Abstract
S-(2-Hydroxy-3,4-epoxybutyl)glutathione (DEB-GSH conjugate) is formed from the reaction of 1,2:3,4-diepoxybutane (DEB) with glutathione (GSH), and the conjugate is considerably more mutagenic than several other butadiene-derived epoxides—including DEB—in Salmonella typhimurium TA1535 (Cho, S-H. et al., Chem. Res. Toxicol. 23, 1544–1546 (2010)). We previously identified six DNA adducts in the reaction of the DEB-GSH conjugate with nucleosides and calf thymus DNA and two DNA adducts in livers of mice and rats treated with DEB (Cho, S-H., and Guengerich, F.P., Chem. Res. Toxicol. 25, 706–712 (2012)). In order to define the role of GSH conjugation in 1,3-butadiene (BD) metabolism and characterize the mechanism of GSH transferase (GST)-enhanced mutagenicity of DEB, mutation spectra of BD and its metabolites in the absence and presence of GST/GSH and mouse liver microsomes were compared in the rpoB gene of Escherichia coli TRG8. The presence of GST considerably enhanced mutations. The mutation spectra derived from the DEB-GSH conjugate, the DEB/GST/GSH system, and the BD/mouse liver microsomes/GST/GSH system matched each other and were different from those derived from the other systems devoid of GSH. The major adducts in E. coli TRG8 cells treated with the DEB/GST/GSH system, the BD/mouse liver microsomes/GST/GSH system, or the DEB-GSH conjugate were S-[4-(N7-guanyl)-2,3-dihydroxybutyl]GSH, S-[4-(N3-adenyl)-2,3-dihydroxybutyl]GSH, and S-[4-(N6-deoxyadenosinyl)-2,3-dihydroxybutyl]GSH, indicating the presence of the GSH-containing DNA adducts in the systems. These results, along with the strong enhancement of mutagenicity by GST in this system, indicate the relevance of these GSH-containing DNA adducts.
INTRODUCTION
1,3-Butadiene (BD) is an important industrial chemical used in the production of synthetic plastics and rubber, as well as an environmental pollutant found in cigarette smoke and automobile exhaust.1,2 BD is also classified as a “probable human carcinogen” by the International Agency for Research on Cancer (IARC)3 based on the increased lymphatic and hematopoietic cancer risk in occupationally exposed humans4–6 and its carcinogenic effects in laboratory animals, e.g. B6C3F1 mice and Sprague-Dawley rats. The former species is of particular interest regarding tumors and their relevance to human cancer risk, in that tumors are linked to the increased formation of 1,2:3,4-diepoxybutane (DEB) and other DNA-reactive metabolites, due in part to species differences in epoxide hydrolase activity.7–9
Although BD is relatively unreactive itself, it is converted to DNA reactive metabolites by P450 enzymes. The first epoxidation yields 3,4-epoxy-1-butene (EB). EB can then be hydrolyzed to 1-butene-3,4-diol or undergo a second epoxidation to yield DEB, both of which can be further transformed to 3,4-epoxy-1,2-butanediol.7,10,11 EB, DEB, and 3,4-epoxy-1,2-butanediol are capable of alkylating DNA and proposed to be responsible for the mutagenic properties of BD.12,13 Among the three epoxide metabolites of BD, DEB is the most mutagenic and, because of its bis electrophilic properties, also yields DNA-DNA and DNA-protein crosslinks. 14–17
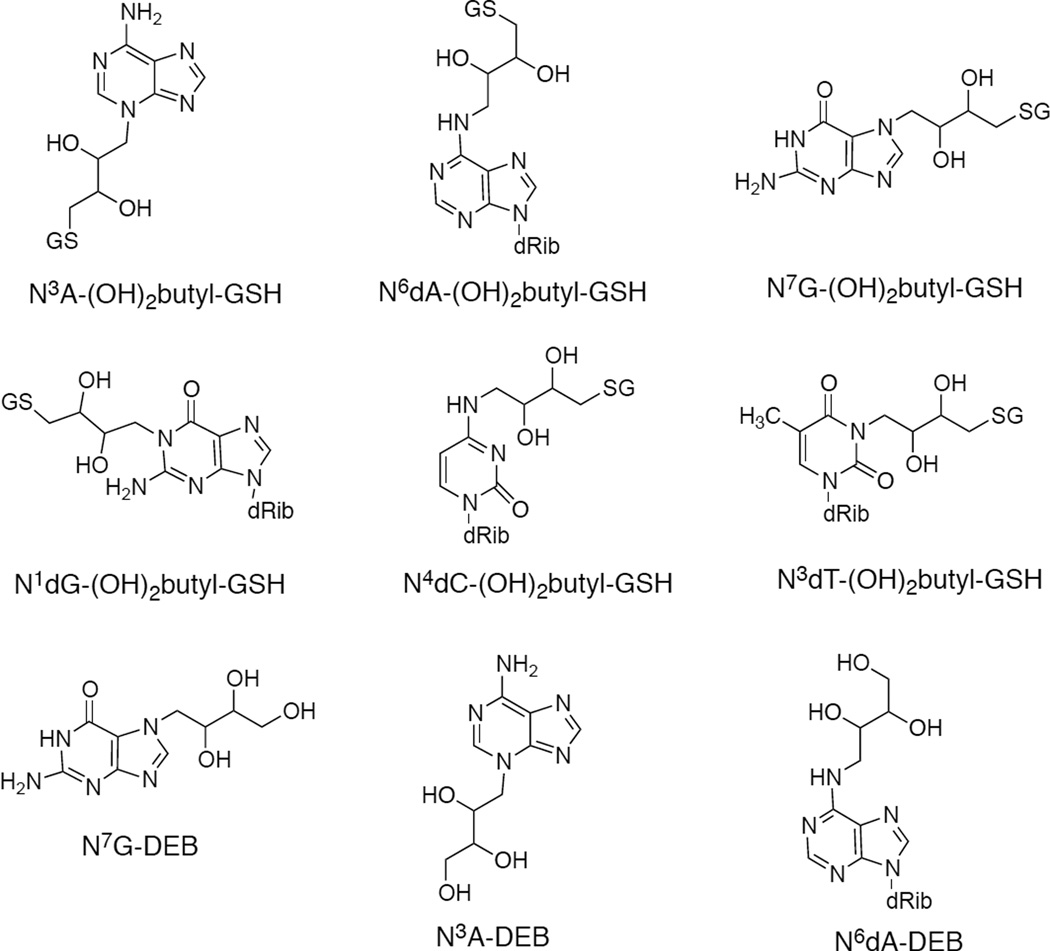
Although GSH conjugation is generally considered to be a detoxication process in the metabolism of xenobiotic chemicals,18 we previously reported enhanced mutagenicity of DEB in Salmonella typhimurium TA1535 due to the cellular expression of rat GSH S-transferase (GST) 5-519 or human GST T1-1.20 Significantly higher mutagenicity of the synthetic S-(2-hydroxy-3,4-epoxybutyl)GSH (DEB-GSH) conjugate was measured compared to several other butadiene-derived epoxides, including DEB, in S. typhimurium TA1535.21 Six DNA adducts—S-[4-(N3-adenyl)-2,3-dihydroxybutyl]GSH (N3A-(OH)2butyl-GSH), S-[4-(N6-deoxyadenosinyl)-2,3-dihydroxybutyl]GSH (N6dA-(OH)2butyl-GSH), S-[4-(N7-guanyl)-2,3-dihydroxybutyl]GSH (N7G-(OH)2butyl-GSH), S-[4-(N1-deoxyguanosinyl)-2,3-dihydroxybutyl]GSH (N1dG-(OH)2butyl-GSH), S-[4-(N4-deoxycytidinyl)-2,3-dihydroxybutyl]GSH (N4dC-(OH)2butyl-GSH,), and S-[4-(N3-thymidinyl)-2,3-dihydroxybutyl]GSH (N3dT-(OH)2butyl-GSH)—were identified in the reaction of the DEB-GSH conjugate with nucleosides and calf thymus DNA, and two of the adducts—N6dA-(OH)2butyl-GSH and N7G-(OH)2butyl-GSH—were identified and quantitated in vivo in the livers of mice and rats treated with DEB.22 These results suggest that a GSH conjugate of DEB reacts with DNA and is a major mutagen with biological activity as great or greater than other BD oxidation products, including DEB, and therefore expected to contribute to the carcinogenicity of DEB.
