Abstract
The CRF (corticotropin releasing factor) system is a key mediator of the stress response. Alterations in CRF signaling have been implicated in drug craving and ethanol consumption. The development of negative reinforcement via activation of brain stress systems has been proposed as a mechanism that contributes to alcohol dependence. Here we isolated a gain-of-function allele of seb-3, a CRF receptor-like GPCR in C. elegans, providing an in vivo model of a constitutively activated stress system. We also characterized a loss-of-function allele of seb-3 and showed that SEB-3 positively regulates a stress response that leads to an enhanced active state of locomotion, behavioral arousal, and tremor. SEB-3 also contributed to acute tolerance to ethanol and to the development of tremor during ethanol withdrawal. Furthermore, we found that a specific CRF1 receptor antagonist reduced acute functional tolerance to ethanol in mice. These findings demonstrate functional conservation of the CRF system in responses to stress and to ethanol in vertebrates and invertebrates.
Keywords: Stress, CRF receptor, locomotion, ethanol tolerance
Introduction
Animals respond to environmental stimuli or stress by modulating their behavioral state. For instance, environmental stressors cause physiological arousal and anxiety, manifest as altered behavior, such as increased locomotor activity or compulsive drug seeking. Such modulation of behavioral states is mediated by conserved stress response systems. (Lee and Tsai, 1989; Goeders and Guerin, 1994; Piazza and Le Moal, 1998).
Stress and reward pathways have been studied as key elements of neural systems that drive drug dependence. In drug addiction studies, it has been suggested that the brain stress system is activated and sensitized during repeated withdrawal. As a consequence, a negative emotional state induced by activation of stress system promotes drug addiction (Koob, 2008). The CRF (corticotropin-releasing factor) system plays a pivotal role in mediating stress responses in the brain from amphibians to primates (Winslow et al., 1989; Lowry and Moore, 1991; Johnson et al., 1994; Clements et al., 2002). CRF and the related urocortin peptides are known to mediate stress responses through two CRF receptor subtypes, CRF1 and CRF2 (Chen et al., 1993; Hauger et al., 2006; Liaw et al., 1996).
Here, we identify and characterize seb-3, a gene predicted to encode a GPCR related to CRF receptors in C. elegans. We initially identified seb-3 as a gene that mediates the expression of a distinct locomotory state. Further characterization demonstrated that it also regulates behavioral responses to environmental stress and ethanol. We isolated a seb-3 gain-of-function allele (gf) eg696 that induces behaviors resembling those elicited by stress. This mutant provides an in vivo genetic tool to investigate how persistent activation of the stress system in C. elegans influences responses to ethanol.
In this study, we also found that non-physiological heat exposure, an environmental stress, causes wild-type animals to display tremor in liquid environments. This tremor was abolished by a seb-3 loss-of-function (lf) mutation, whereas the seb-3 (gf) mutant exhibited the defect even under physiological temperatures. Wild-type animals also exhibited tremor during withdrawal from prior exposure to ethanol, while seb-3 (lf) animals showed impaired development of tremor under this condition. In addition, seb-3 mediated the development of acute tolerance to the locomotor-impairing effects of ethanol. Pharmacological blockade of CRF1 receptors identified a similar role for the CRF1 receptor in mediating acute functional (behavioral) tolerance (AFT) to ethanol in mice. These data demonstrate conservation of the CRF system in mediating responses to stress and ethanol in nematodes and mammals.
Materials and Methods
Strains and Genetic screens
Wild-type animals were of the Bristol N2 strain. Worms were cultured at 20°C using standard methods (Brenner, 1974). The seb-3 (tm1848 lf) strain was obtained from S. Mitani (Tokyo Women’s Medical University, Tokyo, Japan) and backcrossed twice with N2. seb-3 (eg696 gf) was isolated from a genetic screen for an enhanced active state of locomotion as a che-2 mutant suppressor and out crossed three times with N2. che-2 (e1033) animals were mutagenized by exposure to ethane methyl sulfonate (EMS, 47 mM) for 4 h. L4 stage, single F2 progeny of mutagenized worms were transferred to NGM plates with abundant OP50, and left undisturbed for 17 h. che-2 (e1033) animals generated a distinct track pattern only in a restricted region. Worms that suppressed abnormal locomotion of che-2 (e1033) were selected for further analysis and mapping. Amphid and phasmid neurons were stained with DiI (Molecular Probes). N2, che-2 (e1033), and seb-3 (eg696 gf) animals were incubated in M9 buffer containing 10 μg of DiI for 2 h at 20°C, transferred to a fresh plate, and allowed to crawl on a bacterial lawn for 1h to remove excess dye (Fujiwara et al., 1999). We used snip-SNPs identified by Wicks et al. (2001) and Davis et al. (2005) for mapping. The eg696 was mapped to locus between two SNPs, C27C12 and F09B12 on LG X. Sequencing identified the eg696 mutation from 46 predicted genes in the interval of approximately 0.31 map units (based on estimated genetic distance from these two SNPs). The genomic region containing the eg696 mutation was introduced into che-2 (e1033) for phenocopy of eg696. che-2 (e1033) seb-3 (eg696 gf) Ex[pofm-1:gfp] was crossed to che-2 (e1033) and F1 progenies with the GFP expression in coelomocyte were selected as heterozygous eg696 animals.
Molecular Biology
For introduction of the eg696 mutation into che-2 (e1033) and N2 worms, 7739 bp of genomic DNA including 3082bp of the promoter region was amplified from eg696 mutant lysate by nested PCR (primers: pseb-3F1-catttctctggctagaagattgtcttgt, pseb-3F2-ccagttcaaattaactctacccaactac, seb-3B1-gaaagatgattgaccaaacaaagtgga, seb-3B2-ttttgctttgggctgttgacgggt) and used for microinjection with pofm-1:gfp as an injection marker. The transformation suppressed the roaming defect of che-2 in 3 independent transgenic lines. For the construct of pseb-3:gfp, 3082bp of 5′ upstream promoter region was amplified (primers: pseb-3F1-catttctctggctagaagattgtcttgt, pseb-3::GFPB1-agtcgacctgcaggcatgcaagtgagcaacaagtttctgaaaggttg) from cosmid C18B12 (provided by A. Coulson, Sanger Center, U.K.), and the gfp coding sequence and the 3′ UTR were amplified (primers: gfpF1-agcttgcatgcctgcaggtcgact, gfpB1-aagggcccgtacggccgactactagg) from the Fire vector pPD95.75. Two primary products were fused (primers: pseb-3F2-ccagttcaaattaactctacccaactac, gfpB2-ggaaacagttatgtttggtatattggg) as described previously (Hobert, 2002) and the fusion product was confirmed by sequencing before microinjection. The full-length translational fusion construct, pseb-3:SEB-3::GFP was generated in the same manner (primers: pseb-3F1- catttctctggctagaagattgtcttgt, seb-3::gfpB1-agtcgacctgcaggcatgcaagagatttcgtagacaccgagtagat, gfpF1-agcttgcatgcctgcaggtcgact, gfpB1-aagggcccgtacggccgactactagg, pseb-3F2-ccagttcaaattaactctacccaactac, gfpB2-ggaaacagttatgtttggtatattggg). The coding region of GFP and the 3′ UTR was fused at the C-terminus of SEB-3. Genomic DNA containing 3082bp of promoter region was amplified from cosmid C18B12 and this construct was used to rescue seb-3 (tm1848 lf) animals. The same genomic region was also amplified from the eg696 mutant lysate for phenocopy experiments. The transformation rescued the roaming defect of seb-3 (tm1848 lf) in 2 independent transgenic lines and successfully phenocopied eg696 in 3 independent transgenic lines.
Behavioral assays
Tracking assays and DIAS analyses were conducted as described previously (Fujiwara et al., 2002). Well-fed L4 animals were transferred to NGM plates (3.5cm for the genetic screen and 9cm for further analysis) with abundant OP50. A single worm was allowed to explore for 17 h without disturbance and then track patterns were analyzed. Results for number of squares (5mm grid; 5×5) traveled was obtained from 10 or more animals and mean values from 4 trials were analyzed by oneway ANOVA with Tukey’s multiple comparison test. For DIAS analysis, the movement of a single young adult animal was recorded using Open Lab ver. 5.0. software, 30 min after transfer. Movements were recorded for 100 min (1 frame / 5 sec) and analyzed using DIAS software (Solltech Inc., Oakdale, IA). The percentages of time roaming and dwelling were defined according to previous methods (Fujiwara et al., 2002) and then were analyzed by a chi-square test.
The octanol avoidance assay was conducted according to Raizen et al. (2008). To assay the response to 1-octanol, an eyelash taped to a Pasteur pipette was dipped in octanol and placed in front of an animal’s nose. The latency to initiate backward movement was measured. Octanol was diluted fresh each day in pure ethanol (Gold Shield, Hayward, CA) to a concentration of 5, 10 or 30%. Single L4 worms were transferred to each NGM plate with abundant OP50, and 15 h later the assay was conducted. Non-quiescent control worms were gently stimulated by a flat worm pick before exposure to octanol. Behavioral quiescence was identified by lack of movement and foraging. An avoidance index was calculated as the number of stimulus presentations (octanol or worm pick) that were avoided, divided by the total number of presentations. Lack of response after 20 seconds was considered a negative response. Mean latency to respond was calculated for all positive responses. Assays were conducted under blind conditions and were not performed more than once on any individual animal. Data were obtained from 10 or more animals and mean values from 3 or more trials were analyzed by one-way ANOVA with a post-hoc Dunnett’s test or two-tailed t-test.
