Abstract
Objective
The skeletal muscle of obese humans is characterized by an inability to appropriately respond to alterations in substrate availability. The purpose of the current study was to determine if this metabolic inflexibility with obesity is retained in mitochondria of human skeletal muscle cells raised in culture (HSkMC) and to identify potential mechanisms involved.
Design
Mitochondrial respiration was measured in permeabilized myotubes cultured from lean and obese individuals before and after a 24 h lipid incubation.
Results
Mitochondrial respiration (State 3) in the presence of lipid substrate (palmitoyl carnitine) increased by almost 2-fold after lipid incubation in HSkMC from lean, but not obese subjects, indicative of metabolic inflexibility with obesity. The 24 h lipid incubation increased mitochondrial DNA (mtDNA) copy number in HSkMC from lean subjects by +16% (P<0.05); conversely, mtDNA copy number decreased in myotubes cultured from obese individuals (−13%, P=0.06). When respiration data were normalized to mtDNA copy number and other indices of mitochondrial content (COX-IV protein content and CS activity), the significant treatment effects of lipid incubationpersisted in the lean subjects, suggesting concomitant alterations in mitochondrial function; no similar adjustment was evident in HSkMC from obese individuals.
Conclusions
These data indicate that the skeletal muscle of obese individuals inherently lacks metabolic flexibility in response to lipid exposure, which consists of an inability to increase mitochondrial respiration in the presence of lipid substrate and perhaps by an inability to induce mitochondrial proliferation.
Keywords: Skeletal Muscle, Lipid Oxidation, Mitochondria
INTRODUCTION
Lipid oxidation in skeletal muscle is reported to be reduced in obese and insulin resistant populations (1-4). Likewise, the skeletal muscle of obese humans typically displays a phenotype characterized by intracellular lipid accumulation (5) and reduced overall oxidative capacity (3, 6-8). Some suggest that this reduced oxidative capacity is primarily attributable to a reduction in mitochondrial content with obesity and/or type 2 diabetes (9-11); however, there are also findings indicating that the existing mitochondria are unable to effectively oxidize substrates (8, 12). Emerging evidence suggests that an inability to adapt to a metabolic challenge, such as insulin stimulation or lipid exposure (metabolic flexibility) may be an important aspect of the oxidative impairment in the obese/diabetic phenotype ((3, 13, 14) for review see (15)). More importantly, chronic lipid exposure may exacerbate these phenotypic differences (13, 16).
We have observed that lipid oxidation is reduced in skeletal muscle and primary human skeletal muscle cell cultures (HSkMC) from obese individuals (1, 2, 4, 17) and others have reported reduced lipid oxidation in HSkMC derived from obese, type 2 diabetics (18, 19). Corpeleijn et al. (20) recently demonstrated that 2 days of lipid incubation induced a large increase in 14CO2 production from 14C-labeled oleate in myotubes cultured from lean, insulin sensitive humans, whereas increases in cells cultured from obese, type 2 diabetic humans were 50% lower. Others have shown that a similar lipid-induced increase in HSkMC 14CO2 production is correlated with in vivo metabolism (14). These findings suggest that metabolic inflexibility is preserved in HSkMC, although the effect of obesity alone and the role of mitochondrial function is not yet evident. Providing such data by measuring respiration in intact mitochondria could reveal whether mitochondrial function per se plays a role in the phenotypic differences in oxidative metabolism observed in the skeletal muscle of obese, insulin resistant populations.
In the present study, by permeabilizing the cell membrane of primary human myotubes undergoing lipid pre-incubation (24 h), we measured mitochondrial respiration in skeletal muscle cells from obese and lean humans in the presence of a lipid substrate under various physiological and supra-physiological conditions (basal, ADP-stimulated, and chemically uncoupled), while leaving the mitochondrial structure and membrane intact. Data were also normalized to indices of mitochondrial content to determine if differences were due to mitochondrial function as opposed to overall content. We hypothesized that HSkMC harvested from obese, insulin resistant subjects would display an inability to appropriately regulate mitochondrial respiration in response to lipid pre-incubation.
SUBJECTS AND METHODS
Materials
All chemical reagents and substrates were purchased from Sigma (St. Louis, MO), unless otherwise stated. Fetal bovine serum, heat-inactivated horse serum, gentamicin, 0.05% trypsin EDTA, and Hank’s balanced salt solution were obtained from Invitrogen (Carlsbad, CA). Growth media (GM) and differentiation media (DM) consisted of low glucose (5 mmol/L) Dulbecco’s Modified Eagles Medium from Invitrogen. Tissue culture plates were obtained from Becton Dickinson (Franklin Lakes, NJ). PCR reagents were purchased from Applied Biosystems (Foster City, CA).
Human Subjects
Skeletal muscle was obtained by needle biopsy under local anesthesia (0.1% lidocaine) from the vastus lateralis of 7 lean (BMI <25 kg/m2) and 7 obese (BMI>30 kg/m2) Caucasian males (ages 18-27 y). Participants were free from disease, nonsmokers, and not taking medications known to alter metabolism. All participants had maintained a constant body mass (±2 kg) in the 6 months prior to the biopsy. The protocol was approved by the East Carolina University Policy and Review Committee on Human Research, and informed consent was obtained. Subject characteristics are presented in Table 1.
TABLE 1.
Subject characteristics
| Demographics | Lean (n=7) | Obese (n=7) |
|---|---|---|
| Age (y) | 22.3 ± 0.9 | 21.8 ± 1.1 |
| Stature (cm) | 181.6 ± 2.8 | 182.8 ± 2.0 |
| Mass (kg) | 71.7 ± 3.1 | 130.2 ± 7.9a |
| BMI (kg/m2) | 21.7 ± 0.8 | 39.0 ± 2.0a |
| Glucose (mmol/l) | 4.9 ± 0.2 | 5.0 ± 0.2 |
| Insulin (μIU/l) | 5.3 ± 1.1 | 15.9 ± 1.4a |
| HOMA | 1.2 ± 0.3 | 3.6 ± 0.3a |
| Cholesterol (mg/dl) | 176.7 ± 9.0 | 168.5 ± 18.6 |
| Triglycerides (mg/dl) | 147.9 ± 36.8 | 137.2 ± 35.8 |
Data are presented as mean ± SEM; fasting blood values are presented for glucose, insulin, HOMA, cholesterol and triglyceride.
