Abstract
Most mammals possess a tail, humans and the Great Apes being notable exceptions. One approach to understanding the mechanisms and evolutionary forces influencing development of a tail is to identify the genetic factors that influence extreme tail length variation within a species. In mice, the Tailless locus has proven to be complex, with evidence of multiple different genes and mutations with pleiotropic effects on tail length, fertility, embryogenesis, male transmission ratio, and meiotic recombination. Five cat breeds have abnormal tail length phenotypes: the American Bobtail, the Manx, the Pixie-Bob, the Kurilian Bobtail, and the Japanese Bobtail. We sequenced the T gene in several independent lineages of Manx cats from both the US and the Isle of Man and identified three 1-bp deletions and one duplication/deletion, each predicted to cause a frameshift that leads to premature termination and truncation of the carboxy terminal end of the Brachyury protein. Ninety-five percent of Manx cats with short-tail phenotypes were heterozygous for T mutations, mutant alleles appeared to be largely lineage-specific, and a maximum LOD score of 6.21 with T was obtained at a recombination fraction (Θ) of 0.00. One mutant T allele was shared with American Bobtails and Pixie-Bobs; both breeds developed more recently in the US. The ability of mutant Brachyury protein to activate transcription of a downstream target was substantially lower than wild-type protein. Collectively, these results suggest that haploinsufficiency of Brachyury is one mechanism underlying variable tail length in domesticated cats.
Introduction
Tails in vertebrates are used for a wide variety of functions, including balance, locomotion, and communication. Both within and among species, variation in tail length is common and phylogenetic analyses of mammals suggest that shortening or loss of the tail may have occurred multiple times during evolution, independently (e.g., cats, dogs, monkeys). The evolutionary mechanism(s) that explains this observation is, as of yet, unknown. However, mice heterozygous for mutations of the canonical T-box gene, T, which encodes the transcription factor Brachyury, a key regulator of notochord differentiation in all vertebrates, have tail length variation. To date, eight different T alleles that cause a short-tailed phenotype in mice have been described (Wu et al. 2010). These alleles include both point mutations and frameshifts that result in truncation of either the amino or carboxy terminal end of Brachyury. Similarly, T has been associated with a short-tail phenotype in multiple breeds of dogs (Haworth et al. 2001; Hytonen et al. 2009).
Tail length variation is particularly common worldwide in two breeds of domestic cat (Felis silvestris catus), the Japanese Bobtail and the Manx. Perhaps the most widely known example is the Manx cat, putatively from the Isle of Man. The origin of the short-tail phenotype of the Manx cat has been the fodder of both popular myth and investigation by classical geneticists and evolutionary biologists, including Darwin (1868). The variable tail length of the Manx recapitulates that of tail variation in mice and can be categorized into four specific tail-length phenotypes. These range from absence of the tail (anury) (i.e., rumpy), a minimal tail (i.e., rumpy-riser) that is apparent only by palpation, a short tail (i.e., stumpy), to a full tail (i.e., longie) (Fig. 1) (Howell and Siegel 1963; Robinson 1993; Todd 1963). The length of the tail is proportional to the number of caudal vertebrae, with complete absence of caudal vertebrae found only in cats with the rumpy phenotype. Shortened tails in Manx pedigrees segregate in an autosomal dominant pattern (Howell and Siegel 1963; Todd 1964), and litters of crosses between short-tailed parents are often smaller than typical (Deforest and Basrur 1979). Accordingly, homozygosity is presumed to result in early embryonic lethality (Basrur and Deforest 1979).
Fig. 1.
Variable tail length phenotypes of the Manx cat. Manx cats exhibit varied tail lengths, including absence of the tail or rumpy (a), a minimal tail or rumpy-riser (b), a short tail or stumpy (c), and a full tail or longie (d). Cats pictured in a and b are Pedigree A2, V:3 and Pedigree A1, IV:4, respectively in Fig. 3
Taillessness is just one characteristic of a spectrum of congenital anomalies observed in Manx cats. Abnormalities associated with Manx taillessness have been noted for over 100 years. Approximately 20 % of Manx cats with shortened tails have at least one additional congenital anomaly of which about 90 % are found in the cats with the rumpy phenotype. These defects include sacral agenesis/dysgenesis, absence of the sacral spinal cord, diastematomyelia, tethered cord, intradural lipomas, and imperforate anus (Deforest and Basrur 1979; Howell and Siegel 1966; James et al. 1969; Leipold et al. 1974; Martin 1971; Plummer et al. 1993). Manx cats with shortened tails can also exhibit functional abnormalities, including hindlimb paresis/paralysis and incontinence of urine/feces (James et al. 1969).
Herein is described the sequencing of the feline T homolog and an analysis of T in five breeds of cats with varied short-tailed phenotypes. Four T alleles, i.e., three 1-bp deletions and a small duplication/deletion, are associated with reduction of tail length in Manx cats. Each allele is predicted to cause a frameshift resulting in premature truncation of Brachyury, suggesting that shortened tail length is caused by haploinsufficiency.