To assay heat stress-induced tremor, temperature controlled NGM buffer solution (22°C, 37°C) was prepared in a tissue embedding base mold (Allegiance Healthcare Corporation Cat. M7307-2) floated in a temperature-controlled water bath. Well-fed young adult animals (17 to 20 h after mid L4) were gently transferred to the preheated NGM buffer solution. Worms exhibiting tremor were counted 5 min later. The assay was conducted under blind conditions and a Fisher’s exact test was used to compare animals showing tremor vs. those not showing tremor. Similar results were observed using M9 buffer solution.
To assay ethanol withdrawal-induced tremor, OP50 was seeded to a half region of a NGM plate 1 day prior ethanol treatment. Ethanol was added to the unseeded region and allowed to diffuse for 2 h; the plates were sealed with parafilm to keep the ethanol from evaporating. Young adult N2 animals were incubated on a 300 mM ethanol plate with OP50 for 4h, and then transferred to M9 buffer after washing. 5 min later, swimming was recorded for 1min. If the animals showed one event within a minute, they were scored positive (with tremor). Assays were performed by an observer blind to the genotype of the animals and the numbers of animals with or without tremor were compared by a Fisher’s exact test. Comparison of ethanol withdrawal-induced tremor between wild type and seb-3 mutant animals was performed using worms exposed to 400 mM ethanol for 4 h.
N2 worms were also incubated for 3 d on ethanol (300 mM) at the embryo stage and ethanol withdrawal-induced tremor was measured in the same manner (N2 animals were grown to adult stage 3 d later on 300 mM ethanol). To determine whether ethanol withdrawal-induced tremor is abolished or reduced by re-exposure to ethanol, ethanol was added back to the group of N2 animals incubated for 3 d. Five min later, after ethanol withdrawal tremors were observed, ethanol was added to achieve a concentration of 300 mM. Swimming was recorded and worms exhibiting tremor were counted 5 min later.
Acute ethanol tolerance and internal concentrations of ethanol were measured as described (Davies et al., 2003; Davies et al., 2004 and Lee et al., 2009). Data were obtained from 10 animals and mean values from 4 trials were analyzed by two-way repeated ANOVA with Boferroni post hoc test.
For studies of swimming, the assay plate (1.6 % BBL-Agar, 5mM potassium phosphate,pH 6.0, 1mM CaCl2 and 1mM MgSO4) was prepared 6 h before assay and ethanol was added to a concentration of 400 mM. Then M9 buffer solution containing ethanol (400 mM) was added to the assay plate and 10 well-fed young adult animals were transferred. We obtained similar results using NGM buffer. Swimming on ethanol or M9 (for untreated control) was recorded for 1 min (30 frames / sec) at 10, 30 and 50 min. The number of body bends per 40 seconds was analyzed to calculate values for relative thrashing (treated / untreated x 100). Differences between mean values were analyzed by two-way repeated measures ANOVA with Bonferroni post-hoc tests.
Rodent Care
Eight-week old male C57BL/6J mice were obtained from The Jackson Laboratory (Bar Harbor, ME) and group housed in a 12:12 hour light-dark cycle (lights on at 7 AM and off at 7 PM) with ad libitum access to food and water. All procedures were conducted in accordance with the Gallo Center Institutional Animal Care and Use Committee and were in accordance with National Institutes of Health Guide for the Care and Use of Laboratory Animals (Committee on Care and Use of Laboratory Animals, 1985). Mice were randomly assigned to experimental groups and a different group of animals was used for each behavioral test.
Pharmacology
The CRF-R1 antagonist, R121919 (3-[6-(dimethylamino)-4-methyl-pyrid-3-yl]-2,5-dimethyl-N,N-dipropyl-pyrazolo[2,3-a]pyrimidin-7-amine), was dissolved in 1M hydrochloric acid before addition of 25% hydroxypropyl-beta-cyclodextrin (HBC) to make a 10 mg/ml solution in a 20% final concentration of HBC, pH 4.5. The vehicle was prepared as above without R121919. This compound was provided by K. Rice (NIDDK, Bethesda, MD). R121919 crosses the blood-brain barrier and blocks the peripheral and central effects of CRF (Zorrilla and Koob, 2004). Pharmacologically significant brain and plasma levels of R121919 have been reported with doses used in this study (Chen et al., 2004), and receptor occupancy has also been shown (Heinrichs et al., 2002).
Rotarod Ataxia and blood ethanol levels
The latency to fall on a constant velocity rotarod was measured and the difference between mean values were analyzed by two way repeated measures ANOVA with Bonferroni post hoc test. Acute functional tolerance was measured using a stationary dowel test as described previously (Dixon, 1965; Findlay et al., 2002; Wallace et al., 2007) and statistical significance was determined by two-tailed t-tests. Blood samples were stored at −80°C until ethanol concentrations were determined using an NAD-ADH enzymatic assay (Carnicella et al., 2009). Ethanol clearance was determined using ethanol-naïve mice administered 4.0 g/kg ethanol by intraperitoneal injection, as in previous work (Choi et al., 2008).
Results
The C. elegans seb-3 encodes a CRF receptor-like GPCR that modulates locomotory and arousal states
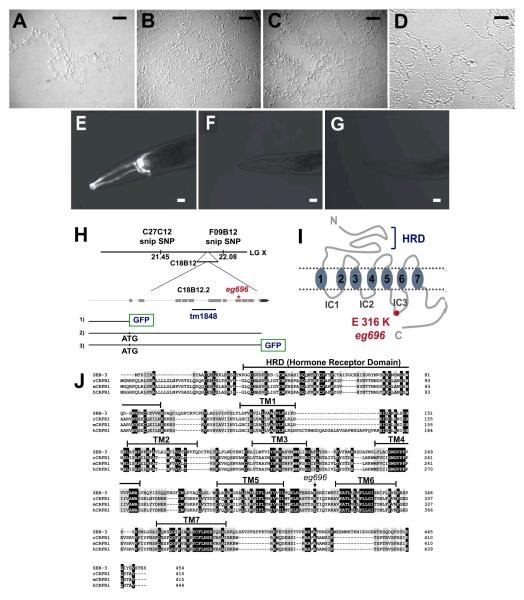
Sensory perception allows an animal to respond properly to its environment and to adapt to changing conditions. To better understand how the nervous system responds to environmental stimuli, we conducted a genetic study to identify genes that regulate behavioral states downstream of sensory perception. C. elegans exhibits two alternative states of locomotory behavior on food: roaming, an active locomotory state of infrequent turns and rapid movement, and dwelling, an inactive locomotory state of frequent stops and reversals. Expression of these behavioral states is modulated by environmental stimuli (Fujiwara et al., 2002). che-2 mutants, which have defects in sensory perception due to abnormal cilium structure (Fujiwara et al., 1999), exhibit a marked decrease in roaming. Although che-2 mutants can move like wild-type animals, they generally dwell in a restricted area. Previously, it was reported that loss of function mutations in egl-4, a cGMP-dependent protein kinase (PKG), suppress decreased roaming of che-2 mutants (Fujiwara et al., 2002). Here we conducted a genetic screen using chemical mutagenesis to identify additional mutations that suppress the roaming defect of che-2 mutants. The most striking phenotype was observed in animals with the eg696 allele (Fig. 1). Heterozygous eg696 animals also showed enhanced roaming (Fig. 1D), indicating that the mutation is dominant. This enhanced roaming was not due to restoration of cilium structure because visualization of sensory neurons using DiI showed persistence of abnormal cilia like che-2 mutants (Fig. 1E-G). These results indicate that the eg696 suppressor mutation lies in a gene that functions downstream of initial sensory perception mediated through cilia expressed by sensory neurons.
Figure 1.
Identification of seb-3, a CRF receptor-like GPCR in C. elegans. A-D, A mutant that exhibits an enhanced active locomotory state was isolated as a suppressor of increased dwelling in che-2 mutant worms. Shown are representative tracks generated over 17 h by single animals A, che-2(e1033), B, che-2 seb-3(eg696), and C, che-2(e1033); egEx[seb-3(eg696)], D, che-2 seb-3(eg696 +/-). Scale bar: 1 mm. E-G, cilia of head sensory neurons were visualized with DiI; E, N2, F, che-2(e1033) and G, che-2(e1033)seb-3(eg696). Scale bar: 10μm. H, The locus for eg696 was mapped to the right arm of X chromosome, 0.31 m.u. between C27C12 and F09B12 SNP markers. tm1848 indicates 765bp of deletion and asterisk indicates the location of eg696. Also displayed are constructs used for further characterization: 1) promoter region of 3082bp fused with GFP; 2) full-length genomic region containing 3082bp of promoter; and 3) full-length genomic region fused with GFP. I, Diagram of full-length SEB-3 as a 7 transmembrane GPCR with an HRD (hormone receptor domain). The amino acid substitution (E316K) generated by the eg696 mutation is represented by a red circle in third intracellular loop. J, Sequence alignment of SEB-3, rat CRFR1, mouse CRFR1 and human CRFR1. Conserved residues are shaded in black and similar residues are shaded in gray. HRD and TM (transmembrane) regions are identified above the SEB-3 sequence. Arrow indicates the missense mutation encoded by eg696.
