Abstract
Bacteria have recently revealed an unexpectedly complex level of intracellular organization. Magnetotactic bacteria represent a unique class of such organization through the presence of their magnetosome organelles, which are organized along the magnetosome filament. Although the role of individual magnetosomes-associated proteins has started to be unraveled, their interaction has not been addressed with current state-of-the-art optical microscopy techniques, effectively leaving models of the magnetotactic bacteria protein assembly arguable. Here we report on the use of FLIM-FRET to assess the interaction of MamK (actin-like protein) and MamJ, two magnetosome membrane associated proteins essential to the assembly of magnetosomes in a chain. We used a host organism (E. coli) to express eGFP_MamJ and MamK_mCherry, the latest expectedly forming a filament. We found that in the presence of MamK the fluorescence of eGFP_MamJ is distributed along the MamK filament. FRET analysis using the fluorescence lifetime of the donor, eGFP, revealed a spatial proximity of MamK_mCherry and eGFP_MamJ typical of a stable physical interaction between two proteins. Our study effectively led to the reconstruction of part of the magnetotactic apparatus in vivo.
1. Introduction
The discovery of bacterial actin-like protein in the past decade has fundamentally changed our understanding of the subcellular organization of bacteria.1 Like eukaryotic cells, bacteria have organized substructures. The bacterial actin- and tubulin-like proteins such as MreB, FtsZ, and ParM are homologues of the eukaryotic cytoskeletal proteins. They are involved in many important processes for the viability of the cell. For instance, MreB is an actin-like protein forming helical filaments which is involved in chromosome segregation.2 It colocalizes underneath the cell membrane where it is associated with MbI another filamentous protein.3 MreB and Mbl are mutually essential in the stability of the cell shape.
Bacterial protein–protein interactions in vitro is typically studied using tandem affinity purification (TAP),4 immunoprecipitation,5 surface plasmon resonance (SPR),6 quartz crystal microbalance (QCM),7 circular dichroism,8 fluorescence spectroscopy,9 and Förster resonance energy transfer (FRET).10 These techniques require a high level of purity and a high concentration of the protein. Furthermore, in vitro experiments are typically performed in solution, eventually offering very different physicochemical conditions to those found in living micro-organisms (e.g., protein concentration, physiological control, confinement, folding machinery). In order to overcome these shortcomings, researchers have used yeast two-hybrid (Y2H),11,12 bacterial two-hybrid,13,14 and FRET.15,16 For example, Y2H has been used to study the interaction between FtsZ and FtsA, two bacterial proteins involved in cell division.17 A variant of Y2H is the bacterial two-hybrid. This is considered more appropriate to the study of protein–protein interactions in cellular compartment (i.e., outside nuclei); it offers the possibility of using the system in “mutation-driven structure-function studies” and for applications in which proteins transiently interact.18,19 Bacterial two-hybrid assay has been applied for the interaction of proteins such as the cytochrome c2 and cytochrome c peroxidase in Rhodobacter capsulatus,19 the proteins involved in the transduction signal cascade of Bordetella pertussis,13 and the interaction between MamJ and LimJ with MamK of the magnetotactic bacteria Magnetospirillum magneticum.20 However, two-hybrid assays are prone to false positives arising from the possible binding of the prey protein to the bait protein in the two-hybrid assay even though this would not happen in the normal in vivo context or from artifacts related to transcriptional activities independent of protein interactions.21,22
FRET is superior to yeast and bacterial two-hybrid because it does not rely on the signal amplification that occurs when protein–protein interactions initiate transcriptional activation. Hence, it is significantly less prone to false positives.11,22 Furthermore, as opposed to two-hybrid assay, FRET can be used to image the system studied.
FRET is a mechanism by which an excited fluorescent molecule (donor) transfers nonradiatively some energy to a neighboring ground-state molecule (acceptor). The efficiency of this transfer (E) is inversely dependent on the sixth power of the distance between the donor and the acceptor. Upon energy transfer, the acceptor molecule typically goes to an excited state from which it undergoes de-excitation through paths typical of fluorescent molecules. The efficiency of FRET (eq 4) is described by the Förster radius, R0 (eq 1), which is the distance between a donor and an acceptor, r, at which the efficiency is 50%. Since its efficiency inversely decreases with the sixth power of the distance, FRET does not occur when two fluorophores are more than ca. 10 nm apart. Therefore, this technique is useful for the study of phenomena, such as protein–protein interactions, which happen within distances typically smaller than 10 nm. Since the fluorescence of the donor competes with the transfer of energy to the acceptor, FRET can be monitored by measuring the fluorescence properties of the donor. This can be done using steady-state fluorescence measurement by monitoring the decrease in florescence intensity of the donor and/or the increase in fluorescence intensity of the acceptor. However, intensity-based FRET requires a high degree of control (e.g., to take into account photobleaching of the donor) and, for mapping applications, is prone to unwanted variations such as local changes in the fluorophore concentration, local environmental effects, and a range of optical artifacts. In turn, fluorescence lifetime imaging microscopy (FLIM) is insensitive to local changes in the fluorophore concentration.16 In FRET experiments, FLIM is thus used to map the fluorescence lifetime of the donor which reflect changes in the environment of the fluorophore (e.g., proximity of an acceptor molecule). In bacteria, FLIM-FRET has been used solely for the in vivo study the interaction pathway of FtsZ the tubulin-like bacterial proteins involved into the cell division.23
One of the most complex prokaryotic protein scaffolds observed to date is the protein assembly found in magnetotactic bacteria. This assembly is based on the actin-like protein MamK,24 which forms a filamentous structure extending from pole to pole of the cell and contributes to the mechanical stability of the magnetic chain arrangement.25,26 Scheffel et al. and Katzmann et al. showed that in order to obtain a fully stable chain in Magnetospirillum gryphiswaldense (MSR-1) MamK requires the presence of another protein, MamJ, which works as a biomolecular linker mediating a physical interaction between the filament and the magnetosomes.14,26,27 Although experimental indications suggest the direct interaction between MamJ and MamK in MSR-1, this has not been undoubtedly demonstrated to date, since the interactions studied using the yeast 2-hybrid (Y2H) method are not fully demonstrative of a real interaction.14
In order to assess the interaction between MamK filaments and MamJ of MSR-1 in vivo, fusion proteins with respectively mCherry and eGFP were generated and overexpressed in E. coli. The bacterial cultures were then imaged using fluorescence confocal microscopy, and FLIM images were generated in order to calculate the FRET characteristics between mCherry and eGFP and hence to evaluate the interaction between MamK and MamJ.