Indicates significant difference between the lean and obese subjects (P< 0.05)
Primary Human Skeletal Muscle Cell Culture (HSkMC)
Satellite cells were isolated from 50-100 mg of fresh muscle tissue and cultured as previously described to ensure purity (1, 17, 21). For experiments, cells were thawed and subcultured to confluence into T-75 flasks. At 80–90% confluence, differentiation was induced by changing to low-serum DM containing 2% heat-inactivated horse-serum. Starting on day 6 of differentiation, cells were incubated for 24 hours in DM supplemented with either 0.1% BSA (CONTROL) or 100 μM oleate:palmitate (1:1 ratio) bound to 0.1% BSA plus 2 mM carnitine (LIPID), after which the cells were permeabilized with digitonin and the respirometry experiments were performed. Separate aliquots of the same passage number were grown and treated similarly, then harvested for the analysis of protein content and mtDNA copy number.
Respirometry Experiments
Myotubes were lifted from culture flasks with 0.05% trypsin EDTA, centrifuged for 10 minutes at 1000 rpm at room temperature, and re-suspended in warmed DMEM. An aliquot of these cells was used for normalization. Harvested myotubes were centrifuged for an additional 5 minutes at 500 rpm, the supernatant discarded, and cells resuspended in room temperature respiration buffer (130 mM sucrose, 60 mM potassium gluconate, 3 mM magnesium chloride, 10 mM potassium phosphate, 20 mM HEPES, 0.1% BSA; pH 7.4) supplemented with fresh EGTA (1 mM, pH 7.4) and digitonin (7-10 μg/106 cells) and immediately transferred to a respiration chamber (~1-1.5 × 106 cells/2 mL) (Oroboros Oxygraph-2K, Oroboros Instruments Corp, Innsbruck, Austria). Once oxygen concentration flux stabilized, substrates were added as described in Table 2 allowing for flux stabilization between each addition. Side-by-side experiments for CONTROL and LIPID were performed on cells from each subject. The most stable portion of the oxygen concentration slope was determined for each condition and normalized to cell count as in previous cell culture respiration studies (22-24), and to separate indices of mitochondrial content as previously reported in permeabilized muscle fiber experiments (6, 9, 12). Both methods were chosen to best reference our data to existing reports in terms of ‘per cell’ and ‘per mitochondrion’ respiration rates.
Table 2.
Respirometry Protocol
| Step | Substrate | Notation | Concentration |
|---|---|---|---|
| 1 | Digitonin | Baseline | 7-10 μg/106 cells |
| 2 | Palmitoyl Carnitine | PC | 5 μmol/l |
| 2 | Malate | M | 1 mmol/l |
| 3 | ADP | D | 2 mmol/l |
| 4 | Cytochrome C | CytC | 10 μmol/l |
| 5 | Glutamate | G | 2 mmol/l |
| 6 | Succinate | S | 3 mmol/l |
| 7 | Oligomycin | State 2 | 2.5 μg/ml |
| 8 | FCCP | Uncoupled | 2 μmol/l |
|
| |||
| PC, M: PCM4 | |||
| PC, M, D: PCM3 | |||
| PC, M, G, D: PCMG3 | |||
| PC, M, G, S, D: PCMGS3 | |||
| S, M, D: SM3 | |||
Specific substrate additions allowed for measurement of State 4 (substrate only, no ADP added), State 3 (+ 2mM ADP), State 2 (+ oligomycin) and chemically uncoupled (+ carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone [FCCP]) respiration rates. Respiration supported exclusively by lipid (palmitoyl carnitine, PC) under state 4 (PCM4), state 3 (PCM3), or under the convergence of both lipid and carbohydrate substrates through mitochondrial Complexes I & II (PCMGS3) were also measured (See Table 2).
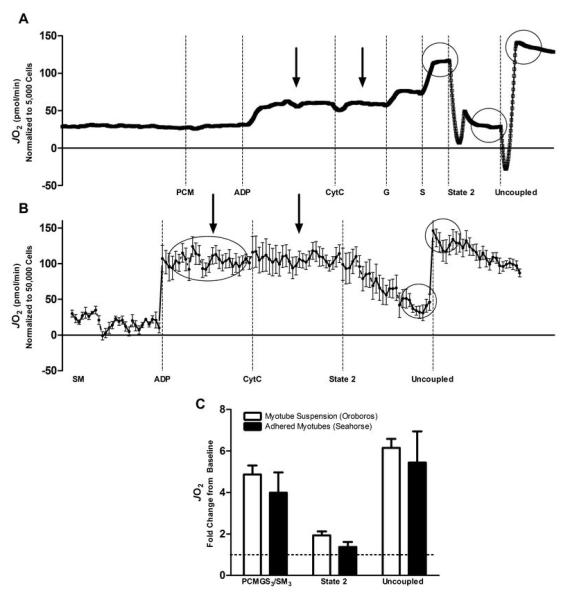
Substrate concentrations were chosen to produce a maximal substrate-specific stimulus. In preliminary experiments, we determined that 5 μM palmitoyl carnitine was sufficient to induce maximal responses in respiration; and titrations up to 100 μM did not induce additional oxygen consumption. Substrates were prepared at pH 7.4 and frozen (−20°C) in single use aliquots. Preliminary experiments were also conducted to assure permeabilized cell integrity. Addition of cytochrome C (10 μM, Table 2, step 4) was performed to determine intactness of the outer mitochondrial membrane and no change in respiration was noted following cytochrome C addition for either group (all data ± 5% from previous measure). To assess possible cell death as a result of trypsinization we measured whole cell viability using trypan blue (all samples were 95-100% viable) and data were normalized to these numbers. To determine the possible ramifications of permeabilizing differentiated myotubes in a suspension, we also repeated similar permeabilization experiments on adhered cells using the same digitonin and respiration buffer parameters with the Seahorse Flux Analyzer XF-24 (Seahorse Bioscience, Billerica, MA). Comparable representative tracings from suspended (n=16 repeats) and adhered (n=6 repeats) permeabilized cell preparations are shown in Figure 1. Assessments were comparable between the two methodologies and trypsinization did not adversely affect cell permeabilization or integrity. Therefore we chose to use the Oroboros for subsequent experiments because of the ability to assess a greater number of parameters and substrate titrations in succession. In addition, the greater cost of the Seahorse Flux Analyzer may limit the use of these methods while the Oroboros, or other Clark electrodes, are typically more accessible.