We mapped eg696 using the snip-SNP method (Wicks et al., 2001; Davis et al., 2005) to an interval of approximately 0.31 map units on LG X (Fig. 1H). The eg696 allele encoded a G to A transition that resulted in a missense mutation in the predicted gene, C18B12.2. The C. elegans genome project has designated C18B12.2 as seb-3, a member of the secretin family (or family B) of GPCRs. To prove this mutation was responsible for the gain of function phenotype of C18B12.2, we amplified the full genomic region of seb-3 including 3082bp of the promoter region from a che-2 (e1033) seb-3 (eg696) worm lysate and then transformed the DNA into che-2 (e1033) mutant animals. As expected, the transformation suppressed the roaming defect of che-2 in transgenic animals (Fig. 1C).
GPCRs have been subdivided into five main families, named glutamate, rhodopsin, adhesion, frizzled/taste2, and secretin (Fredriksson et al., 2003). The secretin family corresponds to Clan B of the A-F classification system (Kolakowski, 1994; Harmar, 2001). Phylogenetic analysis suggests that there are 3 secretin family GPCRs in the C. elegans genome (Harmar, 2001; Cardoso et al., 2006). seb-3 is the GPCR most closely related to CRF receptors of the secretin family (Fig. 1I, 1J; Cardoso et al., 2006) and shares 28 % to 30 % sequence identity and 46 % to 48 % similarity with the mammalian CRF receptor 1 (Fig. 1J). The eg696 mutation changes a conserved glutamic acid 316 to a lysine in the third intracellular loop, where CRF receptors interact with heterotrimeric G-proteins (Fig. 1I). Taken together with the dominant phenotype of eg696, these results suggested that the eg696 is a gain-of-function mutation.
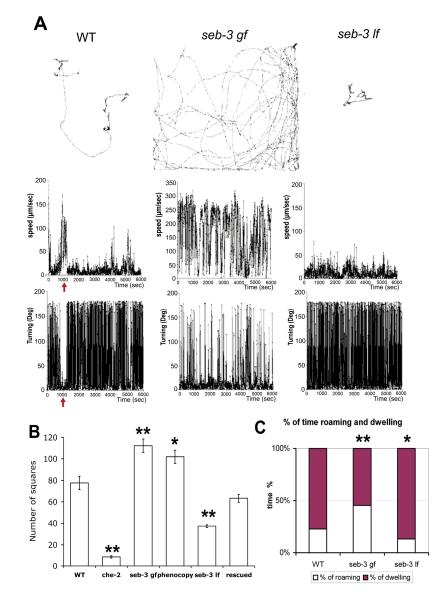
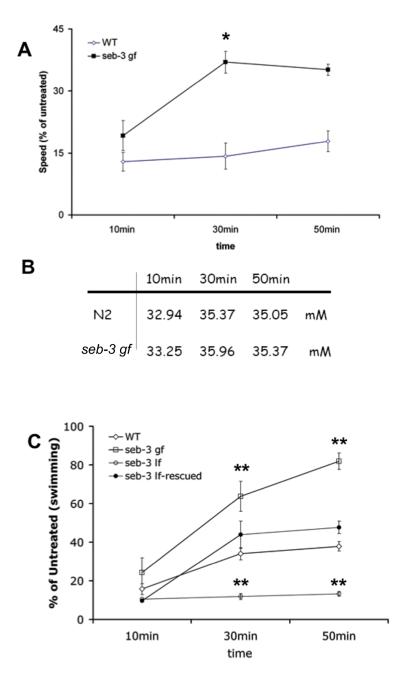
seb-3 (gf) mutant animals showed enhanced exploratory behavior in a single animal tracking assay (F(5, 25) = 74.80, P<0.0001 by one-way ANOVA and ** P<0.001 by Tukey’s multiple comparison test) (Fig. 2B). To determine how roaming versus dwelling is affected, we used a computerized image analysis system (DIAS) to analyze the movements of single worms over 100 min. Figure 2A shows an example of this movement analysis. Roaming was defined as forward movement at high speed with low turning whereas dwelling was defined as low speed movement with high turning and stops (Fujiwara et al., 2002). Movements lasting 44 sec or longer were identified as roaming, whereas movements less than 44 sec in duration were identified as dwelling (Fujiwara et al., 2002). seb-3 gf mutants spent significantly more time roaming, indicating an enhanced active state of locomotion (** P < 0.001 by chi-square analysis, X2 (1, N=10)=29.87) (Fig. 2C).
Figure 2.
Analysis of behavioral states based on changes in speed and direction.
A, Locomotion of single worms on a bacterial lawn was recorded for 100 min then analyzed with DIAS. Shown are examples of speed and turning analyses of N2, seb-3 (eg696 gf) and seb-3 (tm1848 lf) animals. In the top panels, each dot represents the position of the worm at 5 sec intervals in the worm track. Graphs below represent speed (μm/sec) and turning (change of direction, degrees/5sec). Arrow in N2 graphs shows a roaming state characterized by high-speed movements with few turns. B, Tracks generated on food for 17 h (9cm plate) were superimposed on a 5mm grid (5 × 5) and the number of squares that animals traversed was counted. Data are mean ± SEM values. n=78 for N2; 105 for che-2; 65 for seb-3 (eg696 gf); 50 for phenocopy -N2Ex[SEB-3(eg696)]; 55 for seb-3(tm1848 lf); 52 for rescued). C, Percentage of time roaming Total number of minutes and number (n) of animals analyzed were: 1000 min (n=10) for N2; 1000 min (n=10) for seb-3 (eg696 gf); and 1000 min (n=10) for seb-3 (tm1848 lf).
To determine if eg696 increases roaming in the absence of the che-2 mutation, we introduced the mutation into wild-type worms. Genomic DNA containing eg696 was amplified by nested PCR and microinjected into worms of the N2 wild-type strain [Fig. 1H 2)]. These transgenic worms and eg696 mutant animals without a che-2 mutation showed enhanced roaming behavior (Fig. 2B). We next tested if a seb-3 loss-of-function mutation could reduce roaming. The seb-3 (tm1848) allele contains a 765 bp deletion within the seb-3 gene (Fig. 1H) and is a putative null mutation. seb-3 (lf) animals showed reduced roaming on both the tracking assay (F(5, 25) = 74.80, P<0.0001 by one-way ANOVA and * P<0.05 by Tukey’s multiple comparison test) and by movement analysis (*P < 0.05 by chi-square analysis, X2 (1, N=10)=5.647) (Fig. 2). In contrast, their locomotory speed without food (WT, 189.18 μm/sec; seb-3 lf, 193.15 μm/sec; p<0.1) and speed after mechanical stimulation were like wild-type animals (wild type, 262.32 μm/sec; seb-3 lf, 246.30 μm/sec; p<0.1). This reduced roaming state was restored by introduction of wild-type seb-3, suggesting that SEB-3 modulates the locomotory state when animals are placed on food (Fig. 2B).
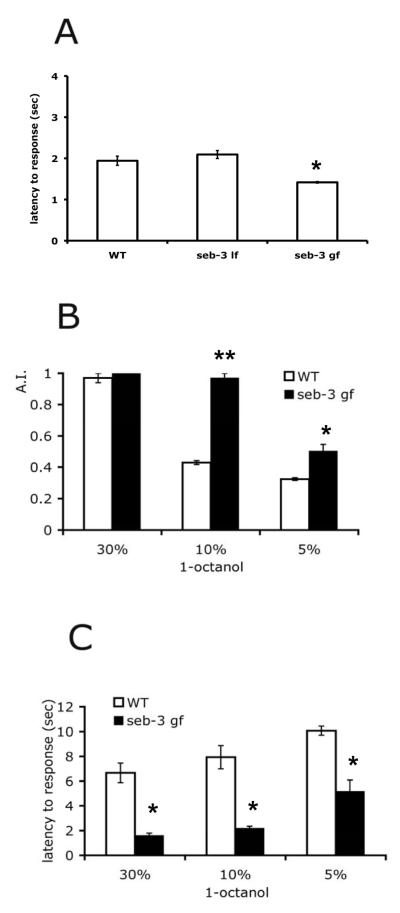
Since seb-3(lf) mutants exhibit altered states of roaming but normal locomotion, we considered if the seb-3 mutation might modulate the state of arousal in these animals. We hypothesized that enhanced roaming of seb-3(gf) animals results from hyper-arousal, while prolonged dwelling of seb-3(lf) animals is due to diminished arousal. C. elegans exhibit a sleep-like lethargus state during which they are quiescent. Quiescent, wild-type animals display a less sensitive olfactory response to repellants such as 1-octanol. However, mutants that have defects in maintaining this quiescent state are more sensitive to 1-octanol, due to hyper-arousal (Raizen et al., 2008). Therefore, to test if seb-3 modulates arousal, mutants were tested for response to 1-octanol. A single worm was transferred to an assay plate with abundant food and left undisturbed to induce behavioral quiescence. In non-quiescent controls, both seb-3 (gf) and seb-3 (lf) mutant animals responded normally, although seb-3 (gf) animals showed more rapid responses (F(2,6)=17.02, P=0.0034 by one-way ANOVA and * P < 0.01 by Dunnett’s multiple comparison test) (Fig. 3A). Behavioral quiescence of seb-3 (gf) animals was more easily interrupted by the olfactory repellant. seb-3 (gf) mutants exhibited marked and rapid avoidance behavior after exposure to even lower concentrations of 1-octanol (Fig. 3B, C). seb-3 (gf) animals were more sensitive than wild-type (WT) worms to 1-octanol as shown by (Fig. 3B) their greater avoidance index (A.I., see methods) at low concentrations (5%; t(4)=4.061, P<0.05 and 10%; t(4)=16.46, P<0.001) of 1-octanol and (Fig. 3C, 30%; t(4)=6.228, P<0.05, 10%; t(4)=6.025, P<0.05, 5%; t(4)=4.980, P<0.05) their shorter latency to respond to 1-octanol at all concentrations tested (* P <0.05 and ** P < 0.001 by two-tailed t-tests). Therefore, seb-3 (gf) animals exhibit an abnormally increased state of arousal.