2. Experimental Section
a. Bacterial Culture and Protein Expression
mamK (MamK MSR-1 accession number: CAE12034) and mamJ (MamJ MSR-1 accession number: CAE12033) genes were fused with mCherry and eGFP genes, respectively. mamK_mCherry and eGFP_mamJ were purchased from Biomatik (Biomatik, Canada). These were subcloned into the expression vector pET28a(+), under the control of the promoter T7 and between the restriction sites NcoI and XhoI (Biomatik, Canada). The fusion of the fluorescence proteins were performed on the C-terminal of MamK and the N-terminal of MamJ. This choice was driven by previously published results showing the formation of a MamK_GFP filament in E. coli(28) and the presence of interacting sites at both extremities of MamJ.14eGFP and mCherry genes were amplified from eGFP_MamJ and MamK_mCherry vectors and subcloned into the expression vectors pET22b(+) (Merck Chemicals) using a restriction-free cloning method.29 The primers used for the PCR amplification are F5′ccgaattcgagctccgtcgacaagcttgcatggtgagcaagggc3′ and R5′ccggatctcagtggtggtggtggtggtgcttgtacagctcgtc3′ for eGFP and F5′cggatccgaattcgagctccgtcgacatggtgagcaagggcgag3′ and R5′atctcagtggtggtggtggtggtgcttgtacagctcgtccat3′ for mCherry. The vectors carrying the different genes were cotransformed in E. coli (Rosetta DE3). Cells were grown in LB (Luria–Bertani) medium containing antibiotics (specifications and concentrations listed with the individual expressions below, Sigma-Aldrich) at 37 °C until the optical density measured at 600 nm reached 0.7. Seven different E. coli cultures were prepared expressing the following proteins: (1) eGFP (100 μg/mL ampicillin); (2) mCherry (100 μg/mL ampicillin); (3) eGFP_MamJ (100 μg/mL kanamycin); (4) MamK_mCherry (100 μg/mL kanamycin); (5) eGFP_MamJ and mCherry (50 μg/mL kanamycin and 50 μg/mL ampicillin); (6) MamK_mCherry and eGFP (50 μg/mL kanamycin and 50 μg/mL ampicillin); (7) MamK_mCherry and eGFP_MamJ (150 μg/mL kanamycin). The level of protein expression was evaluated by SDS-PAGE (Supporting Information Figure S1) for different induction temperature, time, and IPTG concentration and assessed by Western Blot (AP-His Detector, KPL) following the provider instructions (Figure S2). Proteins expression was induced by addition of 1 mM IPTG (isopropylthio-β-galactoside). The cells were grown for 20 h at 25 °C, harvested, and washed by resuspension in a PBS buffer.
Visual screening of the bacterial colonies carrying both vectors expressing MamK_mCherry and EGFP_MamJ was performed using laser-scanning confocal fluorescence microscopy and confirmed the coexpression of both proteins. This is also confirmed by visual inspection of the expression on the Western Blot presented in Figure S2.
b. Sample Preparation
The bacteria are harvested and resuspended in an aqueous solution containing 1% of low-melt agarose at 30 °C and 20 μL of the suspension is sandwiched between a microscope slide and a coverslip (#1.5). The sample is cooled down to room temperature for the agarose to form a gel. This allowed the bacteria to be kept immobilized and alive while imaged.