FIGURE 1.
Representative oxygen consumption tracings from permeabilized primary human myotubes using high resolution respirometry in non-adhered (Panel A; Oroboros Oxygraph 2K) and adhered (Panel B; Seahorse Flux Analyzer XF-24) myotubes. Data for both methodologies are normalized to cell count. Arrows indicate a lack of Cytochrome C response confirming intact mitochondria and circles identify comparable data points for each method. Abbreviations are as presented in Table 2. The relative change over State 4 respiration for the comparable data points is presented in Panel C where the dotted line represents State 4 respiration.
Protein Analysis
Cells were lysed and harvested from control and lipid treated flasks using a 4% SDS solution containing protease and phosphatase inhibitors (Sigma; St. Louis, MO). Total protein concentrations were determined by bicinchoninic acid (BCA) protein assay (Pierce; Rockford, IL). Total protein (20 μg) prepared from cell lysates was separated by either 7.5% or 12.5% SDS-PAGE, transferred to polyvinylidene fluoride (PVDF) membranes (Biorad; Hercules, CA) and then incubated with antibodies diluted in 5% BSA in Tris-buffered saline. Proteins were visualized by horseradish peroxidase conjugated immunoglobulin G. COX-IV and PGC-1α antibodies were purchased from Cell Signaling (Danvers, MA). NRF-1 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Mitochondrial Content
Myotubes were harvested from CONTROL and LIPID using a 10% SDS solution (pH 8.0) containing proteinase K (Qiagen Inc.; Valencia, CA) and rocked overnight at 50°C. DNA was isolated by phenol/chloroform extraction with ethanol precipitation and re-suspended in 10 mM Tris, 0.1 mM EDTA. Mitochondrial DNA copy number was then measured as previously described (17).
For measurement of citrate synthase (CS) activity cells were harvested from control and lipid treated flasks using a 100 mM potassium phosphate solution containing 0.05% BSA. Cells were briefly sonicated and 20 μL of each sample was incubated at 25°C in 200 μL of a freshly-prepared assay solution (72.5 mM Tris, 110 μM 5,5′-Dithiobis(2-nitrobenzoic acid) (DTNB), and 79.5 μM Acetyl CoA; pH 8.3). The reaction was initiated with the addition of 1.75 mM oxaloacetate, and read at 412 nm every 50 seconds for ten cycles. The slope of each sample normalized for a blank control, measurement duration, sample dilution, molar extinction coefficient of DTNB and protein content.
For data normalization to mitochondrial content, three separate indices were used and normalized to baseline oxygen flux (Pre PCM addition). MtDNA copy number was referenced to cell number (mtDNA copy number per cell count). Respiration data were then normalized to this number (PCM3 divided by (mtDNA copy number per cell count)). Respiration data were normalized similarly for COX-IV protein content and CS activity.
Statistical Analyses
Statistical analyses were performed using PASW Statistics 18® Software (SPSS Inc. Chicago, IL) on raw or log-transformed data. For participant characteristics, independent t-tests were performed. Two- way ANOVA with repeated measures was used to compare differences between control and lipid-incubated cells cultured from lean and obese subjects. All data met assumptions of sphericity and homogeneity of variance. Data are presented as the mean ± SEM.
RESULTS
Participant Characteristics
Participant characteristics are presented in Table 1. By design, the obese subjects were heavier and most had a BMI indicative of class II or III obesity (35-39.9 kg/m2 or >40 kg/m2, respectively). Fasting blood glucose and triglyceride values did not differ between the groups, although insulin levels and HOMA values were higher in the obese subjects.
Respirometry
For all measurements, respiration chamber oxygen concentrations were well above values required for normal respiration (~130 mmHg to ~180 mmHg). In Figure 2, mean oxygen flux data are presented for each group following normalization to cell count. Baseline oxygen flux was not different from PCM4 condition (Figure 2). Under CONTROL respiration rates PCM3 did not differ in HSkMC from lean and obese subjects (see Table 2 for notation details). However, State 3 respiration supported by lipid only (PCM3) increased by approximately 2-fold in HSkMC from lean subjects in response to the 24 hr lipid incubation (LIPID). There was a much less robust response in cells from obese subjects (+13%; interaction P<0.05; Figure 2). No increase in State 4 respiration (PCM4) was evident with lipid treatment (LIPID) nor was there any difference between the lean and obese groups. State 2 respiration remained unchanged with the 24 h lipid incubation and was similar between groups (3.83±0.75 and 4.16±1.01 in CONTROL and LIPID in lean; and 3.05±0.49 and 3.31±0.56 in CONTROL and LIPID in obese). Significant interactions were not noted for any other measurement (PCMGS3, Uncoupled; data not shown).
FIGURE 2.