In the present work on the definition of the role of GSH conjugation in BD metabolism and characterization of the mechanism of GST-enhanced mutagenicity of DEB, the mutation spectrum produced by the DEB-GSH conjugate was compared with that of BD and its metabolites in the absence and presence of GST (plus GSH) and mouse liver microsomes (containing an NADPH-generating system) in the rpoB gene of Escherichia coli TRG8. Six major DNA adducts formed from DEB-GSH conjugate and three “direct” DEB DNA adducts— N7-(2,3,4-trihydroxybutyl)guanine (N7G-DEB), N3-(2,3,4-trihydroxybutyl)adenine (N3A-DEB), and N6-(2,3,4-trihydroxybutyl)adenine (N6A-DEB)—were analyzed in E. coli TRG8 cells treated with DEB, DEB/GST/GSH, DEB-GSH conjugate, or BD/mouse liver microsomes/GST/GSH under these conditions. The major adducts identified in the presence of GST were the guanyl N7 and adenyl N3 and N6 adducts previously described.22 The mutation spectra formed in all cases with GST—plus the DEB-GSH conjugate—were similar to each other but distinct from the DEB spectrum. These results, along with the strong enhancement of mutagenicity by GST in this system, indicate the mutagenic significance of the GSH-containing DNA adducts.
EXPERIMENTAL PROCEDURES
Materials
BD, EB, DEB (CAS 1464-53-5, racemic DEB), GSH, a commercial mixture of equine liver GSTs, male mouse (CD-1) liver microsomes (catalog M9441), rifampicin, and enzymes for digestion were purchased from Sigma Chemical Co. (St. Louis, MO). E. coli TRG8 cells were provided by Prof. A. E. Pegg, Pennsylvania Sate Univ., Hershey, PA. Phusion High-Fidelity DNA polymerase was purchased from New England Biolabs Inc. (Ipswich, MA). The DEB-GSH conjugate was enzymatically synthesized and purified as described previously.21 The six major DNA adducts (N3A-(OH)2butyl-GSH, N6dA-(OH)2butyl-GSH, N7A-(OH)2butyl-GSH, N1dG-(OH)2butyl-GSH, N4dC-(OH)2butyl-GSH, and N3dT-(OH)2butyl-GSH) formed with the DEB-GSH conjugate, the three “direct” DEB DNA adducts (N7G-DEB, N3A-DEB, and N6A-DEB), and the internal standards (N6dA-(OH)2butyl-[glycine-13C2,15N]-GSH, N7G-(OH)2butyl-[glycine-13C2,15N]-GSH, and 18O-N7G-(OH)3butane) were synthesized and purified as described previously.22
Measurement of Cell Survival and Mutations in E. coli
Cell survival and mutation assays were performed as previously described.23,24 TRG8 cells were grown in 50 mL of Luria-Bertani (LB) media at 37 °C until the OD600 of bacterial cultures reached 0.5. Cells were pelleted by centrifugation and resuspended in 2 mL of M9 salts (90 mM Na2HPO4, 25 mM KH2PO4, 10 mM NaCl, and 20 mM NH4Cl). Aliquots of cells (0.5 mL) were then exposed to various concentrations of BD (1–30%, v/v, in air, using a dessicator) or its metabolites (0.01–0.3 mM), EB, DEB, and DEB-GSH conjugate, in the absence and presence of 0.1 µM GST (plus 1 mM GSH) and CD-1 male mouse liver microsomes (1 mg protein mL−1, containing an NADPH-generating system)25 at 37 °C for 90 min. Aliquots of cells (0.5 mL) were also exposed to the solvent 2% DMSO (v/v, for generating spontaneous mutations) at 37 °C for 90 min. The cells were washed with M9 salts and resuspended in 0.5 mL of M9 salts. In order to derive rpoB gene mutants, cells (100 µL) were plated on LB media plates supplemented with 100 µg mL−1 rifampicin. In addition, cells (100 µL) were diluted 1:104–106 fold and plated on LB media plates lacking rifampicin to determine the number of viable cells. The plated cells were grown in a 37 °C incubator for 36 h until discrete colonies appeared. The mutation frequency of the rpoB gene in TRG8 cells was expressed as the number of rpoB mutants per 108 survivors.
Analysis of Rifampicin-Resistant Mutants
Rifampicin-resistant cell clones from rifampicin-containing plates were picked, suspended in 100 µL of deionized water, and mixed vigorously with a vortex device. Aliquots (2 µL) of these suspensions were used as the DNA template in PCR. A section of the rpoB gene was amplified by PCR using 5’-TGGCCTGGTACGTGTAGA-3’ (forward primer), 5’-AACCAGCGGCTTATCAGC-3’ (reverse primer), and Phusion High-Fidelity DNA polymerase. The PCR cycling conditions were as follows: initial melting (98 °C, 4 min), 35 cycles of denaturation (98 °C, 30 s), annealing (52 °C, 30 s), and extension (72 °C, 30 s) followed by a last extension step at 72 °C for 5 min. The size of the DNA fragment (about 703 base pairs) was verified by electrophoresis in a 0.1% (w/v) agarose gel in 40 mM Tris-acetate buffer (pH 7.6) containing 1 mM EDTA (150 V). The PCR products were purified using the QiaQuick PCR purification kit (Qiagen, Hilden, Germany) and submitted for sequence analysis in the Vanderbilt DNA Sequencing Facility.
Statistical Analysis
For comparison of mutation spectra induced by DEB-GSH conjugate with those induced by BD or its metabolites in the absence and presence of GST and mouse liver microsomes, Fisher’s exact test was used,26–28 with significance concluded at P < 0.05.
DNA Adduct Analysis in E. coli
TRG8 cells were grown in 50 mL of LB media to an OD600 of 0.5 at 37 °C. Cells were pelleted and resuspended in 2 mL of M9 salts. Aliquots of cells (0.5 mL) were treated with DEB (0.3 mM), DEB (0.3 mM)/GST/GSH, DEB-GSH conjugate (0.3 mM), or BD (20%, in air, v/v)/mouse liver microsomes/GST/GSH at 37 °C for 90 min. The cells were washed with M9 salts and resuspended in 0.5 mL of M9 salts. DNA was isolated using a Wizard plus Minipreps DNA purification System (Promega, Madison, WI), followed by thermal or acid-catalyzed hydrolysis or enzymatic digestion.22 The reactions were filtered through 3K MWCO Centricon filters (3 kDa cut-off, Millipore Corp., Billercia, MA) and spiked with synthesized N6dA-(OH)2butyl-[glycine-13C2,15N]-GSH, N7G-(OH)2butyl-[glycine-13C2,15N]-GSH, and [18O]-N7G-(OH)3butane. The resulting reactions were analyzed by LC-MS/MS. LC-MS/MS analysis was performed using a Waters Acquity UPLC system (Waters, Milford, MA) interfaced to a Thermo-Finnigan LTQ mass spectrometer (ThermoElectron, Sunnyvale, CA) equipped with an ESI source.22 Chromatographic separation was achieved with a Waters Acquity UPLC BEH C18 octadecylsilane column (2.1 mm × 100 mm, 1.7 µm). LC conditions were as follows: Solvent A was 0.1% CH3CO2H in H2O (v/v) and solvent B was 0.1% CH3CO2H in CH3CN (v/v). The following gradient program (v/v) was used with a flow rate of 300 µL min−1: the gradient started with 5% B (v/v), increased to 15% B (v/v) at 2 min, to 30% B (v/v) at 6 min, and held at 30% B (v/v) for 1 min. The column was re-equilibrated for 3 min with 5% B (v/v). The temperature of the column was maintained at 40 °C. The MS conditions were as follows: ion spray voltage, 4.5 kV; capillary voltage, 20 V; capillary temperature, 350 °C; and tube lens voltage, 40 V.