Materials and methods
Sample collection
Buccal swabs and phenotypic data were collected from 196 domestic cats, including 136 Manx, 21 American Bobtail, 14 Japanese Bobtail, 15 Pixie-Bob, and 10 Kurilian Bobtail. Ninety-nine of the Manx cats were sampled from two large, extended pedigrees distributed across four catteries in the US. Thirty-seven Manx cat samples were obtained from an animal shelter on the Isle of Man. American Bobtail and Japanese Bobtail cats were collected from two large pedigrees ascertained from US catteries. The Pixie-Bob samples were collected at cat shows or submitted by owners in the US. The Kurilian Bobtail samples were collected at European cat shows or submitted by owners. DNA samples from eight American Bobcats were also collected.
Tail phenotypes were assessed by physical examination and included visual inspection and palpation by the owner or the investigators. Phenotypes in the Manx were categorized as follows: rumpy, absence of caudal vertebrae on palpation; rumpy-riser, presence of up to several caudal vertebrae by palpation; and stumpy, more than three caudal vertebrae but fewer than present in a normal tail. The latter consisted of tails of varying length but the exact number of caudal vertebrae in each tail was not determined.
DNA sequencing
Genomic DNA was extracted from buccal cells using Gentra Puregene reagents (Qiagen, Valencia, CA) with standard protocols. Whole-genome amplification (WGA) was performed with the RepliG Mini Kit (Qiagen), and WGA products were used for all experiments on Manx samples collected from the Isle of Man, the Pixie-Bob samples, and the Kurilian Bobtail samples. Using the partial T gene sequence available in Ensembl (ENSFCAT00000000777), several strategies were used to isolate the coding regions of T, including standard PCR, genome walking (Clontech Laboratories, Mountain View, CA), heat-pulse extension PCR (Orpana et al. 2012), and PCR with primers designed to conserved regions of a multispecies alignment (Supplementary Table 1). PCR amplicons were Sanger sequenced in both forward and reverse directions with BDTv3.1 reagents (Life Technologies Corporation, Carlsbad, CA) and products were resolved on a 3130xl Genetic Analyzer (Applied Bio-systems, Foster City, CA). Electropherograms were manually inspected with CodonCode Aligner (Dedham, MA).
Linkage analysis
Linkage of the T locus to the short-tailed phenotype in Manx cats was tested using parametric linkage analysis in MERLIN (Abecasis et al. 2002). All four T mutations identified were collapsed into a single meta-allele representing the presence of any T mutation. To facilitate analysis, the pedigrees were split into smaller subpedigrees (Supplementary Fig. 1). The LOD score was calculated under a dominant model with reduced penetrance (assumed penetrances of 0.02, 0.95, and 1.0 were used for cats with genotypes 0/0, 0/1, and 1/1, respectively, in which “1” represents a mutant T allele) and a population short-tail allele frequency of 0.01.
Functional studies of mutant T alleles
PCR from a mouse T transcript was used to produce truncated fragments that were analogous to three 1-bp deletions, c.998delT, c.1169delC, and c.1199delC. Fragments were inserted into a pcDNA3.1 expression vector with a V5 tag using a TOPO Cloning Kit (Invitrogen) and sequenced to verify the presence of the truncated sequence. Mutant constructs, an empty expression vector control, and a wild-type Brachyury construct were transfected via electroporation into four million EL4 T cells (ATTC, Manassas, VA), a mouse lymphoma cell line, using the Amaxa Nucleofector V Kit standard protocol (Lonza, Walkersville, MD). Cells were cultured in RPMI with 10 % FBS. After 18–22 h, equal volumes of cells were harvested from each transfection. A Western blot probed with mouse anti-V5 antibody (Invitrogen) was performed on total cell lysate to measure expression levels of mutant and control proteins. GAPDH was used as a control for protein expression levels in the Western blot analysis.
RNA was extracted from each transfected line using the RNeasy Kit (Qiagen) and total cDNA was generated with the SuperScript First Strand Synthesis System (Invitrogen). Two-step quantitative real-time PCR was performed on total cDNA using the Brilliant SYBR Green QPCR Core Reagent Kit (Stratagene, La Jolla, CA), with primers specific for the Nkg7 gene (forward: GTATGAGATGGAGCC AGACTC, reverse : CACAAGGTTTCATACTCAG), a known target for T (A. Weinmann, unpublished data), as well as primers specific for the constitutively expressed β-actin gene (forward: GCATTGCTGACAGGATGCAG, reverse: GAGTACTTGCGCTCAGGAGG). Expression levels were normalized to β-actin and wild-type T, respectively. Experiments were performed in duplicate, standard deviations were calculated, and average levels of Nkg7 expression induced by each construct were plotted relative to wild-type Brachyury.