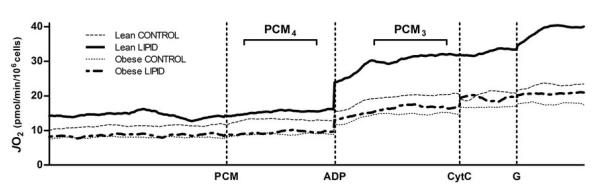
Mean State 4 and State 3 respiration rates in permeabilized myotubes cultured from lean (n=7) and obese (n=7) subjects in the presence of PCM, ADP, CytC, and G (see Table 2 for abbreviation details). Cells were pre-incubated for 24 hours in either CONTROL or LIPID treatments. Data are presented as the mean of the oxygen flux per second normalized to total cell count. There was a significant body type × lipid treatment interaction for PCM3 (P<0.05).
Indices of Mitochondrial Content
As presented in Figure 3A, myotubes cultured from lean and obese individuals responded differently to the 24 h lipid treatment in terms of mtDNA copy number (P<0.05), such that copy number increased in cells cultured from lean subjects and decreased in cells cultured from obese subjects (+16% and −13% in lean and obese, respectively). There was a similar trend for COX-IV protein content (P=0.09) (Figure 3B). Citrate synthase activity (Figure 3C) was not altered.
FIGURE 3.
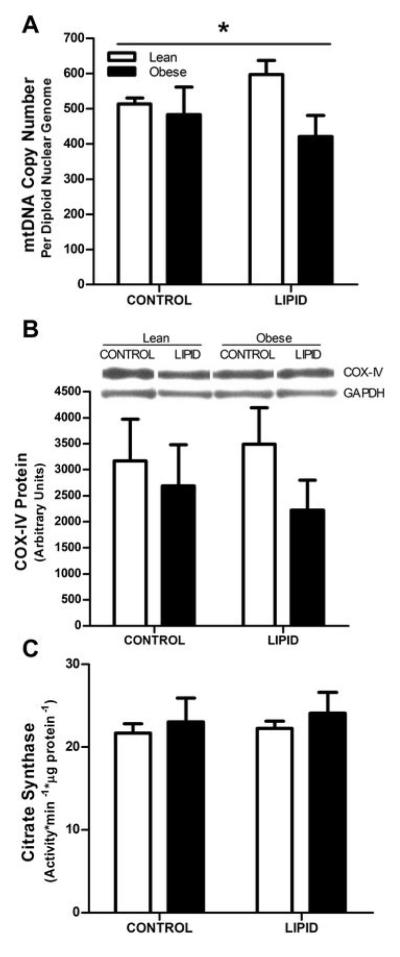
Indices of mitochondrial content in cultured myotubes from lean (open bars) and obese (filled bars) subjects under CONTROL and LIPID treatment conditions. Panel A: mtDNA copy number (n=7 and n=7 in lean and obese, respectively). Panel B: COX-IV protein content (n=5 and n=6 in lean and obese, respectively; P=0.09). Panel C: CS activity (n=7 and n=5 in lean and obese, respectively; n.s.). All values are expressed as mean ± SEM.
* Indicates a significant (P<0.05) interaction effect where the lean and obese subjects responded differently to the LIPID treatment.
PGC-1α protein content was measured in an attempt to discern mechanisms linked with the possible alterations in mitochondrial content with lipid exposure (Figure 3). PGC-1α protein content was not different between cells from lean and obese individuals and was unchanged with lipid incubation (−1% and +3% in cells from lean and obese, respectively; not significant). Changes in nuclear respiratory factor-1 (NRF-1) protein content with lipid incubation were also not significant (+25% and +18% in cells from lean and obese, respectively; not significant) and did not differ between the groups.
Respiration Normalized to Mitochondrial Content
To account for possible differences in respiration relative to mitochondrial content, State 3 respiration rates (PCM3) were normalized to the three indices of mitochondrial content as presented in Figure 4. A significant LIPID treatment effect was noted for data normalized to mtDNA (P<0.05) which appeared to be mainly driven by the increase in PCM3/mtDNA in cells cultured from the lean donors (Figure 4A), as suggested by a trend for a significant interaction effect (P=0.08). No interaction or LIPID treatment effects were noted when data were normalized to COX-IV protein content (Figure 4B, P=0.7). When data were normalized to CS activity there was a significant LIPID treatment effect (P<0.05) and a trend for a significant interaction (P=0.07), again suggesting that the increases in respiration with LIPID were mainly driven by changes in the cells cultured from lean humans (Figure 4C). Together, these data suggest that HSkMC from lean humans exhibit some degree of metabolic flexibility in response to 24 hours of mixed-lipid exposure in terms of increasing respiration on ‘per mitochondria’ basis. This flexibility is dampened or not evident in the cells from the obese humans.
FIGURE 4.
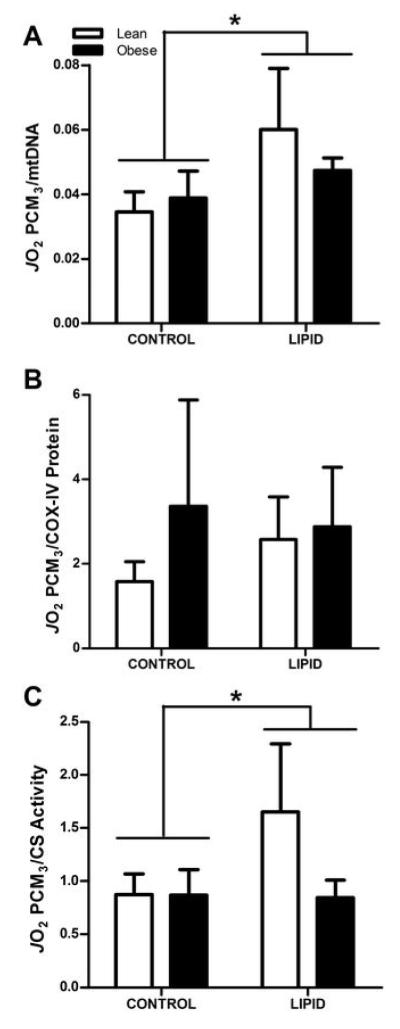
State 3 respiration with palmitoyl carnitine as the substrate normalized to indices of mitochondrial content in myotubes cultured from lean (open bars) and obese (filled bars) subjects under CONTROL and LIPID conditions. Panel A: PCM3 JO2 normalized to mtDNA copy number (n=7 and n=7 in lean and obese, respectively). Panel B: PCM3 JO2 normalized to COX-IV protein content (n=5 and n=5 in lean and obese, respectively). Panel C: PCM3 JO2 normalized to CS activity (n=7 and n=4 in lean and obese, respectively). All values are expressed as mean ± SEM.
