Abstract
Microbial translocation (MT) from the gut is implicated in driving immune activation, increasing morbidity and mortality in HIV. We used bacterial 16S rDNA PCR, Sanger sequencing, and high-throughput sequencing to identify microbial DNA in the bloodstream of HIV-infected children in London, United Kingdom. Blood samples were collected from sequential children attending the HIV clinic at Great Ormond Street Hospital, London. DNA extraction, broad range 16S rDNA PCR, and standard Sanger sequencing were carried out. A subset of positive samples was analyzed by high-throughput sequencing (Roche 454 platform). Of 105 samples collected from sequential children, nine were positive using broad range 16S rDNA PCR (8.6%; 95% CI 4.4–16%). From three amplicons, 16S rDNA sequences were identified as Streptococcus, Propionibacterium acnes, and coagulase-negative Staphylococcus. Four positive samples were analyzed by high-throughput sequencing. In the three samples in which organisms were identified by Sanger sequencing, the same species were identified. Further species, in differing proportions, were identified in all four samples. The identified organisms included known gut orders Bifidobacteriaceae, Lactobacillaceae, Bacteroidales, and Clostridiales. In immunocompetent children of equivalent age, no bacterial DNA was detected in blood using this approach. This is the first study to our knowledge using molecular techniques to identify MT in children in the developed world. Our data indicate that 16S rDNA is detectable in 8.6% of HIV-infected children. Levels of DNA were low and from multiple bacterial species. Further studies are needed to ascertain the importance of MT in HIV-infected children.
Introduction
Immune activation is central to the pathogenesis of HIV infection and is associated with increased mortality and non-AIDS-related morbidity1–10 despite antiretroviral therapy (ART). Microbial translocation (MT) across the gut is one mechanism that may drive immune activation.11–15
Rapid CD4 cell depletion within the gut mucosa early in HIV infection is hypothesized to allow translocation of intestinal microbiotal products into the bloodstream at increased levels compared with uninfected persons.16–22 Translocation may be of viable organisms or of microbial components, which may include lipopolysaccharide (LPS) and bacterial DNA. Intact organisms and microbial components are potential immunostimulants.
MT has been reported to be higher in patients with HIV infection and has been implicated as a cause for increased immune activation and poor CD4 cell count recovery on ART.9,12,17 It has been hypothesized that immune activation is driven by MT as a consequence of HIV-induced gut mucosal damage. If correct, a number of potential treatment strategies could be employed to reduce MT and improve outcomes. There are clinical trials underway with this specific aim.23–25
Most evidence for increased microbial translocation in HIV is based on detection of increased levels of LPS and bacterial 16S rDNA. The latter is detected through a broad-range quantitative polymerase chain reaction (qPCR). However 16S rDNA-PCR is vulnerable to contamination from exogenous and endogenous bacterial DNA.26–29 Without sequencing the amplicons, these results are therefore potentially artifactual and indeed thus far sequencing has largely yielded results compatible with environmental contaminants and not recognized gut commensals.17,30,31
Materials and Methods
We set out to determine if bacterial 16S rDNA was detectable in the bloodstream of HIV-infected children in London, United Kingdom, and if present, to use sequencing to identify the microbial source.
Discard EDTA blood samples were collected from sequential children attending the HIV outpatient clinic at Great Ormond Street Hospital, London. At the time of the study 124 HIV-infected children were being seen regularly, 97 of whom were receiving ART. Of the children 57% were aged 12–17 years and 43% were under 12 years; 78% were of black African ethnic origin, 11% were African/white, and the remainder classified themselves as white, Afrocaribbean, or other. Of those on ART 88% were on triple therapy, with 6% on dual therapy, 4% on monotherapy, and 1% on four drugs. Of the children 47% had a CD4 cell percentage above 30% at the last clinic visit, with 34% having a CD4 cell percentage between 21% and 30%, 16% between 11% and 20%, and 3% under 10%; 64% of children had a viral load <50 copies/ml, with 32% between 50 and 100,000 copies/ml and 4% with a viral load >100,000 copies/ml.