c. Fluorescence Imaging
The experiment is performed on a commercial confocal microscope (SP5, Leica). A laser emitting at 488 nm is used for transmission and confocal fluorescence imaging. For FLIM, a pulsed Ti:sapphire laser (two-photon excitation) is tuned to 927 nm to minimize the excitation of mCherry30 and delivered pulses at 80 MHz. The laser beam is scanned over the sample using the confocal setup through a microscope objective (60×, 1.2 NA, WI, Leica). The fluorescence is collected in an epifluorescence mode at the donor emission wavelength, i.e., between 500 and 550 nm. The time-correlated single photon counting is performed using a photomultiplier tube (PMC-100) equipped with a microchannel channel plate (H5773P, Hammamatsu). The detector (MCP-PMT) is controlled by a PCI card (DCC-100, Becker and Hickl). The TCSPC card (SPC-830, Becker and Hickl) is operated by the SPCM software. This was set to record the arrival time of photons on a 12.5 ns time range in 4095 time bins for 64 × 64 pixels. The laser scanning and intensity settings were controlled by the LASAF (Leica) software. The Ti:S laser power was kept low to minimize photobleaching of the fluorophores during the time of the experiment (typically 10 min.) and to achieve a detection rate of less than 1% of the laser frequency. The setup was tested with a solution of Rhodamine 6G in ethanol. A single-exponential function fitted best the fluorescence decay and a fluorescence lifetime of 4.00 ns was calculated, in agreement with previous publications (3.99 ± 0.03 ns).31 For each imaged bacteria, two fluorescence intensity images, a transmission image, and a fluorescence lifetime image were recorded.
d. Data Analysis
The fluorescence lifetime images were generated by fitting the fluorescence decay at each pixel to a single-exponential decay function using SPCImage (Becker and Hickl). The curves were fitted from the raising edge of the curve to 8 ns after the channel with the most counts using the incomplete decay method. Examples of fitted decay curves are given Figure 3. The goodness of fit was assessed with the Chi2 (χ2) value and visual inspection of the residuals. Using these criteria, a single-exponential decay function fitted best the experimental decay curve (1 < χ2 < 1.3).
Figure 3.
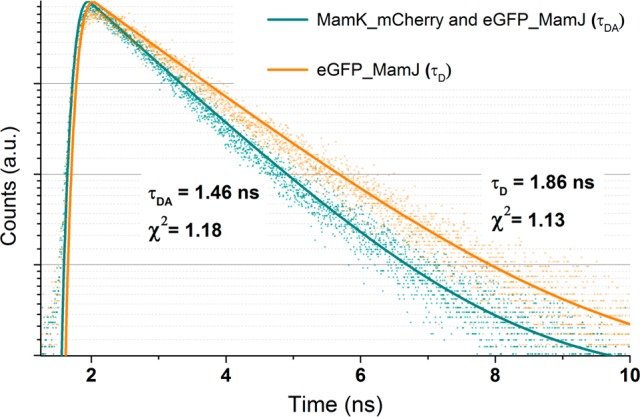
Fluorescence lifetime decay curves of eGFP in E. coli expressing MamK_mCherry and eGFP_MamJ (blue points, bottom) and in E. coli expressing eGFP_MamJ (orange points, top). Single-exponential function best fitting the data points (lines).
An expression for the Förster distance is given by
| 1 |
where κ2 is a factor ranging from 0 to 4 describing the relative orientation of the donor and acceptor transition dipole and is assumed to be equaled to 2/3 when fluorophores are bound to macromolecules;32n is the refractive index of the medium and is assumed equal to 1.39 when calculating R0 in cells (the refractive index of E. coli is 1.384);33QD is the quantum efficiency for fluorescence of the donor in the absence of the acceptor; and J(λ) is a measure of the spectral overlap between the donor emission and the acceptor absorption.32 The Förster distance of the mCherry–eGFP pair is 5.24 ± 0.1 nm.34 This Förster distance is calculated for QD of 60% and a refractive index of 1.33 (water). QD and n vary depending on the physicochemical properties of the system studied. In order to precisely calculate r, it is therefore necessary to recalculate R0. An expression for the quantum yield (QY) for fluorescence is given by32
| 2 |
where kr is the radiative decay rate and τ is the natural fluorescence lifetime of the fluorophore. The fluorescence lifetime decay of eGFP in water is best fitted by a biexponential decay function with an average lifetime of 2.68 ns.35 In cells, the fluorescence decay of eGFP is best fitted to a single-exponential decay with a fluorescence lifetime ranging from 1.6 to 2.5 ns.35−38 Using eqs 1 and 2 and assuming a constant radiative decay rate kr, an expression can be derived in order to correct for the difference between the fluorescence lifetime and the refractive index measured in solution to that in E. coli:
| 3 |
where R0 is the Förster radius of EGFP–mCherry in aqueous solution, τc is the fluorescence lifetime of the fused donor protein (EGFP_MamJ) in the system studied, τ is the natural fluorescence lifetime of EGFP, and nwater, nE coli are the refractive indices of water and E. coli, respectively. An expression for the efficiency of FRET is
| 4 |
This is typically measured using the relative fluorescence of the donor in the absence (ID) and the presence of the acceptor (IDA):32
| 5 |
Since the fluorescence intensity is proportional to the quantum yield, using eqs 2 and 5, the efficiency can also be calculated with the fluorescence lifetime of the donor in the absence (τD) and the presence of the acceptor (τDA):
| 6 |
The distance between donor and acceptor is calculated by rearranging eqs 4 and 6 and is given by
| 7 |
where in the following discussions R0 is the corrected Förster radius of the donor–acceptor pair, τD is the fluorescence lifetime of the donor alone (MamJ_eGFP), and τDA is the fluorescence lifetime of the donor in the presence of an acceptor (MamK_mCherry and eGFP_MamJ).