RESULTS
Toxicity and Mutagenicity of BD and Its Metabolites in E. coli
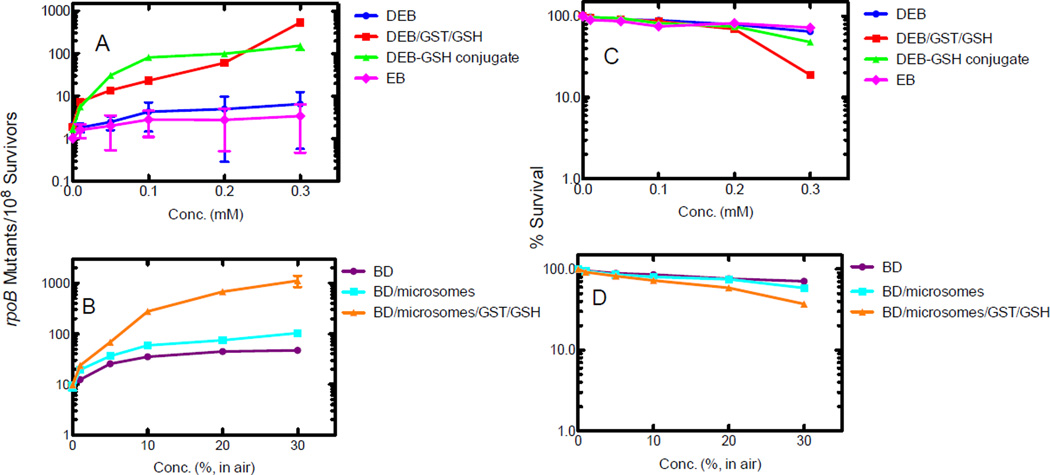
To further define the role of GSH conjugation in BD metabolism, we examined the toxicity and mutagenicity of BD and its metabolites in the absence and presence of GST/GSH and mouse liver microsomes (containing an NADPH-generating system) in E. coli TRG8 cells (Figure 1). Mouse liver was used as a source of P450 because the conversion of BD to DEB is greater than rat.7–9 GSTs have been shown to be rather similar in their abilities to conjugate DEB,22 and therefore a commercial mixture of equine GSTs was used.
Figure 1.
Effects of BD, EB, DEB, and the DEB-GSH conjugate on mutation (A, B) and survival (C, D) in E. coli TRG8 cells with or without GST/GSH and mouse liver microsomes. Rifampicin resistant mutants were obtained on LB plates containing 100 µg mL−1 rifampicin. The mutation frequency of the rpoB gene was corrected with the number of survivors following the treatment. Each point represents the mean of duplicate experiments.
The frequency of rifampicin-resistant mutants produced by BD with mouse liver microsomes or DEB was significantly increased by the presence of GST (Figure 1A, 1B). The frequency of rifampicin-resistant mutants produced by the DEB-GSH conjugate was also higher than with only DBE (Figure 1A). The cell survival observed with 30% BD (in air, v/v) and mouse liver microsomes (59 to 37%) or 0.3 mM DEB (65 to 19%) was decreased by the presence of GST (Figure 1C and 1D), but the presence of GST alone did not significantly enhance cytotoxicity (results not shown). The cytotoxicity induced by the DEB-GSH conjugate was slightly lower than that induced by DEB plus GST (Figure 1C).
Comparisons of Mutation Spectra in the rpoB gene
Comparisons of mutation spectra in the rpoB gene of E. coli TRG8 were performed with the following systems: DEB, DEB/GST/GSH, DEB-GSH conjugate, EB, BD, BD/mouse liver microsomes, and BD/mouse liver microsomes/GST/GSH (Table 1 and Table 2). Mutations at sites within this locus (Table 1) and a summary comparing relative proportions of each class of base-pair substitutions (Table 2) are presented. The spectrum of background mutants (spontaneous mutants) in TRG8 cells did not reveal any specific induction of G:C to T:A or of G:C to A:T mutations and was similar to that reported in a large previous study.29
Table 1.
Frequency and Site of Mutations in the rpoB Gene in E. coli Cells Produced by BD (20%, in air, v/v) and Its Metabolites (0.3 mM)
| base change | codon change |
amino acid | mutation | number of mutations |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| spontaneous (n = 32) |
DEB (n = 43) |
DEB + GST (+ GSH) (n =45) |
DEB-GSH conjugate (n = 48) |
EB (n = 44) |
BD (n = 40) |
BD + microsomes (n = 42) |
BD + microsomes + GST (+ GSH) (n = 41) |
||||
| G:C to T:A | GAC | 516 | D → Y | 0 | 6 | 4 | 3 | 6 | 0 | 5 | 3 |
| TAC | |||||||||||
| CAG | 513 | Q → K | 0 | 3 | 2 | 2 | 0 | 0 | 1 | 2 | |
| AAG | |||||||||||
| CGT | 529 | R → S | 0 | 4 | 8 | 8 | 2 | 0 | 4 | 6 | |
| AGT | |||||||||||
| TCC | 531 | S → Y | 0 | 1 | 3 | 2 | 3 | 0 | 3 | 1 | |
| TAC | |||||||||||
| CGT | 529 | R → L | 0 | 0 | 3 | 4 | 0 | 2 | 0 | 2 | |
| CTT | |||||||||||
| CAC | 526 | H → N | 0 | 0 | 0 | 0 | 4 | 6 | 1 | 1 | |
| AAC | |||||||||||
| TCT | 512 | S → Y | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | |
| TAT | |||||||||||
| G:C to A:T | CAC | 526 | H → Y | 4 | 7 | 2 | 2 | 3 | 3 | 6 | 2 |
| TAC | |||||||||||
| CCT | 564 | P → L | 0 | 4 | 2 | 1 | 5 | 4 | 2 | 1 | |
| CTT | |||||||||||
| CGT | 529 | R → C | 0 | 1 | 0 | 0 | 9 | 0 | 3 | 2 | |
| TGT | |||||||||||
| TCC | 531 | S → F | 0 | 5 | 2 | 2 | 4 | 7 | 5 | 1 | |
| TTC | |||||||||||
| GAC | 516 | D → N | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 2 | |
| AAC | |||||||||||
| A:T to T:A | CAG | 513 | G → L | 2 | 5 | 3 | 1 | 2 | 6 | 5 | 2 |
| CTG | |||||||||||
| GAC | 516 | D → V | 0 | 2 | 1 | 1 | 4 | 2 | 4 | 2 | |
| GTC | |||||||||||
| ATC | 572 | I → F | 0 | 4 | 1 | 2 | 0 | 4 | 3 | 1 | |
| TTC | |||||||||||
| A:T to C:G | ATC | 572 | I → L | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CTC | |||||||||||
| CAG | 513 | Q → P | 0 | 1 | 6 | 8 | 2 | 0 | 0 | 7 | |
| CCG | |||||||||||
| A:T to G:C | GAC | 516 | D → G | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GGC | |||||||||||
| CAC | 526 | H → R | 0 | 0 | 4 | 6 | 0 | 0 | 0 | 4 | |
| CGC | |||||||||||
| CAG | 513 | Q → R | 0 | 0 | 3 | 4 | 0 | 0 | 0 | 2 | |
| CGG | |||||||||||
Table 2.