* Indicates significant LIPID treatment effect (P<0.05).
Flux Control Ratios
The coupled control ration (CCR) was calculated (uncoupled respiration divided by State 3 respiration through Complexes I & II (PCMGS3)) to determine any potential limitation in the phosphorylation system relative to the overall capacity of the electron transport system (25). CCR was not different in cells from lean and obese subjects and did not change as a result of LIPID (Figure 5A). All CCR values were >1.0, indicating that the electron transport capacity was not limiting oxygen consumption in either control or lipid-incubated cells in either group. A substrate control ratio (SCR) was also calculated for PCM (State 3 respiration (PCM3) divided by State 4 respiration (PCM4)) as an index of the metabolic coupling of mitochondrial respiration and oxidative phosphorylation (25). There was a significant interaction between cells cultured from lean and obese subjects (P<0.05, Figure 5B) where cells from lean subjects increased in response to lipid incubation. These treatment effects are accounted for by increased PCM3 rather than decreased PCM4, which indicates that mitochondrial coupling was not negatively affected by LIPID.
FIGURE 5.
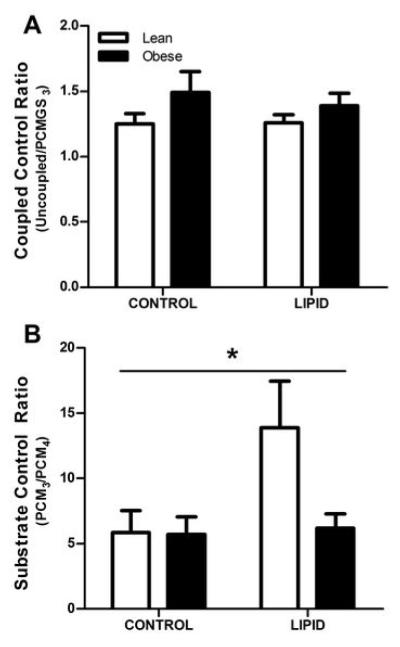
Coupled control ratio (uncoupled respiration divided by PCMGS3; Panel A) and substrate control ratio (PCM3/PCM4; Panel B) in myotubes cultured from lean (open bars) and obese (filled bars) subjects in CONTROL and LIPID treatments. All values are expressed as mean ± SEM.
* Indicates a significant (P<0.05) interaction effect where the lean and obese subjects responded differently to the LIPID treatment.
DISCUSSION
The main finding of the present study was that skeletal muscle cells derived from obese humans failed to upregulate mitochondrial respiration in response to a 24 h lipid incubation (LIPID), while myotubes cultured from lean humans increased State 3 respiration using lipid as a substrate (Figure 2). This impairment in the ability to increase lipid oxidation to a similar extent to that observed in the HSkMC from the lean subjects persisted in the obese subjects when respiration was normalized to mtDNA and CS activity (Figure 4). Cells from the lean subjects exhibited increased mtDNA copy number following LIPID treatment, though a similar response was not evident in HSkMC from obese subjects. A similar trend was noted for COX-IV protein content (P=0.09) (Figure 3). The current data therefore indicate that there is a metabolic inflexibility in the skeletal muscle of obese individuals in terms of increasing mitochondrial lipid oxidation in response to lipid exposure. A novel aspect of these findings is that this inflexibility appears to result from an impaired ability to intrinsically enhance the capacity for lipid oxidation in response to increased lipid availability and may also result from an impaired ability to increase mitochondrial content.
With HSkMC the processes of cellular proliferation and differentiation take place outside of the in-vivo environment; expression of a given characteristic in HSkMC has thus been interpreted as indicating that innate genetic and/or epigenetic traits are involved (19, 26, 27). In-vivo studies indicate that lean individuals can respond to an increase in dietary fat intake by increasing whole-body lipid oxidation (28, 29) while obese and previously obese subjects do not respond in a similar manner (30, 31). We have recently shown that this metabolic inflexibility occurs at the level of skeletal muscle gene expression following 5 days of a 65% fat diet in a cohort similar to that presented here (13). To our knowledge, relatively few investigators have examined if this metabolic inflexibility with obesity persists in HSkMC. Myotubes cultured from obese, type 2 diabetics exhibit increased lipid oxidation in response to short-term palmitate incubation (4 hours) with evidence of reverse Randle-Cycle activity (32); though a more prolonged period of lipid incubation (2 days) in a similar diabetic cohort produced a relatively lower increase in total 14CO2 production from 14C-labeled oleate compared to lean, non-diabetic subjects, suggestive of blunted metabolic flexibility (20). The present findings indicate that this inability to respond appropriately (i.e. to the same extent as lean subjects) to lipid presence in HSkMC does not require the overt expression of type 2 diabetes and is present in obese individuals with insulin resistance (Figure 2). In addition, previous measurements of lipid metabolism have only been obtained in whole cells as a result of CO2 production where substrate utilization depends on many factors independent of mitochondrial oxidation (1, 14, 17-20, 33, 34). For example, cellular uptake, intracellular transport, lipid storage capacity, and mitochondrial lipid uptake all play a role in the ability of the mitochondrion to effectively oxidize lipids. Using permeabilized cells and controlled concentrations of palmitoyl carnitine we are able to directly measure the capacity of the mitochondrion to oxidize lipids. In addition, ADP-stimulated oxidation rates represent physiological respiration, rather than basal or uncoupled metabolism as reported previously (1, 14, 17-20, 33, 34). The present findings thus reveal that metabolic inflexibility is evident in mitochondria of skeletal muscle cells derived from insulin resistant, obese, non-diabetic individuals when energy demand is increased to a physiological range (State 3 respiration; Figure 2).