Results
Identification of mutant feline T alleles
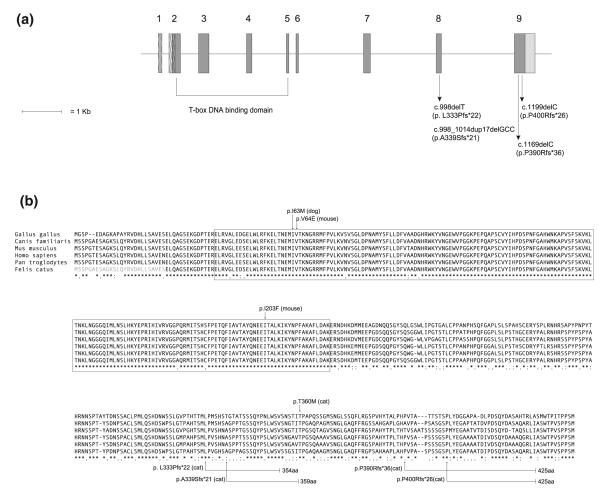
The genomic structure of T was characterized with the exception of the 5′ untranslated region and the first 78 bp of coding sequence of exon 2, which failed repeated sequencing attempts (Supplementary Table 2; Fig. 2). The 3′ end of exon 2 and exons 3–9 of T (Fig. 2) were sequenced in a discovery cohort of 15 Manx cats from a US cattery, including three rumpy, four rumpy-riser, two stumpy, and six full-tailed cats (Fig. 3).
Fig. 2.
Diagram of the feline T gene and a multispecies protein alignment, including inferred protein sequence for feline Brachyury. a Genomic structure of T with untranslated regions (light gray), coding exons (dark gray), and nonsequenced regions (hatched). Arrows indicate the relative positions of the four mutations identified in Manx cats with short tails. In brackets are the exons that encode the T-box DNA binding domain. b A multispecies protein alignment constructed using MUSCLE (Multiple Sequence Comparison by Log-Expectation, http://www.ebi.ac.uk/Tools/msa/muscle/). Amino acids in gray text are consensus sequence. Residues of the T-box DNA binding domain are boxed. The positions of select missense mutations that are associated with short-tailed phenotypes in dog, mouse, and cat are indicated above with arrows. The four frameshift mutations identified in Manx cats are shown, with arrows indicating the residue where each frameshift begins. The connecting black bars designate the amino acids that are translated out of frame, and the residue number where each mutant protein is predicted to terminate is specified. Asterisks mark positions in the alignment where the amino acids are identical across species, colons represent conserved substitutions, and periods indicate semi-conserved substitutions
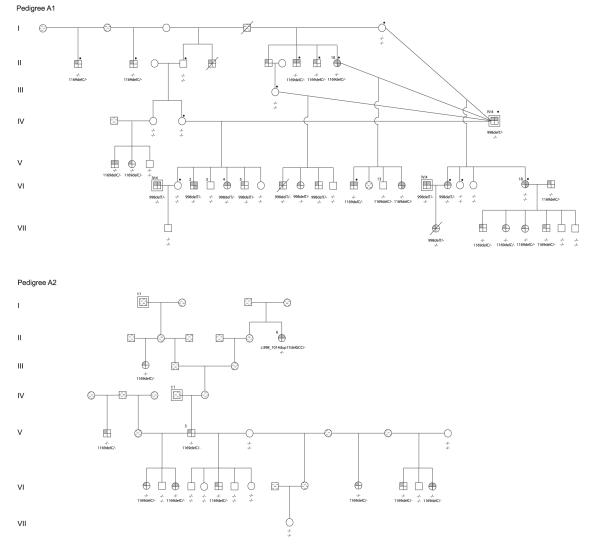
Fig. 3.
Extended pedigree illustrating relationships among Manx cats used for linkage and mutation analysis. Squares represent male cats, circles represent females, and a diagonal line indicates cats that are deceased. Cats with reduced tail lengths are denoted by varied darkened symbols: rumpy, quarter filled; rumpy-riser, half filled; stumpy, three-quarter filled. Full-tailed cats are indicated with open symbols and patterned symbols denote individuals for whom no phenotypic data were available. The 15 cats in the discovery cohort are indicated by a dot. Males represented more than once in the pedigree are indicated by a double square labeled with the generation and individual number of the first time they appear in the pedigree. Genotypes for the c.998delT and c.1169delC or c.1199delC mutants in T are indicated below each symbol. A “–” indicates the mutation was absent
Two 1-bp deletions (c.998delT and c.1169delC), each predicted to cause premature truncation of the Brachyury protein, were identified in exons 8 and 9, respectively. Targeted sequencing of exons 8 and 9 in the remaining 121 Manx samples and in the 21 American Bobtail, 14 Japanese Bobtail, 15 Pixie-Bob, and 10 Kurilian Bobtail samples revealed two additional mutations, a small duplication/deletion in exon 8 (c.998_1014dup17delGCC) and another 1-bp deletion in exon 9 (c.1199delC) only in Manx cats (Table 1, and Supplementary Table 3). For four Manx (three from the Isle of Man and for which no information about tail docking was available), seven Pixie-Bob, seven American Bobtail, all 14 Japanese Bobtail, and all ten Kurilian Bobtail, no mutations were identified in exons 8 and 9. All remaining exons of T were sequenced for these cats but no additional causal variants were identified.