Figure 3.
seb-3 (eg696 gf) mutant animals exhibit hyper-arousal. A, Octanol avoidance during non-quiescence. n=30 for N2, seb-3 (tm1848 lf) and seb-3 (eg696 gf)]. B-C, Octanol avoidance during behavioral quiescence. n=36 for N2 and seb-3 (eg696 gf) (BC). Data are mean ± SEM values.
seb-3 is expressed in the nervous system
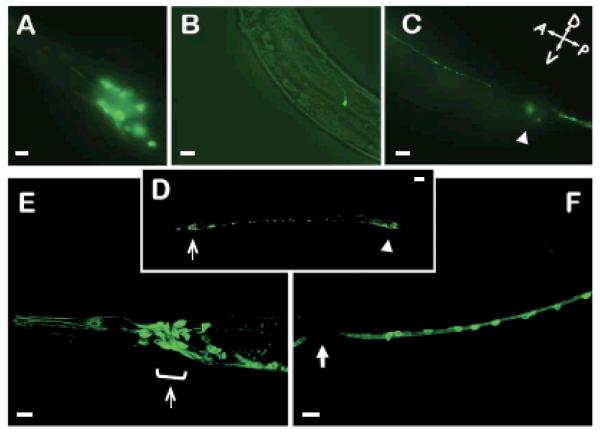
To examine the pattern of seb-3 expression, a seb-3 promoter was fused with a green fluorescent protein (GFP) reporter gene [Fig. 1H 1)]. As shown in Figure 5, GFP was observed mostly in neurons. We also fused GFP to the C-terminal tail of wild-type SEB-3 [Fig. 1H (3)]. This SEB-3::GFP fusion protein was expressed mainly in neurons. It was expressed predominantly in head neurons, dorsal and ventral nerve cords, and tail ganglia (Fig. 4D-F). Faint expression was also observed in posterior body wall muscles and the rectal gland. It was localized to the cytosol, showing strongest expression near the cell membrane throughout the cell body and axon. This SEB-3::GFP fusion protein successfully rescued seb-3 (lf) animals suggesting that the GFP pattern represents proper endogenous expression (Fig. 2B, 6C).
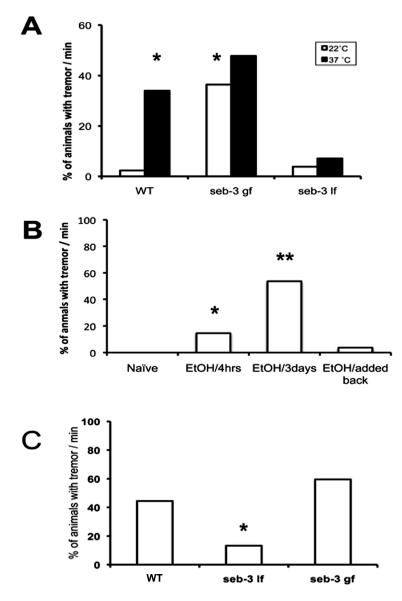
Figure 5.
SEB-3 mediates heat stress-induced and ethanol-withdrawal-induced tremor. A, Heat evoked tremor in N2, but not in seb-3 (lf) animals (*P < 0.0001 by Fisher’s exact test). seb-3 (eg696 gf) displayed tremor without heat stress. [* P < 0.0001 by Fisher’s exact test; n=75 (N2, 22°C), 95 (N2, 37°C), 94 (seb-3 (eg696 gf), 22°C), 80 (seb-3 (eg696 gf), 37°C), 85 (seb-3 (tm1848 lf), 22°C), and 97 (seb-3 (tm1848 lf), 37°C)]. B, Acute ethanol withdrawal (300mM) induced a tremor in N2 animals (n=50) and the incidence of tremor was greater after 3 days (n=54) of ethanol exposure than after 4 h (n=55) of exposure (* P<0.001, ** P <0.0001 by Fisher’s exact test). Reapplication of ethanol completely abolished tremor (n=54). C, Ethanol withdrawal (400mM) induced a tremor in seb-3 mutants but seb-3 (lf) animals (n=91) showed much less tremor than wild type, N2 animals (n=60) (* P < 0.0001 by Fisher’s exact test).
Figure 4.
seb-3 is expressed in the nervous system. A-C, pseb-3:GFP was expressed in head ganglia (A), SDQL (B), and in dorsal cord and tail ganglia (C). D-F, Expression of pseb-3:SEB-3 (WT)::GFP in seb-3 (tm1848 lf) animals. The rescued seb-3 (tm1848 lf) transgenic animals showed SEB-3::GFP fluorescence predominantly in the nervous system. Open arrows indicate the nerve ring and arrowheads indicate tail ganglia. (F) shows ventral nerve cords and solid arrow points to vulva. Scale bar: 10μm.
Figure 6.
seb-3 regulates development of acute tolerance to ethanol.
A, Acute tolerance on locomotion (500mM ethanol). Data are mean ± SEM values. B, Internal concentrations of ethanol during continuous exposure to 500 mM ethanol in the medium were similar in N2 and seb-3 (eg696 gf) worms at all time points measured. C, Acute tolerance on swimming. Data are mean ± SEM values.
SEB-3 regulates tremor induced by heat stress or ethanol withdrawal
Since seb-3 shows sequence homology with mammalian CRF receptors, we hypothesized that like CRF receptors, SEB-3 mediates responses to stress. In liquid environments, C. elegans exhibits swimming behavior, which is distinct from crawling behavior (Pierce-Shimomura et al., 2008). Certain facets of locomotory behavior are more obvious during swimming than crawling (Schreiber et al., 2010). While swimming, seb-3 (gf) animals, exhibited pronounced body tremor (sup movie). A sporadic tremor was observed in distilled water, M9 salt buffer, and NGM buffer solutions. This movement was observed occasionally and lasted 2-20 seconds. Shorter duration tremor (approximately < 3 seconds) affected head and tail movement while longer duration tremor (approximately > 3 seconds) affected the whole body. Since tremor can be evoked by stress in mammals, we explored the effects of several external stressors on tremor in wild type and seb-3 mutant animals. Acute heat caused tremor in wild-type animals and increased the incidence of this behavior among seb-3 (gf) animals (Fig. 5A). The percentage of wild-type animals that displayed tremor dramatically increased after heat stress (P < 0.0001 by Fisher’s exact test) to a level similar to that observed for seb-3 (gf) animals not subjected to heat stress (P < 0.0001 by Fisher’s exact test). In contrast, seb-3 (lf) animals did not show increased tremor after heat stress, indicating that acute heat stress-induced tremor is modulated by SEB-3 (Fig. 5A).
In mammals, up-regulation of CRF signaling occurs with alcohol dependence and contributes to the negative reinforcing effects of ethanol withdrawal (Koob, 2008). Several behavioral manifestations of ethanol withdrawal, such as anxiety and tremor, resemble behavioral responses to stress. Therefore, we investigated if withdrawal from ethanol exposure provokes tremor in C. elegans (Fig. 5B). Animals were exposed to ethanol at media concentrations (300-400 mM) that produce internal concentrations (22 ± 0.8 mM internal concentration on 400 mM ethanol media) associated with intoxication in mammals (Davies et al., 2003). Treatment of wild-type animals for 4 h was long enough to induce tremor upon withdrawal of ethanol (P<0.001 by Fisher’s exact test). The percentage of the animals exhibiting tremor was drastically increased after 3 d of ethanol exposure (P <0.0001 by Fisher’s exact test). Furthermore, the populationexhibiting tremor was dramatically decreased after re-administration of ethanol (Fig. 5B). The severity and incidence of tremor were greater in seb-3 (gf) animals than wild-type animals (Fig. 5C). By contrast, seb-3 (lf) animals showed less withdrawal-induced tremor (P <0.0001 by Fisher’s exact test). Together, these data indicate that SEB-3 regulates tremor induced by ethanol withdrawal.