Summary of Mutations in the rpoB Gene in E. coli Cells Produced by BD (20%, in air, v/v) and Its Metabolites (0.3 mM)
| base change | number of mutations (%) |
|||||||
|---|---|---|---|---|---|---|---|---|
| spontaneous | DEB | DEB + GST (+ GSH) |
DEB-GSH conjugate |
EB | BD | BD + microsomes | BD + microsomes + GST (+ GSH) |
|
| G:C to T:A | 0 (0) | 14 (32.6) | 20 (44.4) | 19 (39.6) | 15 (34.1) | 14 (35.0) | 14 (33.3) | 15 (36.6) |
| G:C to A:T | 4 (12.5) | 17 (39.5) | 8 (17.8) | 7 (14.6) | 21 (47.7) | 14 (35.0) | 16 (38.0) | 8 (19.5) |
| A:T to T:A | 2 (6.3) | 11 (25.6) | 4 (8.9) | 4 (8.3) | 6 (13.6) | 12 (30.0) | 12 (28.6) | 5 (12.2) |
| A:T to C:G | 12 (37.5) | 1 (2.3) | 6 (13.3) | 8 (16.7) | 2 (4.6) | 0 (0) | 0 (0) | 7 (17.1) |
| A:T to G:C | 14 (43.7) | 0 (0) | 7 (15.6) | 10 (20.8) | 0 (0) | 0 (0) | 0 (0) | 6 (14.6) |
| Total | 32 (100) | 43 (100) | 45 (100) | 48 (100) | 44 (100) | 40 (100) | 42 (100) | 41 (100) |
For comparison of mutation spectra, statistical analysis using Fisher’s exact test was performed with segregation of base changes (Table 2, Table 3). For G:C to T:A transversions, the mutation spectrum derived from the DEB-GSH conjugate (40% transversions) was similar to the others (DEB, 33%; DEB/GST/GSH, 44%; EB, 34%; BD, 35%; BD/mouse liver microsomes, 33%; BD/mouse liver microsomes/GST/GSH, 37%), with the exception of the spontaneous mutations (0%). However, for G:C to A:T transitions, the mutation spectrum derived from the DEB-GSH conjugate (15% transversions) was similar to those derived in the spontaneous assay (13%), the DEB/GST/GSH system (18%), and the BD/mouse liver microsomes/GST/GSH system (20%). The mutation spectrum derived from the DEB-GSH conjugate was also similar with those derived from the spontaneous assay, the DEB/GST/GSH system, the EB system, and the BD/mouse liver microsomes/GST/GSH system for A:T to T:A transversions (DEB-GSH conjugate, 8% transversions; spontaneous, 6%; DEB/GST/GSH, 9%; EB, 14%; BD/mouse liver microsomes/GST/GSH, 12%) and A:T to C:G transversions (DEB-GSH conjugate, 17% transversions; spontaneous, 38%; DEB/GST/GSH, 13%; EB, 5%; BD/mouse liver microsomes/GST/GSH, 18%). For A:T to G:C transitions, the mutation spectrum derived from the DEB-GSH conjugate (21% transitions) was similar only with those derived from the DEB/GST/GSH system (16% transitions), and the BD/mouse liver microsomes/GST/GSH system (15%). Specifically, mutations derived from the DEB-GSH conjugate, the DEB/GST/GSH system, or the BD/mouse liver microsomes/GST/GSH system in A:T to C:G transversions or A:T to G:C transitions were higher than those derived from the other systems.
Table 3.
Comparison of Mutation Spectra in the rpoB Gene in E. coli Cells Produced by BD (20%, in air, v/v) and Its Metabolites (0.3 mM)
| base change |
P-value |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| spontaneous | DEB | DEB + GST (+ GSH) |
DEB-GSH conjugate |
EB | BD | BD + microsomes | BD + microsomes + GST (+ GSH) |
||
| G:C to T:A | spontaneous | - | 0.0002 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 |
| DEB | 0.0002 | - | NSa | NS | NS | NS | NS | NS | |
| DEB + GST (+ GSH) | 0.0001 | NS | - | NS | NS | NS | NS | NS | |
| DEB-GSH | 0.0001 | NS | NS | - | NS | NS | NS | NS | |
| EB | 0.0001 | NS | NS | NS | - | NS | NS | NS | |
| BD | 0.0001 | NS | NS | NS | NS | - | NS | NS | |
| BD + microsomes | 0.0001 | NS | NS | NS | NS | NS | - | NS | |
| BD + microsomes + GST (+ GSH) | 0.0001 | NS | NS | NS | NS | NS | NS | - | |
| G:C to A:T | spontaneous | - | 0.01 | NS | NS | 0.001 | 0.03 | 0.018 | NS |
| DEB | 0.01 | - | 0.03 | 0.009 | NS | NS | NS | NS | |
| DEB + GST (+ GSH) | NS | 0.03 | - | NS | 0.003 | NS | NS | NS | |
| DEB-GSH | NS | 0.009 | NS | - | 0.0007 | 0.04 | 0.015 | NS | |
| EB | 0.001 | NS | 0.003 | 0.0007 | - | NS | NS | 0.011 | |
| BD | 0.03 | NS | NS | 0.04 | NS | - | NS | NS | |
| BD + microsomes | 0.018 | NS | NS | 0.015 | NS | NS | - | NS | |
| BD + microsomes + GST (+ GSH) | NS | NS | NS | NS | 0.011 | NS | NS | - | |
| A:T to T:A | spontaneous | - | 0.034 | NS | NS | NS | 0.015 | 0.018 | NS |
| DEB | 0.034 | - | 0.048 | 0.045 | NS | NS | NS | NS | |
| DEB + GST (+ GSH) | NS | 0.048 | - | NS | NS | 0.024 | 0.026 | NS | |
| DEB-GSH | NS | 0.045 | NS | - | NS | 0.012 | 0.015 | NS | |
| EB | NS | NS | NS | NS | - | NS | NS | NS | |
| BD | 0.015 | NS | 0.024 | 0.012 | NS | - | NS | NS | |
| BD + microsomes | 0.018 | NS | 0.026 | 0.015 | NS | NS | - | NS | |
| BD + microsomes + GST (+ GSH) | NS | NS | NS | NS | NS | NS | NS | - | |
| A:T to C:G | spontaneous | - | 0.0001 | 0.027 | NS | 0.0005 | 0.0001 | 0.0001 | NS |
| DEB | 0.0001 | - | NS | 0.03 | NS | NS | NS | 0.028 | |
| DEB + GST (+ GSH) | 0.027 | NS | - | NS | NS | 0.027 | 0.027 | NS | |
| DEB-GSH | NS | 0.03 | NS | - | NS | 0.007 | 0.007 | NS | |
| EB | 0.0005 | NS | NS | NS | - | NS | NS | NS | |
| BD | 0.0001 | NS | 0.027 | 0.007 | NS | - | NS | 0.012 | |
| BD + microsomes | 0.0001 | NS | 0.027 | 0.007 | NS | NS | - | 0.005 | |
| BD + microsomes + GST (+ GSH) | NS | 0.028 | NS | NS | NS | 0.012 | 0.005 | - | |
| A:T to G:C | spontaneous | - | 0.0001 | 0.01 | 0.045 | 0.0001 | 0.0001 | 0.0001 | 0.0008 |
| DEB | 0.0001 | - | 0.01 | 0.001 | NS | NS | NS | 0.01 | |
| DEB + GST (+ GSH) | 0.01 | 0.01 | - | NS | 0.01 | 0.013 | 0.012 | NS | |
| DEB-GSH | 0.045 | 0.001 | NS | - | 0.0013 | 0.0016 | 0.0014 | NS | |
| EB | 0.0001 | NS | 0.01 | 0.0013 | - | NS | NS | 0.01 | |
| BD | 0.0001 | NS | 0.013 | 0.0016 | NS | - | NS | 0.026 | |
| BD + microsomes | 0.0001 | NS | 0.012 | 0.0014 | NS | NS | - | 0.012 | |
| BD + microsomes + GST (+ GSH) | 0.0008 | 0.01 | NS | NS | 0.01 | 0.026 | 0.012 | - | |
NS, not significant (P-value ≥ 0.05).
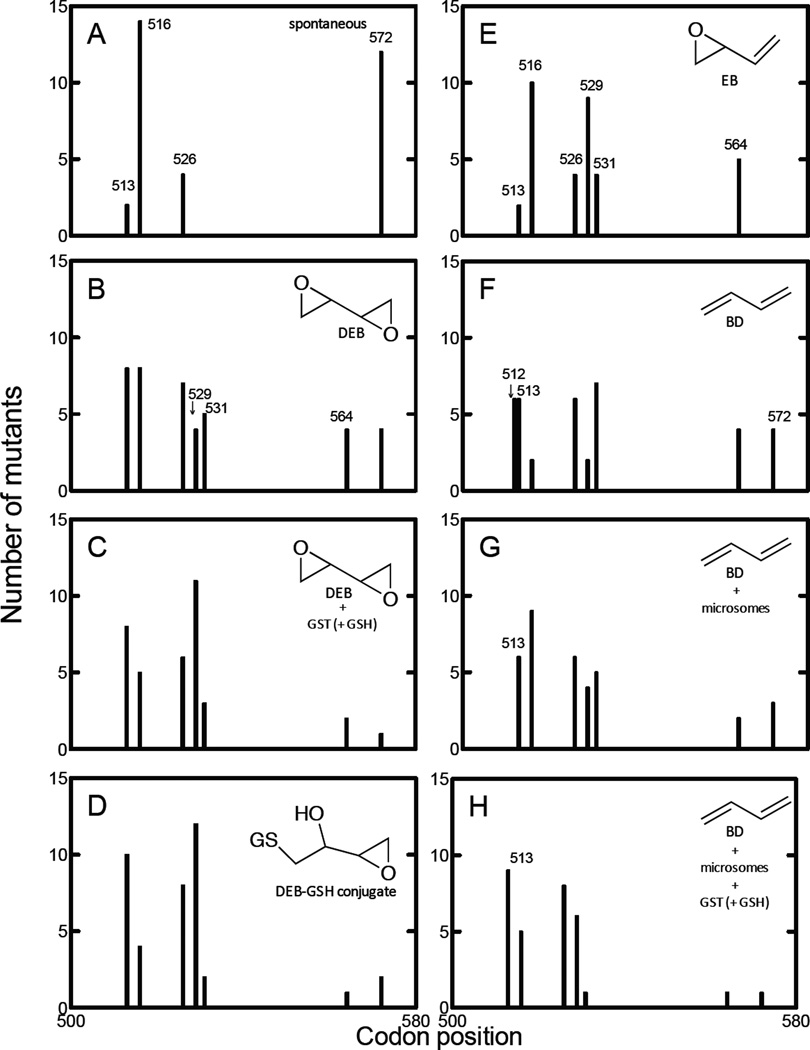
Mutations in the rpoB gene (amino acids 508–574) were compared in E. coli TRG8 cells exposed to DMSO (spontaneous), DEB, DEB/GST/GSH, the DEB-GSH conjugate, EB, BD, BD/mouse liver microsomes, or the BD/mouse liver microsomes/GST/GSH system (Figure 2). Mutations derived from the DEB-GSH conjugate revealed similar patterns with those derived from the DEB/GST/GSH system or the BD/mouse liver microsomes/GST/GSH system (although the codon 529 mutants derived from the BD/mouse liver microsomes/GST/GSH system were fewer than derived from the DEB-GSH conjugate or the DEB/GST/GSH system).