Table 1.
Description of tail lengths and mutations in the Manx cat
| Phenotypea | No. | Mutation |
|||||
|---|---|---|---|---|---|---|---|
| c.998delT | c.998_1014 dup17delGCC |
c.1169delC | c.1199delC | None | Indeterminate | ||
| Full tail | 49 | 0 | 0 | 1 | 0 | 47 | 1 |
| Short tail (all types) | 83 | 26 | 1 | 31 | 20 | 4b | 1 |
| Unknown | 4 | 0 | 0 | 1 | 0 | 1 | 2 |
| Total | 136 | 26 | 1 | 33 | 20 | 52 | 4 |
Tail phenotypes were determined by visual inspection and palpation; no radiographs were available to support the count of the vertebrae
Three cats were from the Isle of Man
Overall, eight coding variants were identified, including both synonymous and nonsynonymous changes (Supplementary Table 4). Variation data were submitted to the European Nucleotide Archive under the accession number HG004538 (http://www.ebi.ac.uk/ena/data/view/HG004538). Each variant was observed in at least one full-tailed cat, with the exception of the exon 9 missense variant p.T360M, which was observed in only three short-tailed cats (one Manx cat from the Isle of Man and two Kurilian Bobtails). This variant is predicted by PolyPhen to be “benign” but it occurs at a highly conserved site and thus could underlie a short-tail phenotype in multiple cat breeds. However, no segregation data were available and the sample size was too small to adequately power a test of association.
Short-tail phenotype in Manx cats is caused by multiple mutant T alleles
Sequencing of T in the primary sire from cattery 1 (Fig. 3, Pedigree A1, IV:4) revealed a c.998delT mutation in exon 8 that is predicted to result in a frameshift and introduction of a termination codon at residue 354 of 437. In three crosses between the primary sire and full-tail females that produced 14 offspring, the c.998delT allele was found in seven of eight offspring with short-tailed phenotypes and in none of the six offspring with a full tail. The primary sire was also bred to an unrelated female (Fig. 3, Pedigree A1, II:10) stumpy who lacked the c.998delT allele. Sequencing of T in this female revealed a different mutant T allele, specifically a c.1169delC mutation in exon 9 that is predicted to result in a frameshift and introduction of a termination codon at residue 425. All of the offspring with short tails produced by this mating were heterozygous for the c.1169delC allele only. None of the cats with a full tail from cattery 1 had a c.998delT allele; however, one full-tailed cat (Fig. 3, Pedigree A1, VI:13) was found to carry a c.1169delC allele. None of the cats with a short-tail phenotype were compound heterozygotes or homozygous for a mutant T allele.
In a putatively independent Manx pedigree from cattery 2, sequencing of the primary sire (a rumpy) (Fig. 3, Pedigree A2, V:3) revealed a c.1169delC mutation, the same variant that had been identified in the short-tailed cats from Pedigree A1. This male was subsequently confirmed to have been bred to a stumpy female (Fig. 3, Pedigree A1, VI:18) in cattery 1, effectively creating a large, extended pedigree of Manx cats across two catteries. All of the offspring with short tails produced by the primary sire (Fig. 3, Pedigree A2, V:3) were heterozygous for the c.1169delC allele. None of the cats with a full tail from cattery 2 had a c.1169delC mutation, and none of the cats with a short-tail phenotype were homozygous for a mutant T allele. An ancestor (Fig. 3, Pedigree A2, II:6) of the primary sire had a rumpy-riser tail phenotype but lacked the c.1169delC allele. This female was found to have a novel 17-bp duplication/deletion c.998_1014dup17delGCC in exon 8. None of her offspring were available for further study.
A single male cat (Supplementary Fig. 2, Pedigree B, I:3) was used as the primary sire in catteries 3 and 4. Sequencing of this male revealed none of the mutant T alleles identified in cats from Pedigree A. Instead, a 1-bp deletion, c.1199delC, was identified in exon 9 of T that is predicted to result in a frameshift and introduction of a termination codon at residue 425, the same residue at which the frameshift caused by the c.1169delC mutation is predicted to terminate. All of the offspring with short tails produced by this sire were heterozygous for the c.1199delC allele and none of the cats with a full tail had a c.1199delC allele. Additionally, four cats with short tails in Pedigree B were found to carry the c.1169delC mutation that was identified in Pedigree A. These cats were descended from a secondary sire (Supplementary Fig. 2, Pedigree B, II:2) for which no sample was available.
Most of the crosses studied were between either a rumpy male or a rumpy-riser male and a full-tailed female. Both rumpy and rumpy-riser males mated to different full-tailed females sired offspring with each of the four tail phenotypes. In one instance, a rumpy-riser male (Fig. 3, Pedigree A1, IV:4) produced kittens with each of the four tail phenotypes in a single litter (Fig. 3, Pedigree A1, VI:2, VI:3, VI:4, VI:5). This observation suggests that additional genetic (e.g., other loci) or environmental factors, or some combination thereof, interact with T to modify the short-tail phenotype of Manx cats.