Using similar permeabilization techniques in fresh skeletal muscle fibers, others have identified impaired State 3 respiration in tissue from obese, type 2 diabetic patients in the presence of various substrates such as malate + pyruvate, glutamate and/or succinate (6, 8, 9, 12). However, no decrement in State 3 respiration with type 2 diabetes was evident with lipid substrate (palmitoyl carnitine + malate [PCM]) (8, 12). Recently we also reported that State 3 respiration in the presence of PCM was similar in permeabilized fibers from lean and obese individuals possessing subject characteristics very similar to those in the present study (6). While these findings of normal PCM3 respiration in permeabilized fibers appear to be at odds with the HSkMC data presented here (Figure 2), it is critical to realize that it is the CONTROL condition in the cell culture experiments that most closely resembles the assessments performed in muscle fibers from diabetic and obese individuals. In light of this comparison, the present HSkMC data corroborate previous reports in permeabilized fibers indicating no difference in PCM3 and extend those findings to indicate that phenotypic differences in skeletal muscle mitochondrial lipid oxidation with obesity are evident in response to a lipid substrate challenge.
Prolonged lipid exposure can lead to lipid accumulation within the cell (18, 19, 33) as a consequence of nutrient supply outpacing metabolic demand. Excess dietary lipid increases skeletal muscle lipid content in both humans (35) and rodents (36); and lipid incubation exacerbates phenotypic differences in lipid metabolism where HSkMC from obese individuals express more CD36 and take up more lipid than cells from lean humans (33). Given the present data, it is plausible that impaired skeletal muscle lipid oxidation may also contribute to the excess intramyocellular lipid content observed in obesity in vivo. In excess, cellular lipids are prone to peroxidation by reactive oxygen species (37) and the ensuing oxidative stress can induce mitochondrial damage, resulting in a cycle of further reductions in substrate oxidation due to reduced mitochondrial numbers and/or altered function.
Mitochondrial biogenesis is a complex process requiring the induction of numerous nuclear and mitochondrial proteins. PGC-1α is a transcriptional co-activator that activates multiple transcription factors, such as NRF-1, which in turn increase the expression of gene networks involved with and contributing to fatty acid oxidation (38). In HSkMC, relatively long-term (20 hr) lipid incubation (0.5 mM oleate) has been reported to increase PGC-1α mRNA content (~2.5-fold) and mitochondrial activity by ~60% (mitochondrial dehydrogenase) (39). We have reported a similar 50% increase in PGC-1a mRNA content in skeletal muscle from lean humans following a 5d high fat diet, though obese, insulin resistant individuals did not respond similarly (~30% decrease) (13). Therefore it is not surprising that in the current study, despite a lower lipid concentration (100 μM), we also observed an increase in mtDNA copy number in cells cultured from lean individuals, while there was a reduction in mtDNA in HSkMC from obese subjects (Figure 3). This pattern in mtDNA copy number tended to be similar for COX-IV protein content but was not evident in CS activity measures (Figure 3). Despite increases in mitochondrial mass there were no concomitant changes in PGC-1α or NRF-1 protein content in cells from either the lean or obese individuals. Further investigation of lipid-induced mitochondrial biogenesis and/or mitoptosis in skeletal muscle cells cultured from obese individuals is needed in order to discern the potential mechanisms involved.
These conflicting data for mitochondrial content (Figure 3) indicate that findings are not consistent among various accepted indices of mitochondrial content and, for this reason, we chose to normalize the respiration data to all three indices to account for the impact of mitochondrial content on the observed increase in State 3 mitochondrial respiration (Figure 2). When respiration data were normalized in this manner it was evident that the LIPID treatment effects observed (Figure 4) were driven by increased mitochondrial respiration in the HSkMC from lean subjects. The severely blunted response observed in HSkMC cultured from the obese subjects is similar to the blunted metabolic flexibility observed by others in HSkMC in response to lipid incubation in whole cells (19, 20, 34); and indicates that the metabolic perturbation observed with obesity in respect to lipid oxidation may not entirely be explained by potential differences in mitochondrial content.
Previous studies on the role of mitochondrial content and function with respect to lipid oxidation in skeletal muscle of obese or insulin resistant humans have produced conflicting results. Many investigators have found that lipid oxidation in muscle is depressed in type 2 diabetic patients (8, 9, 12). This difference, however, was no longer evident when oxidation was normalized to mitochondrial content (9, 11) implying that reduced mitochondrial mass was responsible for any differences in the substrate utilization rate. Others have maintained that there is inherent mitochondrial dysfunction in skeletal muscle of individuals with type 2 diabetes (8, 12). Clouding this issue is that there is no consensus within the scientific community with regard to a standard index of mitochondrial content. In this context we utilized several indices of mitochondrial content and consistently observed a contribution from mitochondrial respiration to the metabolic inflexibility induced by lipid incubation in HSkMC with obesity.
In conclusion, the current data utilizing permeabilized HSkMC indicate that the skeletal muscle of obese individuals inherently lacks metabolic flexibility in terms of increasing lipid oxidation in response to lipid exposure (24 h lipid incubation). A novel aspect of our findings is that this inflexibility appears to result from a dampening of the ability to intrinsically enhance the capacity for lipid oxidation relative to mitochondrial content and, perhaps, in the ability to increase mitochondrial mass in response to increased lipid availability.
ACKNOWLEDGEMENTS
Supported by NIH grants AG025205 and DK56112 (to J.A.H.) and DK073488 (to P.D.N.).
Footnotes
DISCLOSURE The authors declare no conflict of interest.