The commercial QIAmp DNA mini kit (Qiagen, Crawley, UK), with an additional bead-beating step to ensure complete lysis of bacterial cells, was used to extract DNA from 200 μl of EDTA blood within 48 h of collection. Broad range 16S rDNA PCR was carried out as previously described using two primer sets.26,32 16SFa/16SFb (GCTCAGATTGAACGCTGG/GCTCAGGAYGAACGCTGG) and 16SR (TACTGCTGCCT CCCGTA) amplified the V1 and V2 region of the 16S rDNA gene26 and 785F (GGATTAGATACCCBRGTAGTC) and 1175R (ACGTCRTCCCCDCCTTCCTC) amplified the V5 and V6 region. The sensitivity of the broad-range 16S rDNA PCR has previously been demonstrated to be 10–100 colony-forming units per PCR reaction.26 Amplicons derived from positive samples were sequenced using the Big-Dye v.3.1 cycle sequencing kit (Applied Biosystems, Warrington, UK) and analyzed on the 3130 genetic analyzer (Applied Biosystems). The sequences obtained were compared to those on the GenBank database using the BLAST program available at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov). The sequence was classified to species level if there was >98% homology with two or more GenBank sequences from the same species, submitted by independent laboratories, and the percentage identification was lower for all other species. A subset of samples found to be positive by broad range 16S rDNA PCR was analyzed by high-throughput sequencing. Attached to the 16S rDNA primers were standard 454 Titanium adapters and an individual barcode sequence for each sample. The library was pyrosequenced on a 454 FLX Titanium (Roche) platform according to the manufacturer's recommended protocol.
Sequences were processed and analyzed using QIIME.33 16SFa/16SFb/16SR and 785F/1175R reads were discarded if they contained ambiguous bases, if the quality score was <25, if the run of homopolymer bases was >6, if there was a mismatch in primer sequence, and if the barcode could not be corrected. Operational taxonomic units (OTUs) were clustered at 97% sequence similarity and chimeric sequences were removed using USEARCH.34 Representative OTUs were assigned taxonomy using the RDP classifier35 at a minimum support threshold of 80% again the Greengenes database.36
Discard samples were used in accordance with the guidance of the National Patient Safety Agency and National Research Ethics Service, which assessed this project to be within the remit of assay performance evaluation.
Results
Samples were collected from 105 sequential children attending the outpatient clinic representing 85% of the clinic population.
Nine samples were found to be positive using broad range 16S rDNA PCR representing a positivity rate of 8.6% (95% CI 4.4–16%). From three of these amplicons, 16S rDNA sequences were obtained and identified as Streptococcus species, Propionibacterium acnes, and coagulase negative Staphylococcus. Direct Sanger sequencing failed in the other six samples, probably due to a mixture of 16S rDNA sequences from different bacterial species.
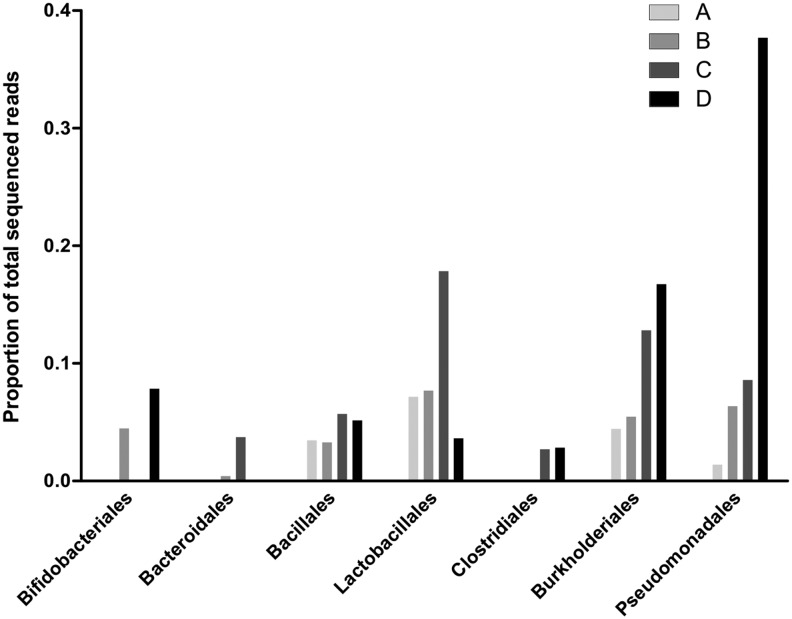
Three samples that were successfully sequenced and one sample on which direct sequencing failed (samples A, B, C, and D) were then analyzed by high-throughput sequencing. This identified several orders of bacteria known to be associated with the human gut.31,30,37 The relative proportions of these bacteria are shown in Fig. 1. Two of four samples were positive for Bifidobacteriales, Bacteroidales, and Clostridiales, whereas all samples were positive for Lactobacillales, Burkholderiales, Bacillales, and Pseudomonadales.