3. Results and Discussion
Images of E. coli expressing eGFP_MamJ (Figure 1a–c), MamK_mCherry (Figure 1d–f), and both eGFP_MamJ and MamK_mCherry (Figure 1g–i) are shown Figure 1. Each expression is presented on a separate row. The columns correspond to the different imaging parameters. The first column (green channel) shows the images recorded with excitation at 488 nm and emission between 500 and 550 nm corresponding to the fluorescence emission of the eGFP protein. The second column (red channel) shows the images recorded with excitation at 561 nm and emission between 570 and 640 nm corresponding to the fluorescence emission of the mCherry protein. The third column is a transmission image of the sample.
Figure 1.
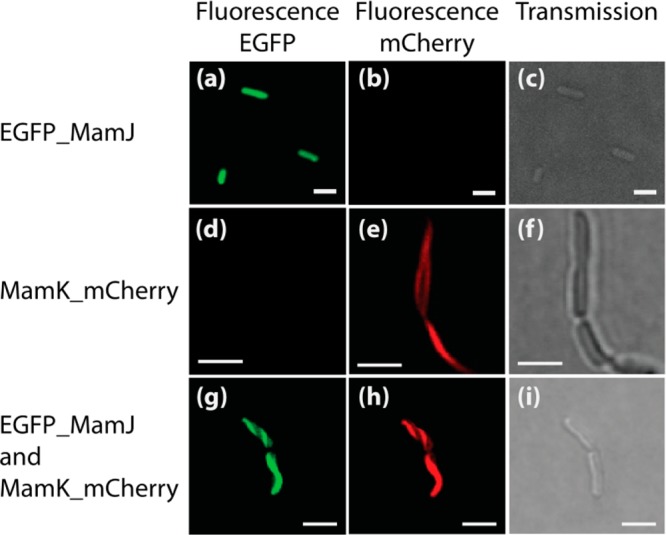
Fluorescence images (a, b) and transmission image (c) of E. coli expressing eGFP_MamJ; fluorescence images (d, e) and transmission image (f) of E. coli expressing MamK_mCherry; and fluorescence images (g, h) and transmission image (i) of E. coli expressing eGFP_MamJ and MamK_mCherry. The excitation and emission wavelengths were 488 and 500–550 nm (a, d, g) and 561 and 570–620 nm (b, e, h). Scale bar 2 μm.
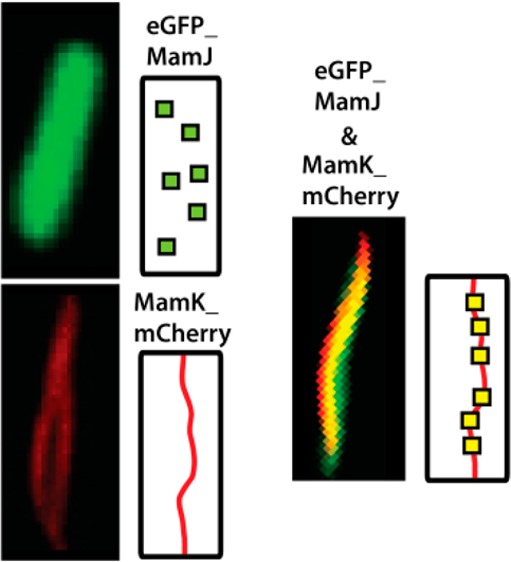
As observed, the fluorescence of eGFP_MamJ protein is in the green region of the spectrum (Figure 1a) and is homogeneously distributed in the entire cells (Figure 1c), whereas the fluorescence of MamK_mCherry is in the red region of the spectrum and its elongated spatial distribution is typical of that of a filament (Figure 1e) extending across cells (Figure 1f) as similar to the pattern observed by Pradel et al.28 and Komeili et al.25 Figure 1g–i shows bacteria from a culture where MamK_mCherry and eGFP_MamJ were coexpressed. All the observed bacteria that were expressing eGFP_MamJ were also expressing MamK_mCherry. These are the bacteria that can be seen in the green and red channels Figure 1g,h. In these, the MamK_mCherry filaments are seen in the red channel. The first evidence for interaction between MamK_mCherry and eGFP_MamJ is found when comparing the localization of the fluorescence signal of the green and red channels. These show that the fluorescence of eGFP_MamJ is no longer homogeneously distributed across the cell as in Figure 1a but rather emanates from the same region as where the MamK_mCherry filament are found, effectively demonstrating the reconstruction in E. coli of part of the protein assembly found in MSR-1. We expected to observe some free unbound eGFP_MamJ in cells where MamK was expressed. This was however not the case. This may result from a relative higher expression of MamK with respect to MamJ as suggested by the Western Blot (Figure S2).
This observation is also evident in Figure 2 that shows the localization of eGFP_MamJ (Figure 2a–c), eGFP with respect to MamK_mCherry in the absence (Figure 2d–f), and in the presence (Figure 2g–i) of MamJ. In the absence of MamJ and/or MamK (Figure 2a–f), the fluorescence from eGFP emanates from the entire cell whereas in the mutual presence of MamJ and MamK, this comes exclusively from the MamK filament.
Figure 2.