REFERENCES
- 1.Hulver MW, Berggren JR, Carper MJ, Miyazaki M, Ntambi JM, Hoffman EP, et al. Elevated stearoyl-CoA desaturase-1 expression in skeletal muscle contributes to abnormal fatty acid partitioning in obese humans. Cell Metab. 2005 Oct;2(4):251–61. doi: 10.1016/j.cmet.2005.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hulver MW, Berggren JR, Cortright RN, Dudek RW, Thompson RP, Pories WJ, et al. Skeletal muscle lipid metabolism with obesity. Am J Physiol Endocrinol Metab. 2003 Apr;284(4):E741–7. doi: 10.1152/ajpendo.00514.2002. [DOI] [PubMed] [Google Scholar]
- 3.Kelley DE, Goodpaster B, Wing RR, Simoneau JA. Skeletal muscle fatty acid metabolism in association with insulin resistance, obesity, and weight loss. Am J Physiol. 1999 Dec;277(6 Pt 1):E1130–41. doi: 10.1152/ajpendo.1999.277.6.E1130. [DOI] [PubMed] [Google Scholar]
- 4.Kim JY, Hickner RC, Cortright RL, Dohm GL, Houmard JA. Lipid oxidation is reduced in obese human skeletal muscle. Am J Physiol Endocrinol Metab. 2000 Nov;279(5):E1039–44. doi: 10.1152/ajpendo.2000.279.5.E1039. [DOI] [PubMed] [Google Scholar]
- 5.Goodpaster BH, Theriault R, Watkins SC, Kelley DE. Intramuscular lipid content is increased in obesity and decreased by weight loss. Metabolism. 2000 Apr;49(4):467–72. doi: 10.1016/s0026-0495(00)80010-4. [DOI] [PubMed] [Google Scholar]
- 6.Anderson EJ, Lustig ME, Boyle KE, Woodlief TL, Kane DA, Lin CT, et al. Mitochondrial H2O2 emission and cellular redox state link excess fat intake to insulin resistance in both rodents and humans. J Clin Invest. 2009 Feb 2; doi: 10.1172/JCI37048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes. 2002 Oct;51(10):2944–50. doi: 10.2337/diabetes.51.10.2944. [DOI] [PubMed] [Google Scholar]
- 8.Mogensen M, Sahlin K, Fernstrom M, Glintborg D, Vind BF, Beck-Nielsen H, et al. Mitochondrial respiration is decreased in skeletal muscle of patients with type 2 diabetes. Diabetes. 2007 Jun;56(6):1592–9. doi: 10.2337/db06-0981. [DOI] [PubMed] [Google Scholar]
- 9.Boushel R, Gnaiger E, Schjerling P, Skovbro M, Kraunsoe R, Dela F. Patients with type 2 diabetes have normal mitochondrial function in skeletal muscle. Diabetologia. 2007 Apr;50(4):790–6. doi: 10.1007/s00125-007-0594-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holloway GP, Thrush AB, Heigenhauser GJ, Tandon NN, Dyck DJ, Bonen A, et al. Skeletal muscle mitochondrial FAT/CD36 content and palmitate oxidation are not decreased in obese women. Am J Physiol Endocrinol Metab. 2007 Jun;292(6):E1782–9. doi: 10.1152/ajpendo.00639.2006. [DOI] [PubMed] [Google Scholar]
- 11.Lefort N, Glancy B, Bowen B, Willis WT, Bailowitz Z, De Filippis EA, et al. Increased reactive oxygen species production and lower abundance of complex I subunits and carnitine palmitoyltransferase 1B protein despite normal mitochondrial respiration in insulin-resistant human skeletal muscle. Diabetes. 2010 Oct;59(10):2444–52. doi: 10.2337/db10-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phielix E, Schrauwen-Hinderling VB, Mensink M, Lenaers E, Meex R, Hoeks J, et al. Lower intrinsic ADP-stimulated mitochondrial respiration underlies in vivo mitochondrial dysfunction in muscle of male type 2 diabetic patients. Diabetes. 2008 Nov;57(11):2943–9. doi: 10.2337/db08-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyle KE, Canham JP, Consitt LA, Zheng D, Koves TR, Gavin TP, et al. A high-fat diet elicits differential responses in genes coordinating oxidative metabolism in skeletal muscle of lean and obese individuals. J Clin Endocrinol Metab. 2011 Mar;96(3):775–81. doi: 10.1210/jc.2010-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ukropcova B, McNeil M, Sereda O, de Jonge L, Xie H, Bray GA, et al. Dynamic changes in fat oxidation in human primary myocytes mirror metabolic characteristics of the donor. J Clin Invest. 2005 Jul;115(7):1934–41. doi: 10.1172/JCI24332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelley DE, Mandarino LJ. Fuel selection in human skeletal muscle in insulin resistance: a reexamination. Diabetes. 2000 May;49(5):677–83. doi: 10.2337/diabetes.49.5.677. [DOI] [PubMed] [Google Scholar]
- 16.Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008 Jan;7(1):45–56. doi: 10.1016/j.cmet.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 17.Consitt LA, Bell JA, Koves TR, Muoio DM, Hulver MW, Haynie KR, et al. Peroxisome proliferator-activated receptor-gamma coactivator-1alpha overexpression increases lipid oxidation in myocytes from extremely obese individuals. Diabetes. 2010 Jun;59(6):1407–15. doi: 10.2337/db09-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaster M. Reduced lipid oxidation in myotubes established from obese and type 2 diabetic subjects. Biochem Biophys Res Commun. 2009 May 15;382(4):766–70. doi: 10.1016/j.bbrc.2009.03.102. [DOI] [PubMed] [Google Scholar]
- 19.Wensaas AJ, Rustan AC, Just M, Berge RK, Drevon CA, Gaster M. Fatty acid incubation of myotubes from humans with type 2 diabetes leads to enhanced release of beta-oxidation products because of impaired fatty acid oxidation: effects of tetradecylthioacetic acid and eicosapentaenoic acid. Diabetes. 2009 Mar;58(3):527–35. doi: 10.2337/db08-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corpeleijn E, Hessvik NP, Bakke SS, Levin K, Blaak EE, Thoresen GH, et al. Oxidation of intramyocellular lipids is dependent on mitochondrial function and the availability of extracellular fatty acids. Am J Physiol Endocrinol Metab. 2010 Jul;299(1):E14–22. doi: 10.1152/ajpendo.00187.2010. [DOI] [PubMed] [Google Scholar]