Since the Manx breed is thought to have originated on the Isle of Man, Manx cats sampled from the island were tested for possession of one or more of the mutant T alleles identified in short-tailed cats from US catteries. Twelve of 16 short-tailed cats were found to be heterozygous for the c.998delT allele. No mutant T allele was identified in three of the four remaining samples from short-tailed Manx, while the fourth sample failed to sequence. None of the full-tailed Manx cats from the Isle of Man (n = 20) were found to have a mutant T allele.
To test the likelihood that mutant T alleles segregated with short-tailed phenotypes by chance, a pairwise LOD score (Zmax) was calculated, collapsing all the mutant alleles into a single meta-allele. A maximum LOD score of 6.21 was obtained with no recombination (Θ = 0.00). This confirmed that the T locus is tightly linked to the short-tailed phenotype in Manx cats. Overall, complete genotype and phenotype data were available for 130 Manx cats. Of 82 cats with a short-tail phenotype, 78 had a mutant T allele, and of 48 cats with a full-tail phenotype, only one possessed a mutant T allele. A one-sided Fisher’s exact test revealed a highly significant association between a mutant T allele and a short-tailed phenotype in all cats (US and Isle of Man, P < 1.8 × 10−29) as well as unrelated cats (Isle of Man only, P < 5.45 × 10−7; n = 35).
Mutant T alleles cause short-tailed phenotypes in the American Bobtail and Pixie-Bob
Twenty of 21 American Bobtail samples collected were from cats with short tails, while only one sample was available from an animal with a full tail. Sequencing of exons 8 and 9 of T in the American Bobtails with a short-tailed phenotype revealed that 12 of 20 (60 %) were heterozygous for a c.998delT allele and one cat was heterozygous for a c.1199delC allele. No T mutations were found in seven (35 %) American Bobtail cats with short-tailed phenotypes. Furthermore, no causal variants in T were found in either Japanese (n = 14) or Kurilian (n = 10) Bobtails (Supplementary Table 5).
Sequencing of exon 8 of T in 15 Pixie-Bobs, including 14 short-tailed cats and one cat with a full tail, identified the c.998delT allele in 8 of 14 short-tailed cats. A Pixie-Bob with a full tail and the six remaining short-tailed Pixie-Bobs did not carry the mutation (Supplementary Table 5). Sequencing of the remaining exons of T in these six short-tailed Pixie-Bobs did not reveal any additional mutations. The origin of the Pixie-Bob is controversial and purportedly resulted from hybridization between a domestic cat and an American Bobcat (Lynx rufus). However, the c.998delT allele was not found in samples from eight American Bobcats.
Functional consequences of mutant T alleles
Each of the mutant T alleles discovered was predicted to result in early termination of translation and impaired ability of T to activate its downstream targets. To assess the functional consequences of the c.998delT, c.1169delC, and c.1199delC mutations, we created expression constructs containing the analogous mutants in mouse T to test their ability to functionally regulate the transcription of a gene that is robustly activated by the wild-type Brachyury protein (Fig. 4a). We transfected EL4 cells with either a control expression vector or expression constructs for wild-type Brachyury or the Brachyury C-terminal deletion mutants and assessed their ability to functionally regulate endogenous Nkg7 expression. Notably, the mutant Brachyury proteins were severely impaired in their ability to activate endogenous Nkg7 transcription in comparison to wild-type Brachyury (Fig. 4a). A Western blot analysis confirmed that all of the Brachyury C-terminal mutant proteins are expressed at similar levels in comparison to the wild-type protein, indicating that the impairments in transcriptional activation potential were not caused by defects in the expression or stability of the mutant Brachyury proteins (Fig. 4b). It is important to note that these functional studies do not rule out the possibility that the Brachyury C-terminal mutant proteins may have different functional capabilities at subsets of target genes depending upon the mechanistic role of Brachyury at individual regulatory regions. Nevertheless, these studies suggest that these natural mutations in Brachyury diminish the transcriptional activation potential of Brachyury in at least some circumstances.
Fig. 4.