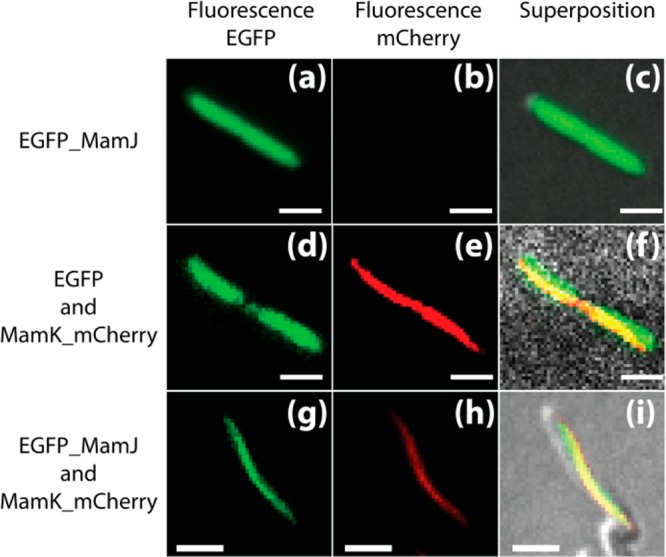
Fluorescence images of E. coli expressing: eGFP_MamJ (a, b, c), MamK_mCherry and eGFP (d, e, f), and MamK_mCherry and eGFP_MamJ (g, h, i). Excitation and emission wavelength of eGFP, i.e., 488 and 500–550 nm (a, d, g); excitation and emission wavelength of mCherry, i.e., 561 and 570–640 nm (b, e, h). Superposition of images (a) and (b) on a transmission image (c); (d) and (e) on a transmission image (f); and (g) and (h) on a transmission image (i). Scale bar 2 μm.
The control intensity imaging experiments performed are all negative, and the colocalization of eGFP and mCherry occurs only when both MamK and MamJ are coexpressed. Most importantly, the coexpression of eGFP and MamK_mCherry does not lead to the arrangement of eGFP along the MamK filament (Figures 1 and 2), a prerequisite to the validation of the FRET experiment shown Figure 4. The control samples were also image by FLIM and their fluorescence lifetime extracted. As summarized Table 1, the fluorescence lifetimes of eGFP of 1.88 ± 0.07, 1.87 ± 0.03, and 1.93 ± 0.03 ns were calculated in E. coli expressing eGFP, eGFP_MamJ, and MamK_mCherry and eGFP, respectively. The fluorescence lifetimes of the three controls are not significantly different; hence, we can conclude that there are no detectable interactions between the coexpressed proteins in those systems. Furthermore, the similar fluorescence lifetime of eGFP and eGFP_MamJ indicates that the fusion of eGFP with MamJ does not alter the fluorescence properties of eGFP (Figure 4b,d and Table 1).
Figure 4.
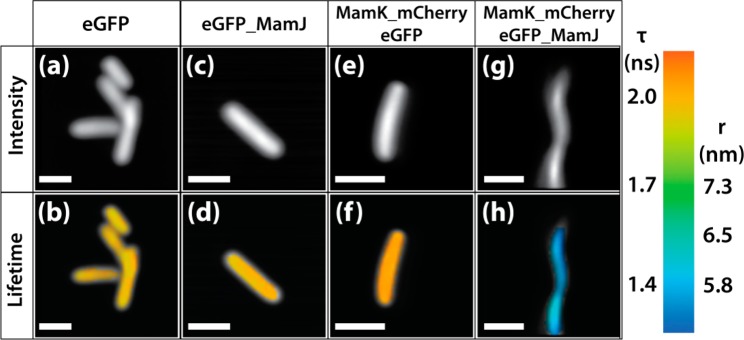
Fluorescence intensity and fluorescence lifetime images of E. coli expressing eGFP (a,b); eGFP_MamJ (c, d); MamK_mCherry and eGFP (e, f); and MamK_mCherry and eGFP_MamJ (g, h). The color-coded scale bar is used to map the fluorescence lifetime of eGFP and the separation distance between eGFP and mCherry calculated. The fluorescence lifetimes range from 1.3 ns (blue) to 2.1 ns (red) and the separation from 5.5 nm (blue) to 7.3 nm (green). Scale bar 2 μm.
Table 1. Mean Fluorescence Lifetime Values and Their Standard Deviations for eGFP in E. coli Expressing eGFP; eGFP_MamJ; MamK_mCherry and eGFP; and MamK_mCherry and eGFP_MamJ.
| strains | τ (ns) | SD (ns) |
|---|---|---|
| eGFP | 1.88 | 0.07 |
| eGFP_MamJ | 1.87 | 0.03 |
| MamK_mCherry and eGFP | 1.93 | 0.03 |
| MamK_mCherry and eGFP_MamJ | 1.49 | 0.09 |
Figure 3 shows the fluorescence decay curves of eGFP in E. coli expressing MamK_mCherry and eGFP_MamJ and in E. coli expressing eGFP_MamJ alone as a comparison. A single-exponential decay function was successfully fitted to the data points (χ2 < 1.3). The distance between donor and acceptor is calculated according to eq 7. The faster decay of eGFP in E. coli expressing MamK_mCherry and eGFP_MamJ results from FRET between mCherry and eGFP, i.e., the physical interaction between MamK and MamJ.