Figure 2.
Mutations in the rpoB gene in E. coli cells exposed to DMSO (spontaneous, A), DEB (B), DEB/GST/GSH (C), DEB-GSH conjugate (D), EB (E), BD (F), BD/mouse liver microsomes (G), and BD/mouse liver microsomes/GST/GSH (H).
Through comparison of mutation spectra, the spectra of mutations derived from the DEB-GSH conjugate, the DEB/GST/GSH system, and the BD/mouse liver microsomes/GST/GSH system matched each other but were quite different from those derived from the other systems.
Quantitation of DNA Adducts in E. coli
For consideration of the significance of the GSH adducts, six major DNA adducts formed from the DEB-GSH conjugate and three “direct” DEB DNA adducts were measured in E. coli TRG8 cells treated with the DEB, the DEB/GST/GSH system, the DEB-GSH conjugate, or the BD/mouse liver microsomes/GST/GSH system using LC-MS/MS methods reported previously (Scheme 1, Table 4).22 In cells treated with DEB, only three “direct” DEB DNA adducts—N7G-DEB (26–32 adducts per 104 bases), N3A-DEB (13 adducts per 104 bases), and N6A-DEB (7–8 adducts per 104 bases)—were measured. In contrast, six major DNA adducts formed from the DEB-GSH conjugate were determined in cells treated with the DEB-GSH conjugate. Among the six DNA adducts, N7G-(OH)2butyl-GSH adducts (6–7 adducts per 105 bases) and N6dA-(OH)2butyl-GSH (5–7 adducts per 105 bases) were highest, followed by N3A-(OH)2butyl-GSH (3–4 adducts per 105 bases), and N3dT-(OH)2butyl-GSH (1.1–1.5 adducts per 105 bases), N4dC-(OH)2butyl-GSH (0.85–1.1 adducts per 105 bases), and N1dG-(OH)2butyl-GSH (0.32–0.57 adducts per 105 bases). In cells treated with the DEB/GST/GSH system or the BD/mouse liver microsomes/GST/GSH system, some (three to six) of the six major DNA adducts formed from the DEB-GSH conjugate and three “direct” DEB DNA adducts were identified.
Scheme 1.
DNA Adducts Measured in This Study
Table 4.
Quantitative Analysis of DNA Adducts in E. coli Cells Treated with DEB, DEB/GST/GSH, DEB-GSH Conjugate, or BD/Mouse Liver Microsomes/GST/GSH
| adduct | adducts per 105 bases |
|||
|---|---|---|---|---|
| concentration 0.3 mM | concentration 20%, in air (v/v) | |||
| DEB | DEB + GST (+ GSH) | DEB-GSH conjugate | BD + mouse liver microsomes + GST (+ GSH) | |
| (N = 2)a | (N = 2) | (N = 2) | (N = 2) | |
| N3A-(OH)2butyl-GSH | NDb | 0.93 ± 0.12 | 3.74 ± 0.59 | 0.10 ± 0.01 |
| N6dA-(OH)2butyl-GSH | ND | 1.55 ± 0.42 | 6.01 ± 0.94 | 0.21 ± 0.04 |
| N7G-(OH)2butyl-GSH | ND | 1.73 ± 0.26 | 6.84 ± 0.69 | 0.23 ± 0.14 |
| N1dG-(OH)2butyl-GSH | ND | ND | 0.45 ± 0.18 | ND |
| N4dC-(OH)2butyl-GSH | ND | ND | 0.98 ± 0.18 | ND |
| N3dT-(OH)2butyl-GSH | ND | 0.62 ± 0.15 | 1.33 ± 0.30 | ND |
| N7G-DEB | 291 ± 46 | 305 ± 35 | ND | 12.2 ± 1.6 |
| N3A-DEB | 130 ± 6 | 135 ± 11 | ND | 6.9 ± 1.1 |
| N6A-DEB | 71 ± 4 | 68 ± 22 | ND | 4.2 ± 1.4 |
Means of duplicate experiments, ± range.
ND, not detected (limit of detection = 0.03 adducts/107 bases).
DISCUSSION
The mutagenicity of the DEB-GSH conjugate system, the DEB/GST/GSH system, and the BD/mouse liver microsomes/GST/GSH system were considerably higher than that of BD or any of its metabolites observed in the absence of GST (Figure 1). The spectra of mutations derived from the DEB-GSH conjugate system, the DEB/GST/GSH system, and the BD/mouse liver microsomes/GST/GSH system matched each other and were quite different from the mutation spectra derived from the other systems (Figure 2, Tables 1–3). The major adducts in E. coli TRG8 cells treated with the DEB/GST/GSH system, the BD/mouse liver microsomes/GST/GSH system, or the DEB-GSH conjugate were N7G-(OH)2butyl-GSH, N6dA-(OH)2butyl-GSH, and N3A-(OH)2butyl-GSH (Scheme 1, Table 4), indicating the presence of these GSH-containing DNA adducts in the systems used for mutational analysis. These results should be considered regarding the role of GSH conjugation in BD metabolism and characterization of the mechanism of GST-enhanced mutagenicity of DEB.
DEB is the most potent mutagenic oxidative metabolite of BD.12,13 Because of its bifunctional nature, it gives rise to DNA-DNA and DNA-protein cross-links.16,17 DNA-protein cross-links can be deleterious to cells because they are bulky, helix-distorting lesions that block the binding and progression of protein complexes and interfere with normal DNA metabolism.30,31 However, there is no evidence that DEB-derived DNA-DNA or DNA-protein cross-links generate mutations aside from the case of these formed with O6-alkylguanine DNA-alkyltransferase.24,26
In order to define the role of GSH conjugation in BD metabolism, the mutagenicity of BD and its metabolites (in the absence and presence of GST/GSH and mouse liver microsomes containing an NADPH-generating system) was measured in E. coli TRG8 cells (Figure 1). E. coli TRG8 cells were previously used for investigation of O6-alkylguanine DNA-alkyltransferase-mediated toxicity of DEB.26 The expression of the DNA repair protein O6-alkylguanine DNA-alkyltransferase in E. coli TRG8 cells (which lack this endogenous activity) significantly increased the mutagenicity of DEB. The frequencies of rifampicin-resistant mutants generated by the BD/mouse liver microsomes/GST/GSH system and the DEB/GST/GSH system, as well as the DEB-GSH conjugate, were higher than those of all other systems absent of GST, in agreement with the higher mutagenicities of DEB following GST expression19,20 and the higher mutagenicity of the DEB-GSH conjugate than DEB or several other butadiene-derived epoxides in S. typhimurium TA1535.21 These results indicate that GSH conjugation enhances the mutagenicity of BD by formation of cross-links between DNA and GSH induced by DEB.