FIG. 1.
Bacteria orders recovered from HIV+ blood using 16S rDNA high-throughput sequencing. Proportions of the total recovered sequences from each sample after quality filtering and assigning taxonomy to reads generated by high-throughput sequencing. Samples are labeled A, B, C, and D.
Of the nine patients in whom microbial DNA was detected, five (55%) had an undetectable viral load; four (45%) had a CD4 percentage above 30% and two (22%) had a CD4 of less than 10%.
Discussion
There has so far been few data generated investigating the question of MT in HIV-infected children,15,22,38,39 with this being the first study to our knowledge using molecular techniques to identify MT in children in the developed world, and the only one to have successfully sequenced bacterial DNA using conventional and high-throughput techniques. In adult populations standard sequencing has been carried out in only a handful of studies17,30 and has yielded results compatible with possible contamination such as Serratia spp. and Rahnella spp.17,31 Previous work in this field has largely relied on the detection of LPS or surrogate markers of MT such as sCD14 (a coreceptor for LPS produced by monocytes) and LPS-binding protein (LBP).9,22,40,41 In view of ongoing debate about optimal methods to detect MT in HIV-infected people,29,42,43 there is an urgent need for further assay development and optimization. This need is compounded by the conflicting data generated in pediatric populations in particular, but also in adult populations. Indeed a major African longitudinal study found no evidence of increasing MT markers during untreated disease progression.44 Detection of known gut commensal microbial DNA in the bloodstream will accelerate our understanding of MT and its potential impact on HIV pathogenesis. These findings are compatible with MT occurring in relatively well children infected with HIV in the United Kingdom, for the most part on ART with reasonably well-controlled disease. In resource-limited settings in the context of malnutrition, enteropathy, and poorly controlled HIV infection, MT has potentially an even greater significance.45,46
It has been difficult to demonstrate evidence of gut-associated organisms using broad-range 16S rDNA PCR and conventional Sanger sequencing techniques.17,30 Although a powerful technique for detection and identification of most bacterial species from culture-negative samples, 16S rDNA PCR does have limitations. One disadvantage of the technique is that typically a single dominant species will be identified with minority species often being outcompeted in the PCR. The addition of a cloning step can resolve mixtures of 16S rDNA sequences and identify minority species. However, the technique is very labor intensive and this limits the maximum number of sequences that can be obtained to somewhere in the order of 100. When cloning has been used in this field, it has demonstrated the presence of mixed sequences, which further emphasizes the need for more sophisticated techniques.30 High-throughput sequencing can generate hundreds of thousands of sequences from a single 16S rDNA amplicon, which, with subsequent bioinformatics input, can provide a comprehensive picture of all bacterial species that have been amplified, even those that are a small minority.47–49 To our knowledge this is the first study to use high-throughput sequencing to investigate microbial translocation in HIV. The potential usefulness of the technique is demonstrated by the detection of 16S rDNA sequences of common gut organisms such as Bifidobacteriales and Lactobacillalese that were not detected by conventional Sanger sequencing of the amplicon.