Figure 4 shows examples of the fluorescence lifetime images of E. coli expressing eGFP (b), MamJ_eGFP (d), MamK_mCherry and eGFP (f), and MamK_mCherry and eGFP_MamJ (h). The decrease of eGFP fluorescence lifetime in the presence of MamK_mCherry and eGFP_MamJ indicates the occurrence of FRET between mCherry and eGFP, hence the interaction of MamJ and MamK.
The distribution of the fluorescence lifetime values calculated from at least five samples of each expression is summarized in Table 1 and plotted in Figure 5.
Figure 5.
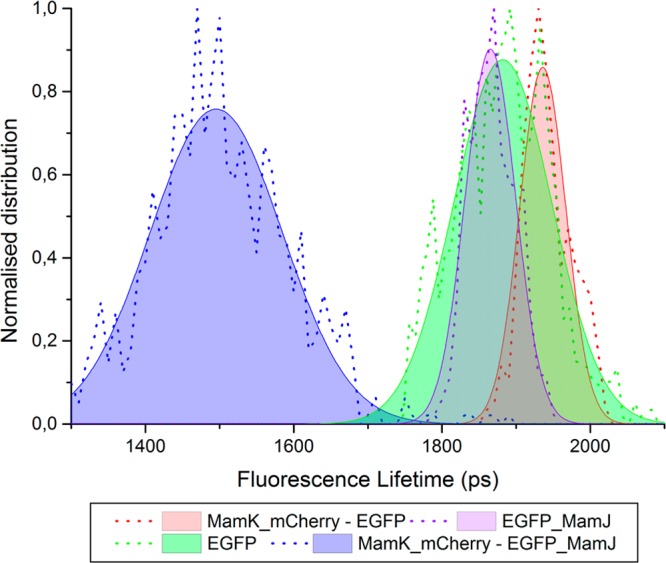
Distribution of the fluorescence lifetime values (dotted lines) fitted to a Gaussian function (filled area delimited by a line) of the E. coli strains expressing eGFP (green); eGFP_MamJ (purple); MamK_mCherry and eGFP (red); and MamK_mCherry and EGFP_MamJ (blue).
Considering a constant radiative decay rate and the fluorescence lifetime of eGFP as measured when expressed in E. coli, we calculated the quantum yield of eGFP_MamJ using eq 2. This is 0.42 in our system. Using eq 3, we calculated that the Förster distance for the eGFP–mCherry pair in our system (n = 1.39 and τD = 1.9 ns) is 4.8 nm. Finally, using eq 7 and the fluorescence lifetime of eGFP in bacteria expressing eGFP_MamJ (τD) and in bacteria expressing MamK_mCherry and eGFP_MamJ (τDA) the separation between eGFP and mCherry r equals 6.1 ± 0.3 nm. Since eGFP and mCherry do not interact, their proximity results unequivocally from the physical interaction of MamK and MamJ.
4. Conclusions
As shown in previous experiments, MamK overexpressed in E. coli readily forms filaments.25,28 The coexpression of MamJ and MamK leads to the colocalization of the two proteins along MamK filaments in vivo, effectively demonstrating the reconstruction of part of the protein assembly found in MSR-1 in a host organism.
The presence of both proteins is essential to the formation of the magnetosome chain27 in MSR-1. Using FLIM-FRET and the coexpression of MamK_mCherry and eGFP_MamJ in E. coli, we have demonstrated the molecular interaction between MamJ and MamK of MSR-1. E. coli expressing eGFP and the different controlled studied exhibit a fluorescence lifetime of ca. 1.9 ns. In the mutual presence of the two fused bacterial proteins, the lifetime of eGFP shortens to 1.49 ± 0.09 ns, corresponding to a separation of 6.1 ± 0.3 nm between mCherry and eGFP. Protein–protein interactions can be transient or stable interactions.39 Since the MamK and MamJ proteins are colocalized and the decrease of eGFP fluorescence lifetime indicates FRET between eGFP and mCherry when MamK_mCherry and eGFP_MamJ are coexpressed, we conclude that these two proteins form a stable interaction, important for the magnetosomes alignment and the chain stability in MSR-1.
Because it is virtually false positive free, can be performed in vivo, and allows high-resolution imaging of the system, FLIM-FRET is an ideal imaging technique for the study of protein–protein interaction.
We believe that following our findings researchers studying bacterial protein interactions will be inclined to use FLIM-FRET as a quantitative imaging method to reconstruct the assembly and quantify the interaction between building blocks of complex protein assembly such as the one found in magnetotactic bacteria. The understanding gained is valuable to the biomimetic design of functional materials. For example, the assembly of nanostructured magnetic inorganic materials might be achievable by assembling MamK, MamJ, and magnetosomes in vitro.
Acknowledgments
The authors are grateful to Dr. Anita Jones and Dr. Giuliano Bellapadrona for the essential scientific discussions, to Dr. Rumiana Dimova and Carmen Remde for providing access and training on the microscopy platform, and to Katharina Tomschek for laboratory work assistance. This research was supported by the Max Planck Society, a priority program of the DFG (SPP1420, FA835/2-1) and the European Research Council (Starting Grant MB2 no. 256915).
Supporting Information Available
Figure S1: SDS-PAGE of MamK_mCherry and eGFP; Figure S2: Western Blot of a strain of E. coli showing the level of coexpression of both EGFP_MamJ and MamK_mCherry; Figure S3: FLIM image of E. coli expressing eGFP_MamJ and mCherry; Figure S4: images of E. coli expressing MamK_mCherry; Note S1: effect of FRET on the fluorescence intensity and localization of the fluorescence proteins. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Author Contributions
† M.A.C. amd M.B. equally contributed to this work.