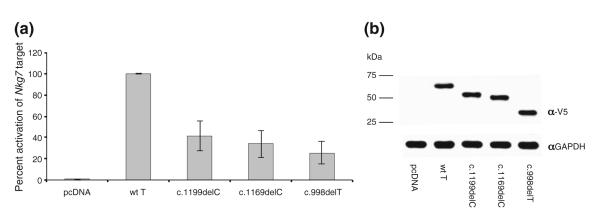
Brachyury C-terminal domain is required for optimal transactivation potential. a T mutants exhibited decreased abilities to activate transcription of the target gene compared to wild-type T. b Western blot of wild-type and mutant Brachyury proteins showing equal transfection levels for all mutant constructs and controls
Discussion
Short-tailed phenotypes in mouse (Wu et al. 2010) and dog (Haworth et al. 2001; Hytonen et al. 2009; Indrebo et al.2008) have been associated with naturally occurring T mutants and induced by disrupting Brachyury expression in model organisms such as zebrafish (Schulte-Merker et al. 1994) and Xenopus (Conlon et al. 1996). Complete absence of the tail is a phenotype found virtually only in the Manx cat. However, several other breeds of cat are noted for short-tail phenotypes, such as the Japanese Bobtail, the Kurilian Bobtail, the Pixie-Bob, and the American Bobtail. The Manx has been shown to have a highly variable presentation, which can also include a variety of health concerns. Commonly, sacral agenesis/dysgenesis and absence of the sacral spinal cord lead to poor innervation of the hind limbs and gut, resulting in lameness, incontinence, and constipation. Thus, the Manx is recognized as a potential model for neural tube development (Green and Green 1987; Kitchen et al. 1972).
Overall, 78 of 82 Manx cats with a shortened tail were heterozygous for a mutant T allele, and 47 of 48 full-tailed Manx cats had wild-type T alleles. These results demonstrate that the short-tailed phenotype in Manx cats is also caused by naturally occurring mutations in T, specifically three 1-bp deletions (c.998delT, c.1169delC, and c.1199delC). A fourth mutation, a small duplication/deletion (c.998_1014dup17delGCC), may also influence tail length in Manx cats but it was discovered in only one individual. No evidence suggested that the presence of a specific mutant allele of T determined the extent of tail length reduction in the Manx cat, which is similar to observations in dog where a single missense in T results in a wide range of tail lengths (Hytonen et al. 2009). Also consistent with reports on other species, no cats were homozygous or compound heterozygous for mutations in T, supporting complete absence of Brachyury results in early embryonic lethality. Genotyping of three fetal cats that died late in gestation found that in all three instances, these individuals were heterozygous for mutant T alleles.
Five Manx cats had phenotypes discordant for their T genotype. There are several possible explanations for the observation that one Manx cat with a c.1169delC allele had a full-length tail. Specifically, the c.1169delC allele could be associated with reduced penetrance, genetic and/or environmental factors could modify the phenotype, or some combination thereof. The tail vertebrae were not counted via a radiographic examination, thus this cat could still have a shortened tail and be misphenotyped. Perhaps more challenging to explain is the observation that four short-tailed Manx did not have a mutant T allele. One possibility is that there is another mutant T allele that was not identified. These cats may possess a mutation in a region of T not sequenced (i.e., introns, noncoding regulatory regions, or the first 78 bp of exon 2). Alternatively, the short tail in these cats could be a phenocopy generated by tail docking, whereby the full tail is surgically shortened to simulate the aesthetic of a stumpy. A majority of these cats were from the Isle of Man, feral cats ascertained by the sanctuary. These cats also could have had shortened tails from injury or from purposeful docking in order to be a more valuable cat for export. To our knowledge, the tails of these cats have not been docked, but no veterinary records or photographs from the early life of these cats were available.
Each of the three deletions in T associated with short-tailed phenotypes impaired the ability of Brachyury to activate transcription of a downstream target. This is in keeping with the general trend of reduced activity with truncation of the C-terminus of Brachyury and suggests that the short-tailed phenotype is caused by this reduced activity. Differences in activity among the various Brachyury mutants were not substantial, which is consistent with the observation that none of the T genotypes were associated with differences in length among short-tailed phenotypes.
In each of the pedigrees studied, males were frequently crossed to females who were closely related to each other such that the matings were not independent. Therefore, it was difficult to make robust inferences about genotype–phenotype relationships. Nevertheless, these pedigrees were sufficient to test whether one or more alleles of T segregated with short-tailed phenotypes in the Manx cat. To this end, in each pedigree assessed, short-tailed phenotypes (i.e., rumpy, rumpy-riser, and stumpy) segregated in a pattern consistent with autosomal dominant inheritance.
The spectrum of short-tailed phenotypes observed in the American Bobtail and the Pixie-Bob is similar to that observed in the stumpy Manx. Mutations in T were also found in the American Bobtail and the Pixie-Bob, but these did not explain all of the short-tailed phenotypes. Both breeds are more recent and less common, with more nebulous breeding records, thus it is not surprising that Manx mutations are segregating within American Bobtails and Pixie-Bobs.
No evidence was found that mutations in T underlie short-tailed phenotypes in the Japanese Bobtail and the Kurilian Bobtail. In these two breeds, the shortened tail is often longer than the tail of a stumpy Manx but appears shorter because it is kinked. In addition, the Japanese Bobtail is an ancient breed that developed in cats of East Asian origin, which are a significantly different population from western European cats such as the Manx (Kurushima et al. 2013; Lipinski et al. 2008). Therefore, yet undiscovered variants in regulatory regions of T or other loci may also influence short-tailed phenotypes in Japanese and Kurilian Bobtails. The latter is perhaps a more likely explanation, as T mutants do not account for all short-tailed phenotypes in other species (Hytonen et al. 2009). The discordant Manx may also be a result of mutations at these as of yet undiscovered loci for the Japanese and Kurilian Bobtail. Moreover, since the American Bobtail and Pixie-Bob are not well-documented breeds, some Japanese Bobtail variants could have been used in breed development, which would explain the nonconcordant short tails in these breeds as well.