The strengths and weaknesses of this approach to identifying bacterial DNA in whole blood are highlighted by our findings. Due to the relative paucity of bacterial DNA in the samples, high-throughput sequencing will identify low levels of multiple organisms including potential contaminants that are present in PCR reagents. 26 However, the depth of coverage afforded by high-throughput sequencing also identifies potentially interesting organisms that could be derived from the GI tract. We cannot rule out contamination as the source of microbial DNA in the samples. However, in 191 blood samples of immunocompetent children previously evaluated using this 16S rDNA PCR technique, none was positive despite some having clinically significant bacteremia.50
The microbial translocation hypothesis needs further investigation in children where data so far are limited and conflicting.15,22,38,51 Our data indicate that MT may consist of low levels of multiple organisms resident within the GI tract. Given the increased survival of HIV-infected children on ART,52,53 investigating the importance of MT in this population and in resource-limited settings is vital for the future rational design of intervention studies. If microbial translocation is indeed responsible for increased levels of immune activation and thus excess morbidity and mortality in those infected with HIV, there is the possibility of rational therapeutic intervention, including probiotics, synbiotics, nonabsorbable antibiotics, and LPS binders.54 There is an urgent need for further investigation using molecular techniques that can comprehensively describe the microbiota detectable in the bloodstream of those with HIV and determine whether this profile changes over time and its relationship to clinical outcome.
Acknowledgments
Dr. Fitzgerald, an MRC clinical training fellow, was funded by an NIHR Academic Clinical Fellowship, and received a grant for consumables from the Institute of Child Health. Dr. Harris is funded by the NHS. Mr. Doyle is funded by the Food and Nutritional Technical Assistance III Project. Professor Klein is funded by the NHS, HEFCE, and GOSH Charity. The work was also supported by funding from an MRC Grant G1001190 and a grant for consumables from the Institute of Child Health.
The study concept was designed by N.K., K.H., D.A., and F.F. Experiments were carried out by F.F., D.A., and K.H., with additional analysis by R.D. The manuscript was drafted by F.F. with additional contributions from each of the authors. The study was overseen by N.K. who is partially funded by Great Ormond Street Hospital Charity.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Massanella M. Negredo E. Perez-Alvarez N, et al. CD4 T-cell hyperactivation and susceptibility to cell death determine poor CD4 T-cell recovery during suppressive HAART. AIDS. 2010;24(7):959–968. doi: 10.1097/QAD.0b013e328337b957. [DOI] [PubMed] [Google Scholar]
- 2.Hunt PW. Martin JN. Sinclair E, et al. T cell activation is associated with lower CD4+ T cell gains in human immunodeficiency virus-infected patients with sustained viral suppression during antiretroviral therapy. J Infect Dis. 2003;187(10):1534–1543. doi: 10.1086/374786. [DOI] [PubMed] [Google Scholar]
- 3.Bofill M. Mocroft A. Lipman M, et al. Increased numbers of primed activated CD8+CD38+CD45RO+ T cells predict the decline of CD4+ T cells in HIV-1-infected patients. AIDS. 1996;10(8):827–834. doi: 10.1097/00002030-199607000-00005. [DOI] [PubMed] [Google Scholar]
- 4.Giorgi JV. Hultin LE. McKeating JA, et al. Shorter survival in advanced human immunodeficiency virus type 1 infection is more closely associated with T lymphocyte activation than with plasma virus burden or virus chemokine coreceptor usage. J Infect Dis. 1999;179(4):859–870. doi: 10.1086/314660. [DOI] [PubMed] [Google Scholar]
