes in transcriptional profiles. Dev. Growth Differ. 53, 567–575 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., Fong J. H., Geer L. Y., Geer R. C., Gonzales N. R., et al. (2011). CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39 Database issue, D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfella C. G., Ohkawa Y., Coles A. H., Garlick D. S., Jones S. N., Imbalzano A. N. (2006a). Mutation of the SNF2 family member Chd2 affects mouse development and survival. J. Cell. Physiol. 209, 162–171 [DOI] [PubMed] [Google Scholar]
- McDaniel I. E., Lee J. M., Berger M. S., Hanagami C. K., Armstrong J. A. (2008). Investigations of CHD1 function in transcription and development of Drosophila melanogaster. Genetics 178, 583–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMains V. C., Liao X. H., Kimmel A. R. (2008). Oscillatory signaling and network responses during the development of Dictyostelium discoideum. Ageing Res. Rev. 7, 234–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramoto T., Chubb J. R. (2008). Live imaging of the Dictyostelium cell cycle reveals widespread S phase during development, a G2 bias in spore differentiation and a premitotic checkpoint. Development 135, 1647–1657 [DOI] [PubMed] [Google Scholar]
- Nishiyama M., Skoultchi A. I., Nakayama K. I. (2012). Histone H1 recruitment by CHD8 is essential for suppression of the Wnt-β-catenin signaling pathway. Mol. Cell. Biol. 32, 501–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagon R. A., Graham J. M., Jr, Zonana J., Yong S. L. (1981). Coloboma, congenital heart disease, and choanal atresia with multiple anomalies: CHARGE association. J. Pediatr. 99, 223–227 [DOI] [PubMed] [Google Scholar]
- Parikh A., Miranda E. R., Katoh-Kurasawa M., Fuller D., Rot G., Zagar L., Curk T., Sucgang R., Chen R., Zupan B., et al. (2010). Conserved developmental transcriptomes in evolutionarily divergent species. Genome Biol. 11, R35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten S. A., Jacobs-McDaniels N. L., Zaouter C., Drapeau P., Albertson R. C., Moldovan F. (2012). Role of Chd7 in zebrafish: a model for CHARGE syndrome. PLoS ONE 7, e31650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson J., Ekwall K. (2010). Chd1 remodelers maintain open chromatin and regulate the epigenetics of differentiation. Exp. Cell Res. 316, 1316–1323 [DOI] [PubMed] [Google Scholar]
- Pleasance E. D., Stephens P. J., O’Meara S., McBride D. J., Meynert A., Jones D., Lin M. L., Beare D., Lau K. W., Greenman C., et al. (2010). A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463, 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2012). R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; [Google Scholar]
- Ryan D. P., Owen-Hughes T. (2011). Snf2-family proteins: chromatin remodellers for any occasion. Curr. Opin. Chem. Biol. 15, 649–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan D. P., Sundaramoorthy R., Martin D., Singh V., Owen-Hughes T. (2011). The DNA-binding domain of the Chd1 chromatin-remodelling enzyme contains SANT and SLIDE domains. EMBO J. 30, 2596–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetz M. P., Bartels C. F., Shastri K., Balasubramanian D., Zentner G. E., Balaji R., Zhang X., Song L., Wang Z., Laframboise T., et al. (2009). Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 19, 590–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetz M. P., Handoko L., Akhtar-Zaidi B., Bartels C. F., Pereira C. F., Fisher A. G., Adams D. J., Flicek P., Crawford G. E., Laframboise T., et al. (2010). CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 6, e1001023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S., Dorighi K. M., Tamkun J. W. (2008). Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. PLoS Genet. 4, e1000217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K., Bloomfield G., MacWilliams A., Ceccarelli A., MacWilliams H., Tsang A. (2012). A retinoblastoma orthologue is a major regulator of S-phase, mitotic, and developmental gene expression in Dictyostelium. PLoS ONE 7, e39914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney K. F., Huang C. H., Devreotes P. N. (2010). Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity. Annu Rev Biophys 39, 265–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun J. W., Deuring R., Scott M. P., Kissinger M., Pattatucci A. M., Kaufman T. C., Kennison J. A. (1992). brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 68, 561–572 [DOI] [PubMed] [Google Scholar]
- Thompson B. A., Tremblay V., Lin G., Bochar D. A. (2008). CHD8 is an ATP-dependent chromatin remodeling factor that regulates beta-catenin target genes. Mol. Cell. Biol. 28, 3894–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T., Palmer J., Landel C. C., Shiloach J., Wu C. (1999). Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 13, 686–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Driessche N., Shaw C., Katoh M., Morio T., Sucgang R., Ibarra M., Kuwayama H., Saito T., Urushihara H., Maeda M., et al. (2002). A transcriptional profile of multicellular development in Dictyostelium discoideum. Development 129, 1543–1552 [DOI] [PubMed] [Google Scholar]
- Van Haastert P. J., Veltman D. M. (2007). Chemotaxis: navigating by multiple signaling pathways. Sci. STKE 2007, pe40 [DOI] [PubMed] [Google Scholar]
- Warnes G. R. (2012). gplots: Various R Programming Tools for Plotting Data. [Google Scholar]
- Wessels D. J., Kuhl S., Soll D. R. (2009). Light microscopy to image and quantify cell movement. Methods Mol. Biol. 571, 455–471 [DOI] [PubMed] [Google Scholar]
- Williams J. G. (2006). Transcriptional regulation of Dictyostelium pattern formation. EMBO Rep. 7, 694–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C. J., Naito T., Arco P. G., Seavitt J. R., Cashman S. M., De Souza B., Qi X., Keables P., Von Andrian U. H., Georgopoulos K. (2004). The chromatin remodeler Mi-2β is required for CD4 expression and T cell development. Immunity 20, 719–733 [DOI] [PubMed] [Google Scholar]
- Yap K. L., Zhou M.-M. (2011). Structure and mechanisms of lysine methylation recognition by the chromodomain in gene transcription. Biochemistry 50, 1966–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner G. E., Layman W. S., Martin D. M., Scacheri P. C. (2010). Molecular and phenotypic aspects of CHD7 mutation in CHARGE syndrome. Am. J. Med. Genet. A. 152A, 674–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner G. E., Tsukiyama T., Henikoff S. (2013). ISWI and CHD chromatin remodelers bind promoters but act in gene bodies. PLoS Genet. 9, e1003317 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.