Across species, both missense and frameshift mutations have been found to cause short-tail phenotypes (Haworth et al. 2001; Schulte-Merker et al. 1994; Wu et al. 2010). These mutations occur in the T-box domain (e.g., mouse and dog) and downstream (e.g., mouse and cat) with little evidence of a relationship between genotype and loss of functional activity or phenotype (Hytonen et al. 2009; Wu et al. 2010). This is not entirely surprising as mutations in T both within and outside of the T-box domain have been demonstrated to abrogate the ability of Brachyury to activate transcription (Haworth et al. 2001; Kispert et al. 1995).
In summary, several alleles of T that reduced the capability of Brachyury to activate transcription were described here and are tightly linked with the short-tailed phenotype of Manx cats. Mutations in T have been associated with short-tailed phenotypes across phylogenetically distant species of vertebrates, and multiple mutant T alleles appear to have arisen independently in domestic cats and in other species such as mice. Variable Brachyury dosage might be a general mechanism for shortening or loss of the tail in vertebrates.
Supplementary Material
Acknowledgments
We are deeply indebted to the community of Manx, American Bobtail, and Japanese Bobtail breeders and the Mann Cat Sanctuary for their assistance and helpful discussion. We also thank Bill Clark and Dawn Reding of Iowa State University for American Bobcat DNA samples. Funding was provided in part by the National Center for Research Resources (R24 RR016094) and research is currently supported by the Office of Research Infrastructure Programs (OD R24OD010928), the Winn Feline Foundation (grant 10-015), and the UC Davis Center for Companion Animal Health George and Phyllis Miller Feline Health Fund (LAL). Research was also supported by grants from the NIAID (AI061061) and the American Cancer Society (RSG-09-045-01-DDC) to ASW.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00335-013-9471-1) contains supplementary material, which is available to authorized users.
Contributor Information
Kati J. Buckingham, Department of Pediatrics, University of Washington School of Medicine, 1959 NE Pacific Street, HSB RR349, Box 356320, Seattle, WA 98195, USA
Margaret J. McMillin, Department of Pediatrics, University of Washington School of Medicine, 1959 NE Pacific Street, HSB RR349, Box 356320, Seattle, WA 98195, USA
Margaret M. Brassil, Department of Immunology, University of Washington, Seattle, WA 98195, USA
Kathryn M. Shively, Department of Pediatrics, University of Washington School of Medicine, 1959 NE Pacific Street, HSB RR349, Box 356320, Seattle, WA 98195, USA
Kevin M. Magnaye, Department of Pediatrics, University of Washington School of Medicine, 1959 NE Pacific Street, HSB RR349, Box 356320, Seattle, WA 98195, USA
Alejandro Cortes, Population Health and Reproduction, School of Veterinary Medicine, University of California, Davis, Davis, CA 95616, USA.
Amy S. Weinmann, Department of Immunology, University of Washington, Seattle, WA 98195, USA
Leslie A. Lyons, Population Health and Reproduction, School of Veterinary Medicine, University of California, Davis, Davis, CA 95616, USA
Michael J. Bamshad, Department of Pediatrics, University of Washington School of Medicine, 1959 NE Pacific Street, HSB RR349, Box 356320, Seattle, WA 98195, USA; Department of Genome Sciences, University of Washington, Seattle, WA 98195, USA; Seattle Children’s Hospital, Seattle, WA 98105, USA
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Basrur PK, Deforest ME. Embryological impact of the Manx gene. Carnivore Genet Newslett. 1979;3:378–384. [Google Scholar]
- Conlon FL, Sedgwick SG, Weston KM, Smith JC. Inhibition of Xbra transcription activation causes defects in mesodermal patterning and reveals autoregulation of Xbra in dorsal mesoderm. Development. 1996;122:2427–2435. doi: 10.1242/dev.122.8.2427. [DOI] [PubMed] [Google Scholar]
- Darwin C. The variation of animals and plants under domestication. John Murray; London: 1868. [Google Scholar]
- Deforest ME, Basrur PK. Malformations and the Manx syndrome in cats. Can Vet J. 1979;20:304–314. [PMC free article] [PubMed] [Google Scholar]
- Green ST, Green FA. The Manx cat: an animal model for neural tube defects. Mater Med Pol. 1987;19:219–221. [PubMed] [Google Scholar]