- 5.Baker JV. Peng G. Rapkin J, et al. CD4+ count and risk of non-AIDS diseases following initial treatment for HIV infection. AIDS. 2008;22(7):841–848. doi: 10.1097/QAD.0b013e3282f7cb76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker JV. Peng G. Rapkin J, et al. Poor initial CD4+ recovery with antiretroviral therapy prolongs immune depletion and increases risk for AIDS and non-AIDS diseases. J Acquir Immune Defic Syndr. 2008;48(5):541–546. doi: 10.1097/QAI.0b013e31817bebb3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Funderburg NT. Mayne E. Sieg SF, et al. Increased tissue factor expression on circulating monocytes in chronic HIV infection: Relationship to in vivo coagulation and immune activation. Blood. 2010;115(2):161–167. doi: 10.1182/blood-2009-03-210179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Epeldegui M. Vendrame E. Martinez-Maza O. HIV-associated immune dysfunction and viral infection: Role in the pathogenesis of AIDS-related lymphoma. Immunol Res. 2010;48(1–3):72–83. doi: 10.1007/s12026-010-8168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ancuta P. Kamat A. Kunstman KJ, et al. Microbial translocation is associated with increased monocyte activation and dementia in AIDS patients. PLoS One. 2008;3(6):e2516. doi: 10.1371/journal.pone.0002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuller LH. Tracy R. Belloso W, et al. Inflammatory and coagulation biomarkers and mortality in patients with HIV infection. PLoS Med. 2008;5(10):e203. doi: 10.1371/journal.pmed.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marques R. Williams A. Eksmond U, et al. Generalized immune activation as a direct result of activated CD4+ T cell killing. J Biol. 2009;8(10):93. doi: 10.1186/jbiol194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gazzola L. Tincati C. Bellistri GM. Monforte A. Marchetti G. The absence of CD4+ T cell count recovery despite receipt of virologically suppressive highly active antiretroviral therapy: Clinical risk, immunological gaps, and therapeutic options. Clin Infect Dis. 2009;48(3):328–337. doi: 10.1086/595851. [DOI] [PubMed] [Google Scholar]
- 13.De Rossi A. Walker AS. Klein N. De Forni D. King D. Gibb DM. Increased thymic output after initiation of antiretroviral therapy in human immunodeficiency virus type 1-infected children in the Paediatric European Network for Treatment of AIDS (PENTA) 5 Trial. J Infect Dis. 2002;186(3):312–320. doi: 10.1086/341657. [DOI] [PubMed] [Google Scholar]
- 14.Thieblemont N. Weiss L. Sadeghi HM. Estcourt C. Haeffner-Cavaillon N. CD14lowCD16high: A cytokine-producing monocyte subset which expands during human immunodeficiency virus infection. Eur J Immunol. 1995;25(12):3418–3424. doi: 10.1002/eji.1830251232. [DOI] [PubMed] [Google Scholar]
- 15.Anselmi A. Vendrame D. Rampon O. Giaquinto C. Zanchetta M. De Rossi A. Immune reconstitution in human immunodeficiency virus type 1-infected children with different virological responses to anti-retroviral therapy. Clin Exp Immunol. 2007;150(3):442–450. doi: 10.1111/j.1365-2249.2007.03526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veazey RS. DeMaria M. Chalifoux LV, et al. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 1998;280(5362):427–431. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- 17.Marchetti G. Bellistri GM. Borghi E, et al. Microbial translocation is associated with sustained failure in CD4+ T-cell reconstitution in HIV-infected patients on long-term highly active antiretroviral therapy. AIDS. 2008;22(15):2035–2038. doi: 10.1097/QAD.0b013e3283112d29. [DOI] [PubMed] [Google Scholar]
- 18.Jiang W. Lederman MM. Hunt P, et al. Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-treated HIV infection. J Infect Dis. 2009;199(8):1177–1185. doi: 10.1086/597476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nazli A. Chan O. Dobson-Belaire WN, et al. Exposure to HIV-1 directly impairs mucosal epithelial barrier integrity allowing microbial translocation. PLoS Pathog. 2010;6(4):e1000852. doi: 10.1371/journal.ppat.1000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofer U. Speck RF. Disturbance of the gut-associated lymphoid tissue is associated with disease progression in chronic HIV infection. Semin Immunopathol. 2009;31(2):257–266. doi: 10.1007/s00281-009-0158-3. [DOI] [PubMed] [Google Scholar]