SEB-3 positively regulates acute ethanol tolerance
C. elegans develops acute tolerance to ethanol similar to that observed in mammals. We previously demonstrated that allelic variation in the npr-1 gene in wild-type animals alters the functional level of NPR-1, a neuropeptide-1-like receptor, and the development of acute tolerance to ethanol (Davies et al., 2004). Animals with decreased npr-1 function exhibit increased exploratory behavior on the tracking assay [N2 (n=54), 71.23; npr-1 (ky13) (n=40), 98.75; npr-1 (ad609) (n=40), 90.12; P <0.05 by t-test]. We therefore investigated if seb-3 (gf) animals, which show increased roaming, similarly exhibit enhanced ethanol tolerance. We observed greater acute tolerance to ethanol in seb-3 (gf) mutants compared with wild-type animals (Fig. 6A). Compared with wild type (n=40), seb-3 (eg696 gf) animals (n=30) recovered more rapidly from the depressive effect of 500 mM ethanol on locomotion. Two-way, repeated measures ANOVA showed main effects of time [F(2,10)=55.29; P < 0.0001] and genotype [F(1,10)=22.88; P < 0.005] with a significant interaction between genotype and time [F(2,10)=31.06; P < 0.0001] (* P<0.05 by Bonferroni post hoc test). This adaptation was not due to altered ethanol clearance, since the internal ethanol concentrations were similar in seb-3 (gf) mutants and wild-type animals over the time course of the assay (Fig. 6B). To address whether seb-3 (lf) animals show defective acute tolerance, an acute tolerance assay was performed based on swimming behavior rather than crawling behavior because wild-type animals show a reduced degree of acute tolerance when crawling compared with swimming (Fig. 6A). Although seb-3 (lf) animals became intoxicated, unlike wild-type animals, they failed to develop acute tolerance (Fig. 6C). Exposure to 400 mM ethanol suppressed swimming in all animals but seb-3 mutants showed differences in development of acute tolerance to this effect of ethanol. Two-way, repeated measures ANOVA showed main effects of time [F(2,22)=124.70; P < 0.0001] and genotype [F(3,22)=45.07; P < 0.0001] with a significant interaction between genotype and time [F(6,22)=18.14; P < 0.0001]. Swimming recovered more rapidly in seb-3 (eg696 gf) worms (n=30) than in wild type (P < 0.01 by Bonferroni post hoc test), N2 worms (n=50), while it failed to recover over 50 min in seb-3 (tm1848 lf) animals (n=40) (P < 0.01 by Bonferroni post hoc test). Introduction of a wild-type copy of the seb-3 gene into seb-3 (lf) animals restored acute tolerance to ethanol. seb-3 (tm1848 lf)-rescued animals (n=30) showed a rate of recovery that was similar to wild-type animals (Fig. 6C). The rescued animals developed acute tolerance slightly more rapidly than wild-type animals, and some of them exhibited tremor like seb-3 gf animals, possibly due to over-expression of seb-3. These results suggest that SEB-3 positively regulates the development of acute tolerance to ethanol.
Conservation of SEB-3 and CRFR1 in development of acute tolerance to ethanol
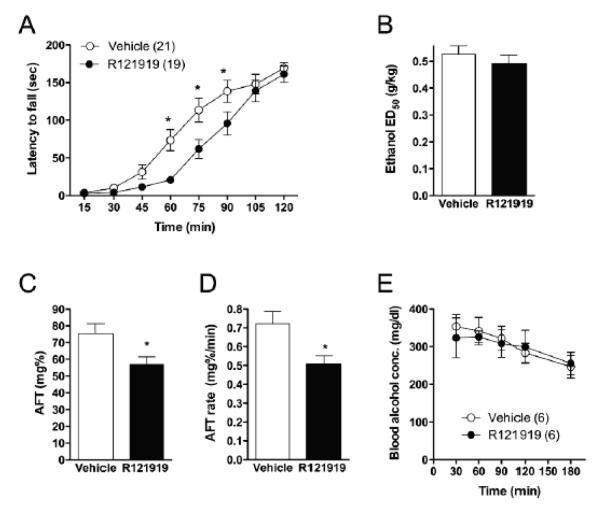
Involvement of seb-3 in acute tolerance to ethanol in C. elegans raises the possibility that orthologs of this receptor regulate acute functional (behavioral) tolerance (AFT) to ethanol in mammals. Since SEB-3 most closely resembles the mammalian CRF1 receptor, we investigated if the CRF1 receptor regulates AFT in mammals. Mice were examined for ataxia on a fixed-speed rotarod after pretreatment with R121919, a selective CRF1 receptor antagonist, followed by administration of 2g/kg ethanol. At 15, 30, and 45 min after ethanol administration, the latencies to fall from the rotarod were similar between R121919- and vehicle-treated groups. However, at 60, 75, and 90 min after ethanol administration, the R121919-treated group showed a decreased ability to stay on the rotarod compared with the vehicle-treated group Two-way, repeated measures ANOVA showed main effects of treatment [F(1, 266) = 6.741; P = 0.0133] and time [F(7, 266) = 104.1 ; P < 0.0001], with an interaction between these factors [F(7, 266) = 3.047; P = 0.0042] (*P < 0.05 compared with R121919-treated mice by Bonferroni post hoc test) (Fig. 7A). These results suggest that though initial sensitivity to ethanol-induced ataxia was similar, the rate of recovery, or the development of AFT, was impaired when the CRF1 receptor antagonist was present. To confirm the lack of difference in acute initial sensitivity, we measured the ED50 (ethanol dose required to cause initial ataxia in 50% of mice) for inducing rotarod ataxia using the “up-down” method and found it to be similar between groups (Fig. 7B).
Figure 7.
Acute tolerance is mediated by CRF1 receptor in mice. A, R121919-treated mice (N = 19) were less able to remain on the rotarod than vehicle-treated mice (N = 21) for 60-90 minutes after receiving 2 g/kg ethanol. B, The ethanol dose required to cause initial loss of righting reflex in 50% of mice (ED50) for ethanol-induced ataxia was similar for R121919- and vehicle-treated mice (n = 6 per group). Data are mean values ± 95% CI. C, AFT magnitude was lower in R121919-treated mice (n = 22) than in vehicle-treated mice (n = 23). *P < 0.05 by two-tailed t-test. D, The rate of development of AFT was also lower in R121919-treated mice (n = 22) than in vehicle-treated mice (n = 23). *P < 0.05 by two-tailed t-test. E, Ethanol clearance was similar for R121919- and vehicle-treated mice (n = 6 per group). Data are mean ± SEM values.
To further examine the role of the CRF1 receptor in AFT to ethanol, we measured AFT using a stationary dowel test, which compares responses to two sequential doses of ethanol (Erwin et al., 2000; Wu et al., 2001; Wallace et al., 2007). We found that the magnitude of AFT and the rate at which AFT developed after administration of 1.5 mg/kg ethanol were lower in mice pre-treated with R121919 compared with vehicle (Fig. 7C; t(43)=2.407, P<0.05, D; t(43)=2.720, P < 0.05 by two-tailed t-test). These differences were not due to altered ethanol clearance, which was similar in R12919- and vehicle-treated mice (Fig. 7E). Together, these data indicate that the CRF1 receptor, like SEB-3, regulates acute tolerance to ethanol.
Discussion
We report the molecular cloning and functional characterization of seb-3, a CRF receptor-like GPCR gene in C. elegans. Two major CRF receptor subtypes, CRF1 and CRF2, have been cloned from vertebrates (Chen et al., 1993; Liaw et al., 1996), and SEB-3 is most closely related to the CRF1 receptor based on sequence homology and similarity in function. The CRF system is present not only in vertebrates, but is also conserved in invertebrates. For example, CRF-like peptides are found in insects where they mediate osmotic homeostasis (Lovejoy and Jahan, 2006; Li et al., 2008). We found that SEB-3 mediates responses to stress in C. elegans like the CRF1 receptor which mediates responses to stress in mammals (Grammatopoulos et al., 1999; Timpl et al., 1998; Smith et al., 1998). In addition, we found that SEB-3 is expressed and functions mainly in the nervous system like the CRF1 receptor. These data support the conclusion that SEB-3 is a CRF receptor-like GPCR in C. elegans.
GPCRs exist in equilibrium between active and inactive conformational states (Lefkowitz et al., 1993). GPCRs couple to and activate G proteins when in their active state, whereas they remain uncoupled in the inactive state. Binding of an agonist to the extracellular region of the receptor induces the active state. However, GPCRs can also adopt an active conformation in the absence of agonist and exhibit constitutive activity. GPCR activation and signaling can be strongly affected by single point mutations, some of which result in human disease (Nishihara et al., 2007; Smit et al., 2007). Point mutations in the third intracellular loop, in a binding region for G proteins, can result in constitutive activation, indicating that the third intracellular loop participates in GPCR activation (Kjelsberg et al., 1992; Nishihara et al., 2007; Chee et al., 2008). Members of the CRF receptor family show the highest homology in the third intracellular loop (Arai et al., 2001; Dautzenberg et al., 2001). The eg696 allele of seb-3 that we identified encodes a mutation that changes a conserved residue in the third intracellular loop of SEB-3 and results in a gain of function, suggesting that this allele encodes a constitutively active form of SEB-3. Indeed, eg696 homozygous and heterozygous mutant animals show phenotypes opposite to those of a seb-3 deletion mutant in locomotory state, stress response and development of acute tolerance to ethanol. The gain-of-function mutant that we have identified should prove very useful for further understanding the structural determinants involved in activation of secretin family GPCRs and also for investigating the function of CRF-like receptors in vivo.
In C. elegans, egl-4, which encodes a PKG, regulates roaming and dwelling on food (Fujiwara et al., 2002). This function is conserved in other invertebrates, such as fruit flies, in which distinct foraging behaviors on food correlate with PKG activity (Osborne et al., 1997) and in honeybees, where PKG is up-regulated during the transition from nurse to foraging bee (Ben-Shahar et al., 2003). Consistent with a role for EGL-4/PKG in roaming and dwelling, egl-4 also regulates behavioral quiescence and arousal in C. elegans (Raizen et al., 2008). egl-4 mutants with reduced quiescence are more sensitive to aversive stimuli (1-octanol) due to hyper-arousal. Consistent with the idea that seb-3 regulates an arousal state in C. elegans, we observed that seb-3 (gf) animals are more sensitive to aversive 1-octanol stimuli during quiescence. Furthermore, activation of the CRF system during stress is characterized by increased arousal (Koob and Thatcher-Britton, 1985) and genetic studies in fruit flies and mice have shown that downstream activation of protein kinase A and CREB signaling promotes wakefulness (Mackiewicz et al., 2008). Taken together, these results indicate an important conserved role for CRF1-like receptors in increasing arousal in vertebrates and invertebrates.