To obtain a more complete mutational analysis than the single site system of S. typhimurium TA1535, the occurrence of rifampicin resistant mutants in the rpoB gene was examined, where predominant base substitution missense mutations yield a phenotype. At least 69 base substitutions at 24 coding positions yield phenotypic changes.29 The rpoB gene has been widely used as a marker to examine the spectra of mutants arising endogenously or through induction by exogenous reagents because it encodes the β-subunit of RNA polymerase II, known to accumulate rifampicin-resistant mutants: cluster I (amino acids 140–148) and cluster II (amino acids 508–574), with the latter being the major mutational hot spot.29,32,33 Comparison of mutation spectra was performed in cluster II of the rpoB gene.
G:C to T:A transversions were observed at higher than 33% levels in all systems except spontaneous mutations (Tables 1, 2) and may be due to formation of a labile N7-guanine adduct susceptible to spontaneous depurination. Error-prone bypass and subsequent preferential misincorporation of an adenine opposing the abasic site (“A-Rule”) are likely to be responsible for the G:C to T:A transversions.34,35 No definitive mechanism for the G:C to A:T transitions has been provided; G:C to A:T transitions could arise from DNA adducts at either the O6 or the N2 atom of guanine, in that these are the only other adducts identified.23,26 A:T to T:A transversions could be generated through the labile N3-adenine adduct, producing abasic sites and adenine incorporation.26,36 The N6-adenine adduct of DEB could generate A:T to C:G transversions or the A:T to G:C transitions, although no mechanism has been identified.37
DEB-induced G:C to T:A and A:T to T:A transversions (33% and 27% of the total, respectively, Table 2) may be caused by labile N7-guanine and N3-adenine adducts (i.e., depurination would be expected to lead to adenine incorporation). Although tissue-specific differences in mutation spectra derived from BD and its metabolites were reported following in vivo exposure to BD and following in vitro exposures to EB and DEB, A:T to T:A transversion was the most consistent mutation.27 In the rpoB gene of E. coli TRG8 cells, A:T to T:A transversions induced by BD and its metabolites in the absence of GST/GSH were higher than those induced by the DEB-GSH conjugate or the systems in the presence of GST/GSH (Tables 2, 3). DEB preferentially reacts at the N7 atom of guanine to yield the 2-hydroxy-3,4-epoxybutane adduct, which can either be hydrolyzed to a 2,3,4-trihydroxybutane adduct or, less frequently, form cross-links with other nucleophiles.38 N7G-DEB and N3A-DEB (mono-adducts)36,38, bis-(guan-7-yl)-2,3-butanediol (the most abundant DNA-DNA cross link adduct),39 and 1-(guan-7-yl)-4-(aden-3-yl)-2,3-butanediol (an adenine-guanine cross-linked adduct)40,41 have been identified in calf thymus DNA treated with DEB, and N7G-DEB and N3A-DEB adducts were identified here in E. coli TRG8 cells treated with DEB (Table 4). The N1A-DEB adduct was identified in lymphocyte DNA of humans exposed to BD,42,43 and the levels of the N1A-DEB adduct in lymphocyte DNA from workers lacking GST M1 were significantly higher than in that from GST M1-positive workers after BD exposure.43 The mutation spectrum derived from DEB was similar to that derived from BD + mouse liver microsomes (Table 2). On the other hand, the A:T to C:G transversions and the A:T to G:C transitions induced by the DEB-GSH conjugate, the DEB/GST/GSH system, and the BD/mouse liver microsomes/GST/GSH system were significantly increased compared with those induced by other systems absent of GST/GSH (Tables 2, 3). These increases may be due to N6dA-(OH)2butyl-GSH adduct formed from the DEB-GSH conjugate, in that mutations at A:T pairs were increased the most by the presence of the GST system (Tables 1, 2).
Mutations in the rpoB gene of E. coli TRG8 cells induced by BD and its metabolites in the absence and presence of GST occurred in eight codon positions (amino acids 512, 513, 516, 526, 529, 531, 564, and 572) (Figure 2). Mutation spectra derived from the DEB-GSH conjugate revealed similar patterns with those derived from the DEB/GST/GSH system and the BD/mouse liver microsomes/GST/GSH system (Figure 2, Table 3). Neither the DEB-GSH conjugate, the DEB/GST/GSH system, nor the BD/mouse liver microsomes/GST/GSH system produced any mutations in codon 512, and mutations in codon 513 were higher than those in codon 516. Among codons 526, 529, and 531, mutations in codon 529 were highest, followed by codon 526 and then codon 531 following treatment with the DEB-GSH conjugate, the DEB/GST/GSH system, or the BD/mouse liver microsomes/GST/GSH system, although some mutations of codon 529 were found in the rpoB gene of E. coli treated with the BD/mouse liver microsomes/GST/GSH system due to the effect of other BD-derived oxidation products in BD metabolism. Mutations in codons 564 and 572 following treatment with the DEB-GSH conjugate, the DEB/GST/GSH system, or BD/mouse liver microsomes/GST/GSH system were lower than those in other systems.
All of the mutations in the rpoB gene were base-pair substitutions. None were frameshifts or large deletions. The rpoB gene is not sensitive to detection of the latter types of changes. However, we did not find evidence for frameshift mutations in S. typhimurium TA1537 treated with the DEB-GSH conjugate,22 although frameshifts and small deletions caused by DEB have been identified in some other systems.14,15 Whether the GSH-containing adducts produce large deletions is unknown. Also, mutations in the rpoB gene of E. coli alter the β subunit of RNA polymerase, which is highly conserved among many microorganisms.29 The rpoB gene system can be developed in other microorganisms lacking systems for genetic analysis. Therefore this system can provide detailed information on mutagenic specificity of BD and its metabolites in other GST–based systems.29
In E. coli TRG8 cells treated with the DEB-GSH conjugate, N7G-(OH)2butyl-GSH and N6dA-(OH)2butyl-GSH adducts were highest among the six DNA adducts detected from the DEB-GSH conjugate, followed by N3A-(OH)2butyl-GSH, N3dT-(OH)2butyl-GSH, N4dC-(OH)2butyl-GSH, and N1dG-(OH)2butyl-GSH. This result is consistent with that in calf thymus DNA treated with the DEB-GSH conjugate.22
The DEB-GSH conjugate could possibly be processed to mercapturic acids and (probably after hydrolysis) excreted in urine. Therefore, mercapturic acids have been used as biomarkers of BD exposure and metabolic processing. Mutagenicity of a mercapturic acid of hexachloro-1,3-butadiene (N-acetyl-S-pentachlorobutadienyl-L-cysteine) was significantly higher than that of hexachloro-1,3-butadiene in S. typhimurium TA100.44 However, it has not been documented that these results are directly linked to the presence of specific mercapturic acid DNA adducts (as opposed to breakdown products). We previously reported comparison of the DNA-alkylating properties and mutagenic responses of a series of S-(2-haloethyl)-substituted cysteine and glutathione derivatives.45 Although the cysteine compounds (mercapturic acids) yielded higher adduct levels than GSH conjugates, mutagenicities of GSH conjugates were higher than for the mercapturic acids in S. typhimurium TA 100.
In this E. coli study, the level of DNA adducts formed from DEB was ~100-fold higher than that of GSH-containing adducts (Table 4). Two points can be made regarding this: (i) The enhancement of mutagenicity (Figure 1) in the presence of GST/GSH, coupled with the lower level of identified adducts, implies that the GSH-containing adducts should be much more intrinsically likely to cause base-pair mutations. A similar conclusion was revealed with adducts formed from ethylene dibromide, i.e. that GSH-containing adducts have intrinsically high mutagenicity.45 (ii) High levels of the three “direct” DEB adducts (N7G-DEB, N3A-DEB, and N6A-DEB) were formed in this E. coli study (Table 4) but not in rat liver, lung, or kidney,22 suggesting that these might be subject to repair processes. Further studies on DNA adducts and mutagenesis in experimental animals are planned.
In summary, we characterized GST-enhanced mutagenicity of DEB by comparing mutation spectra derived from BD and its metabolites in the absence and presence of GST/GSH and mouse liver microsomes. The mutation spectra derived from BD with mouse liver microsomes, DEB in the presence of GST/GSH, and the synthetic DEB-GSH conjugate were similar to each other but distinct from the DEB spectrum. The presence of GST considerably enhanced mutations. These results are attributed to the enhanced mutagenicity of GSH-containing DNA adducts. Although numerous DNA adducts have been identified following the reaction of BD and its oxidation products with DNA,36,38–41,46–48 our results, coupled with our earlier findings,19–22 show the enhanced mutagenicity due to GSH moiety and—in the present study—a shift in the mutation spectrum, clearly documenting a biological effect of the GSH adducts.