- 21.Brenchley JM. Schacker TW. Ruff LE, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200(6):749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papasavvas E. Azzoni L. Foulkes A, et al. Increased microbial translocation in </=180 days old perinatally human immunodeficiency virus-positive infants as compared with human immunodeficiency virus-exposed uninfected infants of similar age. Pediatr Infect Dis J. 2011;30(10):877–882. doi: 10.1097/INF.0b013e31821d141e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butler SL. Valdez H. Westby M, et al. Disease-modifying therapeutic concepts for HIV in the era of highly active antiretroviral therapy. J Acquir Immune Defic Syndr. 2011;58(3):297–303. doi: 10.1097/QAI.0b013e31822ccfcc. [DOI] [PubMed] [Google Scholar]
- 24.Clerici M GA. Rizzardini G. Richter C, et al. Nutritional Intervention with NR100157 Restores Gut Microbiota in HIV-1-infected Adults Not on HAART and Reduces Systemic Immune Activation. 18th Conference on Retroviruses and Opportunistic Infections; Boston. Feb 27, 2011. – March 2, 2011. Abstract number 383. [Google Scholar]
- 25.Byakwaga H. Kelly M. Purcell DF, et al. Intensification of antiretroviral therapy with raltegravir or addition of hyperimmune bovine colostrum in HIV-infected patients with suboptimal CD4+ T-cell response: A randomized controlled trial. J Infect Dis. 2011;204(10):1532–1540. doi: 10.1093/infdis/jir559. [DOI] [PubMed] [Google Scholar]
- 26.Harris KA. Hartley JC. Development of broad-range 16S rDNA PCR for use in the routine diagnostic clinical microbiology service. J Med Microbiol. 2003;52(Pt 8):685–691. doi: 10.1099/jmm.0.05213-0. [DOI] [PubMed] [Google Scholar]
- 27.Kramski M. Gaeguta AJ. Lichtfuss GF, et al. Novel sensitive real-time PCR for quantification of bacterial 16S rRNA genes in plasma of HIV-infected patients as a marker for microbial translocation. J Clin Microbiol. 2011;49(10):3691–3693. doi: 10.1128/JCM.01018-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Millar BC. Xu J. Moore JE. Risk assessment models and contamination management: Implications for broad-range ribosomal DNA PCR as a diagnostic tool in medical bacteriology. J Clin Microbiol. 2002;40(5):1575–1580. doi: 10.1128/JCM.40.5.1575-1580.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferri E. Novati S. Casiraghi M, et al. Plasma levels of bacterial DNA in HIV infection: The limits of quantitative polymerase chain reaction. J Infect Dis. 2010;202(1):176–177. doi: 10.1086/653215. author reply 178. [DOI] [PubMed] [Google Scholar]
- 30.Merlini E. Bai F. Bellistri GM. Tincati C. d'Arminio Monforte A. Marchetti G. Evidence for polymicrobic flora translocating in peripheral blood of HIV-infected patients with poor immune response to antiretroviral therapy. PLoS One. 2011;6(4):e18580. doi: 10.1371/journal.pone.0018580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suau A. Bonnet R. Sutren M, et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999;65(11):4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris KA. Fidler KJ. Hartley JC, et al. Unique case of Helicobacter sp. osteomyelitis in an immunocompetent child diagnosed by broad-range 16S PCR. J Clin Microbiol. 2002;40(8):3100–3103. doi: 10.1128/JCM.40.8.3100-3103.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caporaso JG. Kuczynski J. Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 35.Wang Q. Garrity GM. Tiedje JM. Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeSantis TZ. Hugenholtz P. Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellis CL. Ma ZM. Mann SK, et al. Molecular characterization of stool microbiota in HIV-infected subjects by panbacterial and order-level 16S ribosomal DNA (rDNA) quantification and correlations with immune activation. J Acquir Immune Defic Syndr. 