Supplementary Material
References
- Carballido-Lopez R. The Bacterial Actin-Like Cytoskeleton. Microbiol. Mol. Biol. Rev. 2006, 704888–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T.; Moller-Jensen J.; Lobner-Olesen A.; Gerdes K. Dysfunctional Mreb Inhibits Chromosome Segregation in Escherichia Coli. EMBO J. 2003, 22195283–5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufo H. J. D.; Graumann P. L. Dynamic Localization and Interaction with Other Bacillus Subtilis Actin-Like Proteins Are Important for the Function of Mreb. Mol. Microbiol. 2006, 6251340–1356. [DOI] [PubMed] [Google Scholar]
- Puig O.; Caspary F.; Rigaut G.; Rutz B.; Bouveret E.; Bragado-Nilsson E.; Wilm M.; Séraphin B. The Tandem Affinity Purification (Tap) Method: A General Procedure of Protein Complex Purification. Methods 2001, 243218–229. [DOI] [PubMed] [Google Scholar]
- Yang-Yen H.-F.; Chambard J.-C.; Sun Y.-L.; Smeal T.; Schmidt T. J.; Drouin J.; Karin M. Transcriptional Interference between C-Jun and the Glucocorticoid Receptor: Mutual Inhibition of DNA Binding Due to Direct Protein-Protein Interaction. Cell 1990, 6261205–1215. [DOI] [PubMed] [Google Scholar]
- Karlsson R.; Falt A. Experimental Design for Kinetic Analysis of Protein-Protein Interactions with Surface Plasmon Resonance Biosensors. J. Immunol. Methods 1997, 2001–2121–133. [DOI] [PubMed] [Google Scholar]
- Rodahl M.; Hook F.; Krozer A.; Brzezinski P.; Kasemo B. Quartz-Crystal Microbalance Setup for Frequency and Q-Factor Measurements in Gaseous and Liquid Environments. Rev. Sci. Instrum. 1995, 6673924–3930. [Google Scholar]
- Greenfield N. J. Circular Dichroism Analysis for Protein-Protein Interactions. Methods Mol. Biol. (Clifton, N.J.) 2004, 261, 55–78. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J.; Snapp E.; Kenworthy A. Studying Protein Dynamics in Living Cells. Nat. Rev. Mol. Cell Biol. 2001, 26444–456. [DOI] [PubMed] [Google Scholar]
- Phizicky E. M.; Fields S. Protein-Protein Interactions - Methods for Detection and Analysis. Microbiol. Rev. 1995, 59194–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S.; Song O. K.; Novel A. Genetic System to Detect Protein Protein Interactions. Nature 1989, 3406230245–246. [DOI] [PubMed] [Google Scholar]
- Wilson T. E.; Padgett K. A.; Johnston M.; Milbrandt J. A Genetic Method for Defining DNA-Binding Domains - Application to the Nuclear Receptor Ngfi-B. Proc. Natl. Acad. Sci. U. S. A. 1993, 90199186–9190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G.; Pidoux J.; Ullmann A.; Ladant D. A Bacterial Two-Hybrid System Based on a Reconstituted Signal Transduction Pathway. Proc. Natl. Acad. Sci. U. S. A. 1998, 95105752–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffel A.; Schueler D. The Acidic Repetitive Domain of the Magnetospirillum Gryphiswaldense Mamj Protein Displays Hypervariability but Is Not Required for Magnetosome Chain Assembly. J. Bacteriol. 2007, 189176437–6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertazzi L.; Arosio D.; Marchetti L.; Ricci F.; Beltram F. Quantitative Fret Analysis with the E(0)Gfp-Mcherry Fluorescent Protein Pair. Photochem. Photobiol. 2009, 851287–297. [DOI] [PubMed] [Google Scholar]
- Millington M.; Grindlay G. J.; Altenbach K.; Neely R. K.; Kolch W.; Bencina M.; Read N. D.; Jones A. C.; Dryden D. T. F.; Magennis S. W. High-Precision Flim-Fret in Fixed and Living Cells Reveals Heterogeneity in a Simple Cfp-Yfp Fusion Protein. Biophys. Chem. 2007, 1273155–164. [DOI] [PubMed] [Google Scholar]
- Wang X. D.; Huang J. A.; Mukherjee A.; Cao C.; Lutkenhaus J. Analysis of the Interaction of Ftsz with Itself, Gtp, and Ftsa. J. Bacteriol. 1997, 179175551–5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. C.; Kornacker M. G.; Hochschild A. Escherichia Coli One- and Two-Hybrid Systems for the Analysis and Identification of Protein-Protein Interactions. Methods—Companion Methods Enzymol. 2000, 20180–94. [DOI] [PubMed] [Google Scholar]
- Borloo J.; De Smet L.; Vergauwen B.; Van Beeumen J. J.; Devreese B.; Beta-Galactosidase-Based A. Bacterial Two-Hybrid System to Assess Protein-Protein Interactions in the Correct Cellular Environment. J. Proteome Res. 2007, 672587–2595. [DOI] [PubMed] [Google Scholar]