NPR-9, which is most similar to mammalian galanin receptors, has been recently found to modulate roaming and dwelling in C. elegans (Bendena et al., 2008). Interestingly, the neuropeptide galanin has been implicated in responses to stress and its receptors are being studied as a drug targets for the treatment of anxiety disorders and alcohol abuse (Morilak et al., 2003; Holmes et al., 2003; Belfer et al., 2006; Belfer et al., 2007), further suggesting that enhanced roaming is a characteristic of the behavioral stress response in C. elegans.
Tremor has not been described previously in C. elegans, probably because it is only observed when worms are in a liquid environment. It is distinct from previously reported muscle movements such as pentylenetetrazole (PTZ)-induced muscle convulsion and twitching (Locke et al., 2006; Moerman and Baillie, 1979). Tremor-like movement is markedly faster than PTZ-induced epileptic convulsion (sup movie, Locke et al., 2006) and twitching of unc-22 (lf) is associated with a structural defect that results in an inability to sustain muscle contraction. Therefore, nicotine-induced muscle contraction in unc-22 (lf) leads to uncoordinated twitching, which is a non-synchronized rapid movement of each body part. However unlike unc-22 (lf), seb-3 (gf) animals are completely paralyzed by nicotine-induced muscle contraction (data not shown) and display a tremor-like behavior without any drug. Stress, anxiety and ethanol withdrawal can induce tremor in humans. Here, we demonstrated that C. elegans exhibits tremor as a stress response and as a sign of ethanol withdrawal. seb-3 (gf) mutant animals exhibited pronounced body tremor even without stress, whereas seb-3 (lf) animals showed impaired development of tremor after stress. Wild-type animals developed tremor soon after ethanol withdrawal, as observed in rodents and humans (Heilig et al., 2010). Withdrawal induced tremor is markedly reduced after retreatment with ethanol (Fig. 5B), suggesting that this withdrawal behavior does not originate from a developmental defect caused by ethanol exposure, such as a previously described ethanol withdrawal behavior (Mitchell et al., 2010).
In mammals, repeated exposure to ethanol leads to ethanol dependence and associated dysregulation of CRF signaling in the central nervous system. Such dysregulation is evident within limbic brain structures that make up the extended amygdala (the central nucleus of the amygdala, the medial amygdala, the bed nucleus of the stria terminalis and the medial shell of the nucleus accumbens) (Koob, 2008). Increased CRF release in the extended amygdala during alcohol withdrawal leads to withdrawal-induced anxiety that is blocked by CRF1 receptor antagonists (Merlo Pich et al., 1995; Huang et al., 2010). Whether dysregulation of CRF signaling in these limbic brain regions also contributes to tremor is not yet known. However, it has been reported that administration of CRF into the cerebral ventricles of the rat can induce tremor (Jones et al., 1998) and tremor evoked by harmaline is associated with increased CRF mRNA in the central nervous system of the cat (Cummings et al., 1994). Thus, our studies with seb-3 mutants suggest that the tremor that appears during ethanol withdrawal in rodents and humans is a manifestation of a withdrawal-induced stress response, mediated in part by activation of the CRF system.
We found that seb-3 regulates acute tolerance to ethanol, measured as the rate of recovery from locomotor suppression induced by acute ethanol exposure. Since these findings indicate that SEB-3 promotes acute tolerance to ethanol in C. elegans, we examined its closest mammalian ortholog, the CRF1 receptor, and found that inhibiting CRF1 receptors reduced AFT to ethanol in mice. Up-regulation of the CRF system in alcohol dependent rodents is proposed to be a major factor producing an emotional state that negatively reinforces excessive ethanol consumption in dependent animals (Koob, 2008). This hypothesis is supported by evidence that CRF1 receptor antagonists reduce ethanol consumption in ethanol dependent rodents (Chu et al., 2007; Roberto et al., 2010). However, recent evidence indicates that CRF1 receptor antagonists also decrease high levels of ethanol self-administration in non-dependent rodents provided limited access to ethanol under conditions that model binge drinking (Sparta et al., 2008; Roberto et al., 2010). In addition, CRF1 receptor antagonists reduce ethanol consumption in non-dependent Marchigian-Sardinian rats selectively bred for high ethanol preference; these rats carry a polymorphism in the crhr1 gene associated with elevated brain CRF1 receptor expression (Hansson et al., 2006; Hansson et al., 2007).
Our finding that CRF1 receptor signaling promotes AFT raises the possibility that CRF1 receptor antagonists limit binge-like drinking, in part, by increasing the extent and duration of ethanol intoxication. Acute exposure to ethanol activates the hypothalamic-pituitary-adrenal (HPA) axis, which leads to secretion of glucocorticoids by the adrenal glands (Rivier et al., 1984). However, CRF receptor regulation of binge-like drinking does not involve the HPA axis, but instead involves extrahypothalamic CRF systems since intracerebroventricular administration of CRF1 receptor antagonists reduces binge drinking, even after adrenalectomy (Lowery et al., 2010). Ethanol is known to exert acute effects on the extended amygdala that are CRF1 dependent and could contribute to binge drinking behavior. For example, ethanol stimulates the release of GABA in the central nucleus of the amygdala through a CRF1 receptor dependent mechanism (Bajo et al., 2008; Roberto et al., 2010).
In conclusion, we have identified behavioral responses that are conserved in C. elegans and mammals, and are regulated by related members of the secretin family of GPCRs, SEB-3 and the CRF1 receptor. Because of this striking conservation in function in regulating arousal, activity, and tremor, our finding that SEB-3 also regulates acute tolerance to ethanol in C. elegans led us to identify a novel role for the CRF1 receptor in promoting AFT in mammals. We postulate that by accelerating recovery from acute intoxication, CRF acting at CRF1 receptors facilitates excessive drinking in subjects who are not alcohol dependent. Our results not only demonstrate that C. elegans is a valuable genetic model system to understand mechanisms that underlie alcohol use disorders, but also provide a mechanistic rationale to support investigating the use of CRF1 receptor antagonists to prevent excessive drinking by individuals who, despite periodically drinking heavily, are not alcohol dependent and do not meet criteria for the diagnosis of alcoholism.
Supplementary Material
Acknowledgments
This work was supported by NIH grant AA017072 and funds provided by the State of California for medical research on alcohol and substance abuse through the University of California, San Francisco. We thank the C. elegans Genetics Center for providing strains through a grant from the National Institutes of Health National Center for Research Support, and Shohei Mitani and the National BioResource Project of Japan for the tm1848 allele. We are grateful to Kenner Rice for providing us with the R121919 compound, Eric Zorrilla for advice on solubilizaion, and Jackie Stecher for technical assistance. We also thank Jon T. Pierce-Shimomura and Matt A. Shreiber for helpful discussions, and Hongkyun Kim and L. René García for comments and editing of the manuscript.
Footnotes
Conflicts of Interest: None
References
- Arai M, Assil IQ, Abou-Samra AB. Characterization of three corticotropin-releasing factor receptors in catfish: a novel third receptor is predominantly expressed in pituitary and urophysis. Endocrinology. 2001;142:446–454. doi: 10.1210/endo.142.1.7879. [DOI] [PubMed] [Google Scholar]
- Bajo M, Cruz MT, Siggins GR, Messing R, Roberto M. Protein kinase C epsilon mediation of CRF- and ethanol-induced GABA release in central amygdala. Proc Natl Acad Sci U S A. 2008;105:8410–8415. doi: 10.1073/pnas.0802302105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfer I, Hipp H, Bollettino A, McKnight C, Evans C, Virkkunen M, Albaugh B, Max MB, Goldman D, Enoch MA. Alcoholism is associated with GALR3 but not two other galanin receptor genes. Genes Brain Behav. 2007;6:473–481. doi: 10.1111/j.1601-183X.2006.00275.x. [DOI] [PubMed] [Google Scholar]
- Belfer I, Hipp H, McKnight C, Evans C, Buzas B, Bollettino A, Albaugh B, Virkkunen M, Yuan Q, Max MB, Goldman D, Enoch MA. Association of galanin haplotypes with alcoholism and anxiety in two ethnically distinct populations. Mol Psychiatry. 2006;11:301–311. doi: 10.1038/sj.mp.4001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shahar Y, Leung HT, Pak WL, Sokolowski MB, Robinson GE. cGMP-dependent changes in phototaxis: a possible role for the foraging gene in honey bee division of labor. J Exp Biol. 2003;206:2507–2515. doi: 10.1242/jeb.00442. [DOI] [PubMed] [Google Scholar]