Acknowledgments
Funding
This work was supported in part by United States Public Health Service Grants R01 ES010546 and P30 ES00267.
1. Abbreviations
- BD
1,3-butadiene
- DEB
1,2:3,4-diepoxybutane
- DEB-GSH conjugate
S-(2-hydroxy-3,4-epoxybutyl)GSH
- EB
3,4-epoxy-1-butene
- GST
GSH S-transferase
- IARC
International Agency for Research on Cancer
- N3A-(OH)2butyl-GSH
S-[4-(N3-adenyl)-2,3-dihydroxybutyl]GSH
- N6dA-(OH)2butyl-GSH
S-[4-(N 6-deoxyadenosinyl)-2,3-dihydroxybutyl]GSH
- N7G-(OH)2butyl-GSH
S-[4-(N 7-guanyl)-2,3-dihydroxybutyl]GSH
- N1dG-(OH)2butyl-GSH
S-[4-(N 1-deoxyguanosinyl)-2,3-dihydroxybutyl]GSH
- N4dC-(OH)2butyl-GSH
S-[4-(N 4-deoxycytidinyl)-2,3-dihydroxybutyl]GSH
- N3dT-(OH)2butyl-GSH
S-[4-(N 3-thymidinyl)-2,3-dihydroxybutyl]GSH
- N7G-DEB
N 7-(2,3,4-trihydroxybutyl)guanine
- N3A-DEB
N 3-(2,3,4-trihydroxybutyl)adenine
- N6A-DEB
N 6-(2,3,4-trihydroxybutyl)adenine
Footnotes
The authors declare no competing financial interest.
References
- 1.Pelz N, Dempster NM, Shore PR. Analysis of low molecular weight hydrocarbons including 1,3-butadiene in engine exhaust gases using an aluminum oxide porous-layer open-tubular fused-silica column. J. Chromatogr. Sci. 1990;28:230–235. doi: 10.1093/chromsci/28.5.230. [DOI] [PubMed] [Google Scholar]
- 2.Morrow NL. The industrial production and use of 1,3-butadiene. Environ. Health Perspect. 1990;86:7–8. doi: 10.1289/ehp.90867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.International Agency for Research on Cancer. 1,3-Butadiene, Vol. 54, IARC Monogr. Evalat. Carcinogen. Risk Chem. Humans. Lyon: International Agency for Research on Cancer; 1992. [Google Scholar]
- 4.Delzell E, Sathiakumar N, Hovinga M, Macaluso M, Julian J, Larson R, Cole P, Muir DC. A follow-up study of synthetic rubber workers. Toxicology. 1996;113:182–189. doi: 10.1016/0300-483x(96)03443-9. [DOI] [PubMed] [Google Scholar]
- 5.Macaluso M, Larson R, Delzell E, Sathiakumar N, Hovinga M, Julian J, Muir D, Cole P. Leukemia and cumulative exposure to butadiene, styrene and benzene among workers in the synthetic rubber industry. Toxicology. 1996;113:190–202. doi: 10.1016/0300-483x(96)03444-0. [DOI] [PubMed] [Google Scholar]
- 6.Santos-Burgoa C, Matanoski GM, Zeger S, Schwartz L. Lymphohematopoietic cancer in styrene-butadiene polymerization workers. Am. J. Epidemiol. 1992;136:843–854. doi: 10.1093/aje/136.7.843. [DOI] [PubMed] [Google Scholar]
- 7.Henderson RF, Thornton-Manning JR, Bechtold WE, Dahl AR. Metabolism of 1,3-butadiene: species differences. Toxicology. 1996;113:17–22. doi: 10.1016/0300-483x(96)03422-1. [DOI] [PubMed] [Google Scholar]
- 8.Melnick RL, Huff JE. 1,3-Butadiene induces cancer in experimental animals at all concentrations from 6.25 to 8000 parts per million. IARC Sci. Publ. 1993:309–322. [PubMed] [Google Scholar]
- 9.Owen PE, Glaister JR, Gaunt IF, Pullinger DH. Inhalation toxicity studies with 1,3-butadiene. 3. Two year toxicity/carcinogenicity study in rats. Am. Ind. Hyg. Assoc. J. 1987;48:407–413. doi: 10.1080/15298668791384959. [DOI] [PubMed] [Google Scholar]
- 10.Malvoisin E, Roberfroid M. Hepatic microsomal metabolism of 1,3-butadiene. Xenobiotica. 1982;12:137–144. doi: 10.3109/00498258209046787. [DOI] [PubMed] [Google Scholar]
- 11.Himmelstein MW, Turner MJ, Asgharian B, Bond JA. Metabolism of 1,3-butadiene: inhalation pharmacokinetics and tissue dosimetry of butadiene epoxides in rats and mice. Toxicology. 1996;113:306–309. doi: 10.1016/0300-483x(96)03462-2. [DOI] [PubMed] [Google Scholar]
- 12.Csanády GA, Guengerich FP, Bond JA. Comparison of the biotransformation of 1,3-butadiene and its metabolite, butadiene monoepoxide, by hepatic and pulmonary tissues from humans, rats and mice. Carcinogenesis. 1992;13:1143–1153. doi: 10.1093/carcin/13.7.1143. [DOI] [PubMed] [Google Scholar]
- 13.Jackson MA, Stack HF, Rice JM, Waters MD. A review of the genetic and related effects of 1,3-butadiene in rodents and humans. Mutat. Res. 2000;463:181–213. doi: 10.1016/s1383-5742(00)00056-9. [DOI] [PubMed] [Google Scholar]
- 14.Cochrane JE, Skopek TR. Mutagenicity of butadiene and its epoxide metabolites: II. Mutational spectra of butadiene, 1,2-epoxybutene and diepoxybutane at the hprt locus in splenic T cells from exposed B6C3F1 mice. Carcinogenesis. 1994;15:719–723. doi: 10.1093/carcin/15.4.719. [DOI] [PubMed] [Google Scholar]
- 15.Cochrane JE, Skopek TR. Mutagenicity of butadiene and its epoxide metabolites: I. Mutagenic potential of 1,2-epoxybutene, 1,2,3,4-diepoxybutane and 3,4-epoxy-1,2-butanediol in cultured human lymphoblasts. Carcinogenesis. 1994;15:713–717. doi: 10.1093/carcin/15.4.713. [DOI] [PubMed] [Google Scholar]
- 16.Goggin M, Swenberg JA, Walker VE, Tretyakova N. Molecular dosimetry of 1,2,3,4-diepoxybutane-induced DNA-DNA cross-links in B6C3F1 mice and F344 rats exposed to 1,3-butadiene by inhalation. Cancer Res. 2009;69:2479–2486. doi: 10.1158/0008-5472.CAN-08-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michaelson-Richie ED, Loeber RL, Codreanu SG, Ming X, Liebler DC, Campbell C, Tretyakova NY. DNA-protein cross-linking by 1,2,3,4-diepoxybutane. J. Proteome Res. 2010;9:4356–4367. doi: 10.1021/pr1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meister A, Anderson ME. Glutathione. Annu. Rev. Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 19.Thier R, Müller M, Taylor JB, Pemble SE, Ketterer B, Guengerich FP. Enhancement of bacterial mutagenicity of bifunctional alkylating agents by expression of mammalian glutathione S-transferase. Chem. Res. Toxicol. 1995;8:465–472. doi: 10.1021/tx00045a019. [DOI] [PubMed] [Google Scholar]
- 20.Thier R, Pemble SE, Kramer H, Taylor JB, Guengerich FP, Ketterer B. Human glutathione S-transferase T1-1 enhances mutagenicity of 1,2-dibromoethane, dibromomethane and 1,2,3,4-diepoxybutane in Salmonella typhimurium . Carcinogenesis. 1996;17:163–166. doi: 10.1093/carcin/17.1.163. [DOI] [PubMed] [Google Scholar]
- 21.Cho S-H, Loecken EM, Guengerich FP. Mutagenicity of a glutathione conjugate of butadiene diepoxide. Chem. Res. Toxicol. 2010;23:1544–1546. doi: 10.1021/tx100304f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho S-H, Guengerich FP. Conjugation of butadiene diepoxide with glutathione yields DNA adducts in vitro and in vivo. Chem Res Toxicol. 2012;25:706–712. doi: 10.1021/tx200471x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu L, Hachey DL, Valadez G, Williams KM, Guengerich FP, Loktionova NA, Kanugula S, Pegg AE. Characterization of a mutagenic DNA adduct formed from 1,2-dibromoethane by O 6-alkylguanine-DNA alkyltransferase. J. Biol. Chem. 2004;279:4250–4259. doi: 10.1074/jbc.M311105200. [DOI] [PubMed] [Google Scholar]