2011;57(5):363–370. doi: 10.1097/QAI.0b013e31821a603c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wallet MA. Rodriguez CA. Yin L, et al. Microbial translocation induces persistent macrophage activation unrelated to HIV-1 levels or T-cell activation following therapy. AIDS. 2010;24(9):1281–1290. doi: 10.1097/QAD.0b013e328339e228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pilakka-Kanthikeel S. Huang S. Fenton T, et al. Increased gut microbial translocation in HIV-infected children persists in virologic responders and virologic failures after antiretroviral therapy. Pediatr Infect Dis J. 2012;31(6):583–591. doi: 10.1097/INF.0b013e31824da0f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cassol E. Malfeld S. Mahasha P, et al. Persistent microbial translocation and immune activation in HIV-1-infected South Africans receiving combination antiretroviral therapy. J Infect Dis. 2010;202(5):723–733. doi: 10.1086/655229. [DOI] [PubMed] [Google Scholar]
- 41.Nowroozalizadeh S. Mansson F. da Silva Z, et al. Microbial translocation correlates with the severity of both HIV-1 and HIV-2 infections. J Infect Dis. 2010;201(8):1150–1154. doi: 10.1086/651430. [DOI] [PubMed] [Google Scholar]
- 42.Sandler NG. Wand H. Roque A, et al. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis. 2011;203(6):780–790. doi: 10.1093/infdis/jiq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Redd AD. Eaton KP. Kong X, et al. C-reactive protein levels increase during HIV-1 disease progression in Rakai, Uganda, despite the absence of microbial translocation. J Acquir Immune Defic Syndr. 2010;54(5):556–559. doi: 10.1097/QAI.0b013e3181e0cdea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Redd AD. Dabitao D. Bream JH, et al. Microbial translocation, the innate cytokine response, and HIV-1 disease progression in Africa. Proc Natl Acad Sci USA. 2009;106(16):6718–6723. doi: 10.1073/pnas.0901983106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Humphrey JH. Child undernutrition, tropical enteropathy, toilets, and handwashing. Lancet. 2009;374(9694):1032–1035. doi: 10.1016/S0140-6736(09)60950-8. [DOI] [PubMed] [Google Scholar]
- 46.Prendergast A. Kelly P. Enteropathies in the developing world: Neglected effects on global health. Am J Trop Med Hyg. 2012;86(5):756–763. doi: 10.4269/ajtmh.2012.11-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Margulies M. Egholm M. Altman WE, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437(7057):376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamady M. Knight R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 2009;19(7):1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Armougom F. Bittar F. Stremler N, et al. Microbial diversity in the sputum of a cystic fibrosis patient studied with 16S rDNA pyrosequencing. Eur J Clin Microbiol Infect Dis. 2009;28(9):1151–1154. doi: 10.1007/s10096-009-0749-x. [DOI] [PubMed] [Google Scholar]
- 50.Green E. Klein N. Hartley J. Diagnosis of sepsis in a paediatric setting comparing blood cultures and the SeptiFast PCR system. Ann Clin Biochem. 2008;45(Suppl 1):32. [Google Scholar]
- 51.Pilakka-Kanthikee S. Royal L. Swaminathan S. Pahwa S. Increased Microbial Translocation in Treatment-Naïve HIV-Infected Children in Chennai, India. 18th Conference on Retroviruses and Opportunistic Infections; Boston. Feb 27, 2011. –April 2, 2011. Abstract 126. [Google Scholar]
- 52.WHO: Children and AIDS, 4th Stock taking report. 2009.
- 53.Heidari S. Mofenson LM. Hobbs CV. Cotton MF. Marlink R. Katabira E. Unresolved antiretroviral treatment management issues in HIV-infected children. J Acquir Immune Defic Syndr. 2012;59(2):161–169. doi: 10.1097/QAI.0b013e3182427029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brenchley JM. Douek DC. Microbial translocation across the GI tract. Annu Rev Immunol. 2012;30:149–173. doi: 10.1146/annurev-immunol-020711-075001. [DOI] [PMC free article] [PubMed] [Google Scholar]