- Haworth K, Putt W, Cattanach B, Breen M, Binns M, Lingaas F, Edwards YH. Canine homolog of the T-box transcription factor T; failure of the protein to bind to its DNA target leads to a short-tail phenotype. Mamm Genome. 2001;12:212–218. doi: 10.1007/s003350010253. [DOI] [PubMed] [Google Scholar]
- Howell JM, Siegel PB. Phenotypic variability of taillessness in Manx cats. J Hered. 1963;54:167–169. doi: 10.1093/jhered/54.4.167. [DOI] [PubMed] [Google Scholar]
- Howell JM, Siegel PB. Morphological effects of the Manx factor in cats. J Hered. 1966;57:100–104. doi: 10.1093/oxfordjournals.jhered.a107474. [DOI] [PubMed] [Google Scholar]
- Hytonen MK, Grall A, Hedan B, Dreano S, Seguin SJ, Delattre D, Thomas A, Galibert F, Paulin L, Lohi H, Sainio K, Andre C. Ancestral T-box mutation is present in many, but not all, short-tailed dog breeds. J Hered. 2009;100:236–240. doi: 10.1093/jhered/esn085. [DOI] [PubMed] [Google Scholar]
- Indrebo A, Langeland M, Juul HM, Skogmo HK, Rengmark AH, Lingaas F. A study of inherited short tail and taillessness in Pembroke Welsh corgi. J Small Anim Pract. 2008;49:220–224. doi: 10.1111/j.1748-5827.2007.00435.x. [DOI] [PubMed] [Google Scholar]
- James CC, Lassman LP, Tomlinson BE. Congenital anomalies of the lower spine and spinal cord in Manx cats. J Pathol. 1969;97:269–276. doi: 10.1002/path.1710970212. [DOI] [PubMed] [Google Scholar]
- Kispert A, Koschorz B, Herrmann BG. The T protein encoded by Brachyury is a tissue-specific transcription factor. EMBO J. 1995;14:4763–4772. doi: 10.1002/j.1460-2075.1995.tb00158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen H, Murray RE, Cockrell BY. Animal model for human disease. Spina bifida, sacral dysgenesis and myelocele. Animal model: Manx cats. Am J Pathol. 1972;68:203–206. [PMC free article] [PubMed] [Google Scholar]
- Kurushima JD, Lipinski MJ, Gandolfi B, Froenicke L, Grahn JC, Grahn RA, Lyons LA. Variation of cats under domestication: genetic assignment of domestic cats to breeds and worldwide random-bred populations. Anim Genet. 2013;44(3):311–324. doi: 10.1111/age.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipold HW, Huston K, Blauch B, Guffy MM. Congenital defects on the caudal vertebral column and spinal cord in Manx cats. J Am Vet Med Assoc. 1974;164:520–523. [PubMed] [Google Scholar]
- Lipinski MJ, Froenicke L, Baysac KC, Billings NC, Leutenegger CM, Levy AM, Longeri M, Niini T, Ozpinar H, Slater MR, Pedersen NC, Lyons LA. The ascent of cat breeds: genetic evaluations of breeds and worldwide random-bred populations. Genomics. 2008;91:12–21. doi: 10.1016/j.ygeno.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AH. A congenital defect in the spinal cord of the Manx cat. Vet Pathol. 1971;8:232–238. doi: 10.1177/030098587100800305. [DOI] [PubMed] [Google Scholar]
- Orpana AK, Ho TH, Stenman J. Multiple heat pulses during PCR extension enabling amplification of GC-rich sequences and reducing amplification bias. Anal Chem. 2012;84:2081–2087. doi: 10.1021/ac300040j. [DOI] [PubMed] [Google Scholar]
- Plummer SB, Bunch SE, Khoo LH, Spaulding KA, Kornegay JN. Tethered spinal cord and an intradural lipoma associated with a meningocele in a Manx-type cat. J Am Vet Med Assoc. 1993;203:1159–1161. [PubMed] [Google Scholar]
- Robinson R. Expressivity of the Manx gene in cats. J Hered. 1993;84:170–172. doi: 10.1093/oxfordjournals.jhered.a111311. [DOI] [PubMed] [Google Scholar]
- Schulte-Merker S, van Eeden FJ, Halpern ME, Kimmel CB, Nusslein-Volhard C. No tail (ntl) is the zebrafish homologue of the mouse T (Brachyury) gene. Development. 1994;120:1009–1015. doi: 10.1242/dev.120.4.1009. [DOI] [PubMed] [Google Scholar]
- Todd NB. Independent assortment of Manx and three coat color mutants in domestic cat. J Hered. 1963;54:266. doi: 10.1093/oxfordjournals.jhered.a107263. [DOI] [PubMed] [Google Scholar]
- Todd NB. The Manx factor in domestic cats. A possible genetic basis for expressivity of taillessness and other associated anomalies. J Hered. 1964;55:225–230. [PubMed] [Google Scholar]
- Wu B, Shao Y, Chen B, Liu C, Xue Z, Wu P, Li H. Identification of a novel mouse brachyury (T) allele causing a short tail mutation in mice. Cell Biochem Biophys. 2010;58:129–135. doi: 10.1007/s12013-010-9097-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.