- 24.Valadez JG, Liu L, Loktionova NA, Pegg AE, Guengerich FP. Activation of bis-electrophiles to mutagenic conjugates by human O 6-alkylguanine-DNA alkyltransferase. Chem. Res. Toxicol. 2004;17:972–982. doi: 10.1021/tx049897u. [DOI] [PubMed] [Google Scholar]
- 25.Guengerich FP, Bartleson CJ. Analysis and characterization of enzymes and nucleic acids. In: Hayes AW, editor. Principles and Methods of Toxciology. 5th ed. Boca Raton, FL: CRC Press; 2007. pp. 1981–2048. [Google Scholar]
- 26.Kalapila AG, Loktionova NA, Pegg AE. Alkyltransferase-mediated toxicity of 1,3-butadiene diepoxide. Chem. Res. Toxicol. 2008;21:1851–1861. doi: 10.1021/tx800178t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Recio L, Steen AM, Pluta LJ, Meyer KG, Saranko CJ. Mutational spectrum of 1,3-butadiene and metabolites 1,2-epoxybutene and 1,2,3,4-diepoxybutane to assess mutagenic mechanisms. Chem.-Biol. Interact. 2001;135–136:325–341. doi: 10.1016/s0009-2797(01)00220-4. [DOI] [PubMed] [Google Scholar]
- 28.Shimmyo T, Okada A, Hashimoto T, Kobayashi Y, Miyagi Y, Ishikawa Y, Nakagawa K, Osada H, Tsuchiya E. Etiologic value of p53 mutation spectra and differences with histology in lung cancers. Cancer Sci. 2008;99:287–295. doi: 10.1111/j.1349-7006.2007.00686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garibyan L, Huang T, Kim M, Wolff E, Nguyen A, Nguyen T, Diep A, Hu K, Iverson A, Yang H, Miller JH. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair (Amsterdam) 2003;2:593–608. doi: 10.1016/s1568-7864(03)00024-7. [DOI] [PubMed] [Google Scholar]
- 30.Barker S, Weinfeld M, Murray D. DNA-protein crosslinks: their induction, repair, and biological consequences. Mutat. Res. 2005;589:111–135. doi: 10.1016/j.mrrev.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 31.Barker S, Weinfeld M, Zheng J, Li L, Murray D. Identification of mammalian proteins cross-linked to DNA by ionizing radiation. J. Biol. Chem. 2005;280:33826–33838. doi: 10.1074/jbc.M502477200. [DOI] [PubMed] [Google Scholar]
- 32.Jin DJ, Zhou YN. Mutational analysis of structure-function relationship of RNA polymerase in Escherichia coli . Methods Enzymol. 1996;273:300–319. doi: 10.1016/s0076-6879(96)73027-6. [DOI] [PubMed] [Google Scholar]
- 33.Reynolds MG. Compensatory evolution in rifampin-resistant Escherichia coli . Genetics. 2000;156:1471–1481. doi: 10.1093/genetics/156.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kunkel TA. Mutational specificity of depurination. Proc. Natl. Acad. Sci. U. S. A. 1984;81:1494–1498. doi: 10.1073/pnas.81.5.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sagher D, Strauss B. Insertion of nucleotides opposite apurinic/apyrimidinic sites in deoxyribonucleic acid during in vitro synthesis: uniqueness of adenine nucleotides. Biochemistry. 1983;22:4518–4526. doi: 10.1021/bi00288a026. [DOI] [PubMed] [Google Scholar]
- 36.Tretyakova N, Sangaiah R, Yen TY, Gold A, Swenberg JA. Adenine adducts with diepoxybutane: isolation and analysis in exposed calf thymus DNA. Chem. Res. Toxicol. 1997;10:1171–1179. doi: 10.1021/tx9700681. [DOI] [PubMed] [Google Scholar]
- 37.Carmical JR, Nechev LV, Harris CM, Harris TM, Lloyd RS. Mutagenic potential of adenine N(6) adducts of monoepoxide and diolepoxide derivatives of butadiene. Environ. Mol. Mutagen. 2000;35:48–56. [PubMed] [Google Scholar]
- 38.Tretyakova N, Sangaiah R, Yen TY, Swenberg JA. Synthesis, characterization, and in vitro quantitation of N-7-guanine adducts of diepoxybutane. Chem. Res. Toxicol. 1997;10:779–785. doi: 10.1021/tx970004q. [DOI] [PubMed] [Google Scholar]
- 39.Park S, Anderson C, Loeber R, Seetharaman M, Jones R, Tretyakova N. Interstrand and intrastrand DNA-DNA cross-linking by 1,2,3,4-diepoxybutane: role of stereochemistry. J. Am. Chem. Soc. 2005;127:14355–14365. doi: 10.1021/ja051979x. [DOI] [PubMed] [Google Scholar]
- 40.Goggin M, Anderson C, Park S, Swenberg J, Walker V, Tretyakova N. Quantitative high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry analysis of the adenine-guanine cross-links of 1,2,3,4-diepoxybutane in tissues of butadiene-exposed B6C3F1 mice. Chem. Res. Toxicol. 2008;21:1163–1170. doi: 10.1021/tx800051y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park S, Hodge J, Anderson C, Tretyakova N. Guanine-adenine DNA cross-linking by 1,2,3,4-diepoxybutane: potential basis for biological activity. Chem. Res. Toxicol. 2004;17:1638–1651. doi: 10.1021/tx0498206. [DOI] [PubMed] [Google Scholar]
- 42.Zhao C, Vodicka P, Sram RJ, Hemminki K. Human DNA adducts of 1,3-butadiene, an important environmental carcinogen. Carcinogenesis. 2000;21:107–111. doi: 10.1093/carcin/21.1.107. [DOI] [PubMed] [Google Scholar]
- 43.Zhao C, Vodicka P, Sram RJ, Hemminki K. DNA adducts of 1,3-butadiene in humans: relationships to exposure, GST genotypes, single-strand breaks, and cytogenetic end points. Environ. Mol. Mutagen. 2001;37:226–230. doi: 10.1002/em.1031. [DOI] [PubMed] [Google Scholar]
- 44.Wild D, Schutz S, Reichert D. Mutagenicity of the mercapturic acid and other S-containing derivatives of hexachloro-1,3-butadiene. Carcinogenesis. 1986;7:431–434. doi: 10.1093/carcin/7.3.431. [DOI] [PubMed] [Google Scholar]
- 45.Humphreys WG, Kim DH, Cmarik JL, Shimada T, Guengerich FP. Comparison of the DNA-alkylating properties and mutagenic responses of a series of S-(2-haloethyl)-substituted cysteine and glutathione derivatives. Biochemistry. 1990;29:10342–10350. doi: 10.1021/bi00497a008. [DOI] [PubMed] [Google Scholar]
- 46.Citti L, Gervasi PG, Turchi G, Bellucci G, Bianchini R. The reaction of 3,4-epoxy-1-butene with deoxyguanosine and DNA in vitro: synthesis and characterization of the main adducts. Carcinogenesis. 1984;5:47–52. doi: 10.1093/carcin/5.1.47. [DOI] [PubMed] [Google Scholar]
- 47.Tretyakova N, Chiang SY, Walker VE, Swenberg JA. Quantitative analysis of 1,3-butadiene-induced DNA adducts in vivo and in vitro using liquid chromatography electrospray ionization tandem mass spectrometry. J. Mass. Spectrom. 1998;33:363–376. doi: 10.1002/(SICI)1096-9888(199804)33:4<363::AID-JMS643>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 48.Zhang XY, Elfarra AA. Reaction of 1,2,3,4-diepoxybutane with 2’-deoxyguanosine: initial products and their stabilities and decomposition patterns under physiological conditions. Chem. Res. Toxicol. 2005;18:1316–1323. doi: 10.1021/tx0500979. [DOI] [PMC free article] [PubMed] [Google Scholar]