- Bendena WG, Boudreau JR, Papanicolaou T, Maltby M, Tobe SS, Chin-Sang ID. A Caenorhabditis elegans allatostatin/galanin-like receptor NPR-9 inhibits local search behavior in response to feeding cues. Proc Natl Acad Sci U S A. 2008;105:1339–1342. doi: 10.1073/pnas.0709492105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso JC, Pinto VC, Vieira FA, Clark MS, Power DM. Evolution of secretin family GPCR members in the metazoa. BMC Evol Biol. 2006;6:108. doi: 10.1186/1471-2148-6-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnicella S, Ahmadiantehrani S, Janak PH, Ron D. GDNF is an endogenous negative regulator of ethanol-mediated reward and of ethanol consumption after a period of abstinence. Alcohol Clin Exp Res. 2009;33:1012–1024. doi: 10.1111/j.1530-0277.2009.00922.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee MJ, Morl K, Lindner D, Merten N, Zamponi GW, Light PE, Beck-Sickinger AG, Colmers WF. The third intracellular loop stabilizes the inactive state of the neuropeptide Y1 receptor. J Biol Chem. 2008;283:33337–33346. doi: 10.1074/jbc.M804671200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Wilcoxen KM, Huang CQ, Xie YF, McCarthy JR, Webb TR, Zhu YF, Saunders J, Liu XJ, Chen TK, Bozigian H, Grigoriadis DE. Design of 2,5-dimethyl-3-(6-dimethyl-4-methylpyridin-3-yl)-7-dipropylaminopyrazolo[1 ,5-a]pyrimidine (NBI 30775/R121919) and structure--activity relationships of a series of potent and orally active corticotropin-releasing factor receptor antagonists. J Med Chem. 2004;47:4787–4798. doi: 10.1021/jm040058e. [DOI] [PubMed] [Google Scholar]
- Chen R, Lewis KA, Perrin MH, Vale WW. Expression cloning of a human corticotropin-releasing-factor receptor. Proc Natl Acad Sci U S A. 1993;90:8967–8971. doi: 10.1073/pnas.90.19.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Lewis KA, Perrin MH, Vale WW. Expression cloning of a human corticotropin-releasing-factor receptor. Proc Natl Acad Sci U S A. 1993;90:8967–8971. doi: 10.1073/pnas.90.19.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DS, Wei W, Deitchman JK, Kharazia VN, Lesscher HM, McMahon T, Wang D, Qi ZH, Sieghart W, Zhang C, Shokat KM, Mody I, Messing RO. Protein kinase Cdelta regulates ethanol intoxication and enhancement of GABA-stimulated tonic current. J Neurosci. 2008;28:11890–11899. doi: 10.1523/JNEUROSCI.3156-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu K, Koob GF, Cole M, Zorrilla EP, Roberts AJ. Dependence-induced increases in ethanol self-administration in mice are blocked by the CRF1 receptor antagonist antalarmin and by CRF1 receptor knockout. Pharmacol Biochem Behav. 2007;86:813–821. doi: 10.1016/j.pbb.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements S, Schreck CB, Larsen DA, Dickhoff WW. Central administration of corticotropin-releasing hormone stimulates locomotor activity in juvenile chinook salmon (Oncorhynchus tshawytscha) Gen Comp Endocrinol. 2002;125:319–327. doi: 10.1006/gcen.2001.7707. [DOI] [PubMed] [Google Scholar]
- Cummings S, Hinds D, Young WS., 3rd Corticotropin-releasing factor mRNA increases in the inferior olivary complex during harmaline-induced tremor. Brain Res. 1994;660:199–208. doi: 10.1016/0006-8993(94)91290-4. [DOI] [PubMed] [Google Scholar]
- Dautzenberg FM, Kilpatrick GJ, Hauger RL, Moreau J. Molecular biology of the CRH receptors-- in the mood. Peptides. 2001;22:753–760. doi: 10.1016/s0196-9781(01)00388-6. [DOI] [PubMed] [Google Scholar]
- Davies AG, Bettinger JC, Thiele TR, Judy ME, McIntire SL. Natural variation in the npr-1 gene modifies ethanol responses of wild strains of C. elegans. Neuron. 2004;42:731–743. doi: 10.1016/j.neuron.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Davies AG, Pierce-Shimomura JT, Kim H, VanHoven MK, Thiele TR, Bonci A, Bargmann CI, McIntire SL. A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans. Cell. 2003;115:655–666. doi: 10.1016/s0092-8674(03)00979-6. [DOI] [PubMed] [Google Scholar]
- Davis MW, Hammarlund M, Harrach T, Hullett P, Olsen S, Jorgensen EM. Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics. 2005;6:118. doi: 10.1186/1471-2164-6-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon WJ. The Up-and-Down Method for Small Samples. JAMA. 1965;60:967–978. [Google Scholar]
- Erwin VG, Gehle VM, Deitrich RA. Selectively bred lines of mice show response and drug specificity for genetic regulation of acute functional tolerance to ethanol and pentobarbital. J Pharmacol Exp Ther. 2000;293:188–195. [PubMed] [Google Scholar]
- Findlay GS, Wick MJ, Mascia MP, Wallace D, Miller GW, Harris RA, Blednov YA. Transgenic expression of a mutant glycine receptor decreases alcohol sensitivity of mice. J Pharmacol Exp Ther. 2002;300:526–534. doi: 10.1124/jpet.300.2.526. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- Fujiwara M, Ishihara T, Katsura I. A novel WD40 protein, CHE-2, acts cell-autonomously in the formation of C. elegans sensory cilia. Development. 1999;126:4839–4848. doi: 10.1242/dev.126.21.4839. [DOI] [PubMed] [Google Scholar]
- Fujiwara M, Sengupta P, McIntire SL. Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cGMP-dependent protein kinase. Neuron. 2002;36:1091–1102. doi: 10.1016/s0896-6273(02)01093-0. [DOI] [PubMed] [Google Scholar]
- Goeders NE, Guerin GF. Non-contingent electric footshock facilitates the acquisition of intravenous cocaine self-administration in rats. Psychopharmacology (Berl) 1994;114:63–70. doi: 10.1007/BF02245445. [DOI] [PubMed] [Google Scholar]
- Grammatopoulos DK, Dai Y, Randeva HS, Levine MA, Karteris E, Easton AJ, Hillhouse EW. A novel spliced variant of the type 1 corticotropin-releasing hormone receptor with a deletion in the seventh transmembrane domain present in the human pregnant term myometrium and fetal membranes. Mol Endocrinol. 1999;13:2189–2202. doi: 10.1210/mend.13.12.0391. [DOI] [PubMed] [Google Scholar]
- Hansson AC, Cippitelli A, Sommer WH, Ciccocioppo R, Heilig M. Region-specific down-regulation of Crhr1 gene expression in alcohol-preferring msP rats following ad lib access to alcohol. Addict Biol. 2007;12:30–34. doi: 10.1111/j.1369-1600.2007.00050.x. [DOI] [PubMed] [Google Scholar]
- Hansson AC, Cippitelli A, Sommer WH, Fedeli A, Bjork K, Soverchia L, Terasmaa A, Massi M, Heilig M, Ciccocioppo R. Variation at the rat Crhr1 locus and sensitivity to relapse into alcohol seeking induced by environmental stress. Proc Natl Acad Sci U S A. 2006;103:15236–15241. doi: 10.1073/pnas.0604419103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmar AJ. Family-B G-protein-coupled receptors. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-12-reviews3013. REVIEWS3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauger RL, Risbrough V, Brauns O, Dautzenberg FM. Corticotropin releasing factor (CRF) receptor signaling in the central nervous system: new molecular targets. CNS Neurol Disord Drug Targets. 2006;5:453–479. doi: 10.2174/187152706777950684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig M, Egli M, Crabbe JC, Becker HC. Acute withdrawal, protracted abstinence and negative affect in alcoholism: are they linked? Addict Biol. 2010;15:169–184. doi: 10.1111/j.1369-1600.2009.00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrichs SC, De Souza EB, Schulteis G, Lapsansky JL, Grigoriadis DE. Brain penetrance, receptor occupancy and antistress in vivo efficacy of a small molecule corticotropin releasing factor type I receptor selective antagonist. Neuropsychopharmacology. 2002;27:194–202. doi: 10.1016/S0893-133X(02)00299-3. [DOI] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Holmes A, Heilig M, Rupniak NM, Steckler T, Griebel G. Neuropeptide systems as novel therapeutic targets for depression and anxiety disorders. Trends Pharmacol Sci. 2003;24:580–588. doi: 10.1016/j.tips.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Huang MM, Overstreet DH, Knapp DJ, Angel R, Wills TA, Navarro M, Rivier J, Vale W, Breese GR. Corticotropin-releasing factor (CRF) sensitization of ethanol withdrawal-induced anxiety-like behavior is brain site specific and mediated by CRF-1 receptors: relation to stress-induced sensitization. J Pharmacol Exp Ther. 2010;332:298–307. doi: 10.1124/jpet.109.159186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RW, von Borell EH, Anderson LL, Kojic LD, Cunnick JE. Intracerebroventricular injection of corticotropin-releasing hormone in the pig: acute effects on behavior, adrenocorticotropin secretion, and immune suppression. Endocrinology. 1994;135:642–648. doi: 10.1210/endo.135.2.8033811. [DOI] [PubMed] [Google Scholar]
- Jones DN, Kortekaas R, Slade PD, Middlemiss DN, Hagan JJ. The behavioural effects of corticotropin-releasing factor-related peptides in rats. Psychopharmacology (Berl) 1998;138:124–132. doi: 10.1007/s002130050654. [DOI] [PubMed] [Google Scholar]