- Draper O.; Byrne M. E.; Li Z.; Keyhani S.; Barrozo J. C.; Jensen G.; Komeili A. Mamk, a Bacterial Actin, Forms Dynamic Filaments in Vivo That Are Regulated by the Acidic Proteins Mamj and Limj. Mol. Microbiol. 2011, 822342–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S. High-Throughput Two-Hybrid Analysis. FEBS J. 2005, 272215391–5399. [DOI] [PubMed] [Google Scholar]
- You X.; Nguyen A. W.; Jabaiah A.; Sheff M. A.; Thorn K. S.; Daugherty P. S. Intracellular Protein Interaction Mapping with Fret Hybrids. Proc. Natl. Acad. Sci. U. S. A. 2006, 1034918458–18463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemse J.; Borst J. W.; de Waal E.; Bisseling T.; van Wezel G. P. Positive Control of Cell Division: Ftsz Is Recruited by Ssgb During Sporulation of Streptomyces. Genes Dev. 2011, 25189–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünberg K.; Wawer C.; Tebo B. M.; Schueler D.; Large Gene A. Cluster Encoding Several Magnetosome Proteins Is Conserved in Different Species of Magnetotactic Bacteria. Appl. Environ. Microbiol. 2001, 67104573–4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A.; Li Z.; Newman D. K.; Jensen G. J. Magnetosomes Are Cell Membrane Invaginations Organized by the Actin-Like Protein Mamk. Science 2006, 3115758242–245. [DOI] [PubMed] [Google Scholar]
- Katzmann E.; Scheffel A.; Gruska M.; Plitzko J. M.; Schueler D. Loss of the Actin-Like Protein Mamk Has Pleiotropic Effects on Magnetosome Formation and Chain Assembly in Magnetospirillum Gryphiswaldense. Mol. Microbiol. 2010, 771208–224. [DOI] [PubMed] [Google Scholar]
- Scheffel A.; Gruska M.; Faivre D.; Linaroudis A.; Plitzko J. M.; Schuler D. An Acidic Protein Aligns Magnetosomes Along a Filamentous Structure in Magnetotactic Bacteria. Nature 2006, 4407080110–114. [DOI] [PubMed] [Google Scholar]
- Pradel N.; Santini C.-L.; Bernadac A.; Fukumori Y.; Wu L.-F. Biogenesis of Actin-Like Bacterial Cytoskeletal Filaments Destined for Positioning Prokaryotic Magnetic Organelles. Proc. Natl. Acad. Sci. U. S. A. 2006, 1034617485–17489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent F.; Lowe J. Rf Cloning: A Restriction-Free Method for Inserting Target Genes into Plasmids. J. Biochem. Biophys. Methods 2006, 67167–74. [DOI] [PubMed] [Google Scholar]
- Drobizhev M.; Makarov N. S.; Tillo S. E.; Hughes T. E.; Rebane A. Two-Photon Absorption Properties of Fluorescent Proteins. Nat. Methods 2011, 85393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magde D.; Rojas G. E.; Seybold P. G. Solvent Dependence of the Fluorescence Lifetimes of Xanthene Dyes. Photochem. Photobiol. 1999, 705737–744. [Google Scholar]
- Lakowicz J.Principles of Fluorescence Spectroscopy; Kluwer Academic: Dordrecht, 1999. [Google Scholar]
- Balaev A. E.; Dvoretski K. N.; Doubrovski V. A. Refractive Index of Escherichia Coli Cells. Proc. SPIE 2002, 4707, 253–260. [Google Scholar]
- Akrap N.; Seidel T.; Barisas B. G. Forster Distances for Fluorescence Resonant Energy Transfer between Mcherry and Other Visible Fluorescent Proteins. Anal. Biochem. 2010, 4021105–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess S. T.; Sheets E. D.; Wagenknecht-Wiesner A.; Heikal A. A. Quantitative Analysis of the Fluorescence Properties of Intrinsically Fluorescent Proteins in Living Cells. Biophys. J. 2003, 8542566–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobs S.; Subramaniam V.; Schonle A.; Jovin T. M.; Hell S. W. Egfp and Dsred Expressing Cultures of Escherichia Coli Imaged by Confocal, Two-Photon and Fluorescence Lifetime Microscopy. FEBS Lett. 2000, 4793131–135. [DOI] [PubMed] [Google Scholar]
- Peter M.; Ameer-Beg S. M.; Hughes M. K. Y.; Keppler M. D.; Prag S.; Marsh M.; Vojnovic B.; Ng T. Multiphoton-Flim Quantification of the Egfp-Mrfp1 Fret Pair for Localization of Membrane Receptor-Kinase Interactions. Biophys. J. 2005, 8821224–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi T.; Wang H.-P.; Kinjo M.; Ohta N. Application of Fluorescence Lifetime Imaging of Enhanced Green Fluorescent Protein to Intracellular Ph Measurements. Photochem. Photobiol. Sci. 2008, 76668–670. [DOI] [PubMed] [Google Scholar]
- Sprinzak E.; Altuvia Y.; Margalit H. Characterization and Prediction of Protein-Protein Interactions within and between Complexes. Proc. Natl. Acad. Sci. U. S. A. 2006, 1034014718–14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