- 21.Berggren JR, Tanner CJ, Houmard JA. Primary cell cultures in the study of human muscle metabolism. Exerc Sport Sci Rev. 2007 Apr;35(2):56–61. doi: 10.1249/JES.0b013e31803eae63. [DOI] [PubMed] [Google Scholar]
- 22.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009 Apr 23;458(7241):1056–60. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008 Nov;8(5):347–58. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 24.Philp A, Perez-Schindler J, Green C, Hamilton DL, Baar K. Pyruvate suppresses PGC1alpha expression and substrate utilization despite increased respiratory chain content in C2C12 myotubes. Am J Physiol Cell Physiol. 2010 Aug;299(2):C240–50. doi: 10.1152/ajpcell.00438.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gnaiger E. Capacity of oxidative phosphorylation in human skeletal muscle: new perspectives of mitochondrial physiology. Int J Biochem Cell Biol. 2009 Oct;41(10):1837–45. doi: 10.1016/j.biocel.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 26.Baar K. Epigenetic control of skeletal muscle fibre type. Acta Physiol (Oxf) 2010 Aug;199(4):477–87. doi: 10.1111/j.1748-1716.2010.02121.x. [DOI] [PubMed] [Google Scholar]
- 27.Gaster M, Petersen I, Hojlund K, Poulsen P, Beck-Nielsen H. The diabetic phenotype is conserved in myotubes established from diabetic subjects: evidence for primary defects in glucose transport and glycogen synthase activity. Diabetes. 2002 Apr;51(4):921–7. doi: 10.2337/diabetes.51.4.921. [DOI] [PubMed] [Google Scholar]
- 28.Cameron-Smith D, Burke LM, Angus DJ, Tunstall RJ, Cox GR, Bonen A, et al. A short-term, high-fat diet up-regulates lipid metabolism and gene expression in human skeletal muscle. Am J Clin Nutr. 2003 Feb;77(2):313–8. doi: 10.1093/ajcn/77.2.313. [DOI] [PubMed] [Google Scholar]
- 29.Peters SJ, Harris RA, Wu P, Pehleman TL, Heigenhauser GJ, Spriet LL. Human skeletal muscle PDH kinase activity and isoform expression during a 3-day high-fat/low-carbohydrate diet. Am J Physiol Endocrinol Metab. 2001 Dec;281(6):E1151–8. doi: 10.1152/ajpendo.2001.281.6.E1151. [DOI] [PubMed] [Google Scholar]
- 30.Astrup A, Buemann B, Christensen NJ, Toubro S. Failure to increase lipid oxidation in response to increasing dietary fat content in formerly obese women. Am J Physiol. 1994 Apr;266(4 Pt 1):E592–9. doi: 10.1152/ajpendo.1994.266.4.E592. [DOI] [PubMed] [Google Scholar]
- 31.Thomas CD, Peters JC, Reed GW, Abumrad NN, Sun M, Hill JO. Nutrient balance and energy expenditure during ad libitum feeding of high-fat and high-carbohydrate diets in humans. Am J Clin Nutr. 1992 May;55(5):934–42. doi: 10.1093/ajcn/55.5.934. [DOI] [PubMed] [Google Scholar]
- 32.Gaster M. Metabolic flexibility is conserved in diabetic myotubes. J Lipid Res. 2007 Jan;48(1):207–17. doi: 10.1194/jlr.M600319-JLR200. [DOI] [PubMed] [Google Scholar]
- 33.Bell JA, Reed MA, Consitt LA, Martin OJ, Haynie KR, Hulver MW, et al. Lipid partitioning, incomplete fatty acid oxidation, and insulin signal transduction in primary human muscle cells: effects of severe obesity, fatty acid incubation, and fatty acid translocase/CD36 overexpression. J Clin Endocrinol Metab. 2010 Jul;95(7):3400–10. doi: 10.1210/jc.2009-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitzmann M, Lantier L, Hebrard S, Mercier J, Foretz M, Aguer C. Abnormal metabolism flexibility in response to high palmitate concentrations in myotubes derived from obese type 2 diabetic patients. Biochim Biophys Acta. 2011 Apr;1812(4):423–30. doi: 10.1016/j.bbadis.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 35.Schrauwen-Hinderling VB, Kooi ME, Hesselink MK, Moonen-Kornips E, Schaart G, Mustard KJ, et al. Intramyocellular lipid content and molecular adaptations in response to a 1-week high-fat diet. Obes Res. 2005 Dec;13(12):2088–94. doi: 10.1038/oby.2005.259. [DOI] [PubMed] [Google Scholar]
- 36.Koves TR, Li P, An J, Akimoto T, Slentz D, Ilkayeva O, et al. Peroxisome proliferator-activated receptor-gamma co-activator 1alpha-mediated metabolic remodeling of skeletal myocytes mimics exercise training and reverses lipid-induced mitochondrial inefficiency. J Biol Chem. 2005 Sep 30;280(39):33588–98. doi: 10.1074/jbc.M507621200. [DOI] [PubMed] [Google Scholar]
- 37.Schrauwen P. High-fat diet, muscular lipotoxicity and insulin resistance. Proc Nutr Soc. 2007 Feb;66(1):33–41. doi: 10.1017/S0029665107005277. [DOI] [PubMed] [Google Scholar]
- 38.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999 Jul 9;98(1):115–24. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 39.Staiger H, Staiger K, Haas C, Weisser M, Machicao F, Haring HU. Fatty acid-induced differential regulation of the genes encoding peroxisome proliferator-activated receptor-gamma coactivator-1alpha and -1beta in human skeletal muscle cells that have been differentiated in vitro. Diabetologia. 2005 Oct;48(10):2115–8. doi: 10.1007/s00125-005-1895-z. [DOI] [PubMed] [Google Scholar]