- Kjelsberg MA, Cotecchia S, Ostrowski J, Caron MG, Lefkowitz RJ. Constitutive activation of the alpha 1B-adrenergic receptor by all amino acid substitutions at a single site. Evidence for a region which constrains receptor activation. J Biol Chem. 1992;267:1430–1433. [PubMed] [Google Scholar]
- Kolakowski LF., Jr. GCRDb: a G-protein-coupled receptor database. Receptors Channels. 1994;2:1–7. [PubMed] [Google Scholar]
- Koob GF. A role for brain stress systems in addiction. Neuron. 2008;59:11–34. doi: 10.1016/j.neuron.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Thatcher-Britton K. Stimulant and anxiogenic effects of corticotropin releasing factor. Prog Clin Biol Res. 1985;192:499–506. [PubMed] [Google Scholar]
- Lee EH, Tsai MJ. The hippocampus and amygdala mediate the locomotor stimulating effects of corticotropin-releasing factor in mice. Behav Neural Biol. 1989;51:412–423. doi: 10.1016/s0163-1047(89)91052-2. [DOI] [PubMed] [Google Scholar]
- Lee J, Jee C, McIntire SL. Ethanol preference in C. elegans. Genes Brain Behav. 2009;8:578–585. doi: 10.1111/j.1601-183X.2009.00513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkowitz RJ, Cotecchia S, Samama P, Costa T. Constitutive activity of receptors coupled to guanine nucleotide regulatory proteins. Trends Pharmacol Sci. 1993;14:303–307. doi: 10.1016/0165-6147(93)90048-O. [DOI] [PubMed] [Google Scholar]
- Li B, Predel R, Neupert S, Hauser F, Tanaka Y, Cazzamali G, Williamson M, Arakane Y, Verleyen P, Schoofs L, Schachtner J, Grimmelikhuijzen CJ, Park Y. Genomics, transcriptomics, and peptidomics of neuropeptides and protein hormones in the red flour beetle Tribolium castaneum. Genome Res. 2008;18:113–122. doi: 10.1101/gr.6714008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw CW, Lovenberg TW, Barry G, Oltersdorf T, Grigoriadis DE, de Souza EB. Cloning and characterization of the human corticotropin-releasing factor-2 receptor complementary deoxyribonucleic acid. Endocrinology. 1996;137:72–77. doi: 10.1210/endo.137.1.8536644. [DOI] [PubMed] [Google Scholar]
- Liaw CW, Lovenberg TW, Barry G, Oltersdorf T, Grigoriadis DE, de Souza EB. Cloning and characterization of the human corticotropin-releasing factor-2 receptor complementary deoxyribonucleic acid. Endocrinology. 1996;137:72–77. doi: 10.1210/endo.137.1.8536644. [DOI] [PubMed] [Google Scholar]
- Locke CJ, Williams SN, Schwarz EM, Caldwell GA, Caldwell KA. Genetic interactions among cortical malformation genes that influence susceptibility to convulsions in C. elegans. Brain Res. 2006;1120:23–34. doi: 10.1016/j.brainres.2006.08.067. [DOI] [PubMed] [Google Scholar]
- Lovejoy DA, Jahan S. Phylogeny of the corticotropin-releasing factor family of peptides in the metazoa. Gen Comp Endocrinol. 2006;146:1–8. doi: 10.1016/j.ygcen.2005.11.019. [DOI] [PubMed] [Google Scholar]
- Lowery EG, Spanos M, Navarro M, Lyons AM, Hodge CW, Thiele TE. CRF-1 antagonist and CRF-2 agonist decrease binge-like ethanol drinking in C57BL/6J mice independent of the HPA axis. Neuropsychopharmacology. 2010;35:1241–1252. doi: 10.1038/npp.2009.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry CA, Moore FL. Corticotropin-releasing factor (CRF) antagonist suppresses stress-induced locomotor activity in an amphibian. Horm Behav. 1991;25:84–96. doi: 10.1016/0018-506x(91)90041-f. [DOI] [PubMed] [Google Scholar]
- Mackiewicz M, Naidoo N, Zimmerman JE, Pack AI. Molecular mechanisms of sleep and wakefulness. Ann N Y Acad Sci. 2008;1129:335–349. doi: 10.1196/annals.1417.030. [DOI] [PubMed] [Google Scholar]
- Merlo Pich E, Lorang M, Yeganeh M, Rodriguez de Fonseca F, Raber J, Koob GF, Weiss F. Increase of extracellular corticotropin-releasing factor-like immunoreactivity levels in the amygdala of awake rats during restraint stress and ethanol withdrawal as measured by microdialysis. J Neurosci. 1995;15:5439–5447. doi: 10.1523/JNEUROSCI.15-08-05439.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Mould R, Dillon J, Glautier S, Andrianakis I, James C, Pugh A, Holden-Dye L, O’Connor V. A differential role for neuropeptides in acute and chronic adaptive responses to alcohol: behavioural and genetic analysis in Caenorhabditis elegans. PLoS One. 2010;5:e10422. doi: 10.1371/journal.pone.0010422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman DG, Baillie DL. Genetic Organization in CAENORHABDITIS ELEGANS: Fine-Structure Analysis of the unc-22 Gene. Genetics. 1979;91:95–103. doi: 10.1093/genetics/91.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morilak DA, Cecchi M, Khoshbouei H. Interactions of norepinephrine and galanin in the central amygdala and lateral bed nucleus of the stria terminalis modulate the behavioral response to acute stress. Life Sci. 2003;73:715–726. doi: 10.1016/s0024-3205(03)00392-8. [DOI] [PubMed] [Google Scholar]
- Nishihara E, Nagayama Y, Amino N, Hishinuma A, Takano T, Yoshida H, Kubota S, Fukata S, Kuma K, Miyauchi A. A novel thyrotropin receptor germline mutation (Asp617Tyr) causing hereditary hyperthyroidism. Endocr J. 2007;54:927–934. doi: 10.1507/endocrj.k07-088. [DOI] [PubMed] [Google Scholar]
- Osborne KA, Robichon A, Burgess E, Butland S, Shaw RA, Coulthard A, Pereira HS, Greenspan RJ, Sokolowski MB. Natural behavior polymorphism due to a cGMP-dependent protein kinase of Drosophila. Science. 1997;277:834–836. doi: 10.1126/science.277.5327.834. [DOI] [PubMed] [Google Scholar]
- Piazza PV, Le Moal M. The role of stress in drug self-administration. Trends Pharmacol Sci. 1998;19:67–74. doi: 10.1016/s0165-6147(97)01115-2. [DOI] [PubMed] [Google Scholar]
- Pierce-Shimomura JT, Chen BL, Mun JJ, Ho R, Sarkis R, McIntire SL. Genetic analysis of crawling and swimming locomotory patterns in C. elegans. Proc Natl Acad Sci U S A. 2008;105:20982–20987. doi: 10.1073/pnas.0810359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen DM, Zimmerman JE, Maycock MH, Ta UD, You YJ, Sundaram MV, Pack AI. Lethargus is a Caenorhabditis elegans sleep-like state. Nature. 2008;451:569–572. doi: 10.1038/nature06535. [DOI] [PubMed] [Google Scholar]
- Rivier C, Bruhn T, Vale W. Effect of ethanol on the hypothalamic-pituitary-adrenal axis in the rat: role of corticotropin-releasing factor (CRF) J Pharmacol Exp Ther. 1984;229:127–131. [PubMed] [Google Scholar]
- Roberto M, Cruz MT, Gilpin NW, Sabino V, Schweitzer P, Bajo M, Cottone P, Madamba SG, Stouffer DG, Zorrilla EP, Koob GF, Siggins GR, Parsons LH. Corticotropin releasing factor-induced amygdala gamma-aminobutyric Acid release plays a key role in alcohol dependence. Biol Psychiatry. 2010;67:831–839. doi: 10.1016/j.biopsych.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber MA, Pierce-Shimomura JT, Chan S, Parry D, McIntire SL. Manipulation of behavioral decline in Caenorhabditis elegans with the Rag GTPase raga-1. PLoS Genet. 2010;6:e1000972. doi: 10.1371/journal.pgen.1000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit MJ, Vischer HF, Bakker RA, Jongejan A, Timmerman H, Pardo L, Leurs R. Pharmacogenomic and structural analysis of constitutive g protein-coupled receptor activity. Annu Rev Pharmacol Toxicol. 2007;47:53–87. doi: 10.1146/annurev.pharmtox.47.120505.105126. [DOI] [PubMed] [Google Scholar]
- Smith GW, Aubry JM, Dellu F, Contarino A, Bilezikjian LM, Gold LH, Chen R, Marchuk Y, Hauser C, Bentley CA, Sawchenko PE, Koob GF, Vale W, Lee KF. Corticotropin releasing factor receptor 1-deficient mice display decreased anxiety, impaired stress response, and aberrant neuroendocrine development. Neuron. 1998;20:1093–1102. doi: 10.1016/s0896-6273(00)80491-2. [DOI] [PubMed] [Google Scholar]
- Sparta DR, Sparrow AM, Lowery EG, Fee JR, Knapp DJ, Thiele TE. Blockade of the corticotropin releasing factor type 1 receptor attenuates elevated ethanol drinking associated with drinking in the dark procedures. Alcohol Clin Exp Res. 2008;32:259–265. doi: 10.1111/j.1530-0277.2007.00575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpl P, Spanagel R, Sillaber I, Kresse A, Reul JM, Stalla GK, Blanquet V, Steckler T, Holsboer F, Wurst W. Impaired stress response and reduced anxiety in mice lacking a functional corticotropin-releasing hormone receptor 1. Nat Genet. 1998;19:162–166. doi: 10.1038/520. [DOI] [PubMed] [Google Scholar]
- Wallace MJ, Newton PM, Oyasu M, McMahon T, Chou WH, Connolly J, Messing RO. Acute functional tolerance to ethanol mediated by protein kinase Cepsilon. Neuropsychopharmacology. 2007;32:127–136. doi: 10.1038/sj.npp.1301059. [DOI] [PubMed] [Google Scholar]
- Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- Winslow JT, Newman JD, Insel TR. CRH and alpha-helical-CRH modulate behavioral measures of arousal in monkeys. Pharmacol Biochem B. 1989;32:919–926. doi: 10.1016/0091-3057(89)90059-2. Pharmacol Biochem B. [DOI] [PubMed] [Google Scholar]
- Zorrilla EP, Koob GF. The therapeutic potential of CRF1 antagonists for anxiety. Expert Opin Investig Drugs. 2004;13:799–828. doi: 10.1517/13543784.13.7.799. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.