Abstract
Background/Aims
A worldwide increase in amoxicillin resistance in Helicobacter pylori is having an adverse effect on eradication therapy. In this study, we investigated the mechanism of the amoxicillin resistance of H. pylori in terms of amino acid substitutions in penicillin-binding protein 1 (PBP1).
Methods
In total, 150 H. pylori strains were isolated from 144 patients with chronic gastritis, peptic ulcers, or stomach cancer. The minimum inhibitory concentrations (MICs) of the strains were determined with a serial 2-fold agar dilution method. The resistance breakpoint for amoxicillin was defined as >0.5 µg/mL.
Results
Nine of 150 H. pylori strains showed amoxicillin resistance (6%). The MIC values of the resistant strains ranged from 1 to 4 µg/mL. A PBP1 sequence analysis of the resistant strains revealed multiple amino acid substitutions: Val16→Ile, Val45→Ile, Ser414→Arg, Asn562→Tyr, Thr593→Ala, Gly595→Ser, and Ala599→Thr. The natural transformation of these mutated genes into amoxicillin-sensitive strains was performed in two separate pbp1 gene segments. A moderate increase in the amoxicillin MIC was observed in the segment that contained the penicillin-binding motif of the C-terminal portion, the transpeptidase domain.
Conclusions
pbp1 mutation affects the amoxicillin resistance of H. pylori through the transfer of the penicillin-binding motif.
Keywords: Helicobacter pylori, Amoxicillin resistance, Penicillin-binding proteins, Amino acid substitution
INTRODUCTION
Helicobacter pylori is a gram-negative spiral bacillus and is a major cause of chronic gastritis and peptic ulcer disease. In addition, there are substantial etiological evidences that H. pylori is a risk factor of gastric cancer and some gastric lymphoma.1,2 The primary eradication regimens comprising proton pump inhibitor, amoxicillin and clarithromycin, or metronidazole have been showing decreased eradication rate with geographic difference. Recent increased reports of eradication failure are mostly associated with emerging antibiotic resistance.3 Antibiotic resistance of H. pylori has been regarded to be mainly related with metronidazole and macrolide such as clarithromycin.4,5 Although amoxicillin is a primary antimicrobial agent for H. pylori eradication, treatment failure with amoxicillin resistance has recently been noted.6-8 The amoxicillin-resistance of H. pylori has been explained by a mutation of penicillin-binding proteins (PBPs).9-11 Especially, the amino acid substitutions of the penicillin-binding motifs adjacent to the transpeptidase domain were known to be related to the development of resistant phenotype.10,11 Although investigations have reported some amino acid substitutions in the PBP1 of resistant isolates and showed transfer of resistance phenotype through the natural transformation, the exact mechanism of amoxicillin resistance and its contribution to eradication failure has not been fully elucidated.10,11 In this study, we assessed the prevalence of amoxicillin resistant H. pylori strains isolated from Korean patient and investigated the relationship between amoxicillin resistance of H. pylori and mutation of PBP1 through the transformation of mutational gene products into amoxicillin sensitive strain.
MATERIALS AND METHODS
1. Bacterial strains
H. pylori were isolated from gastric biopsy specimens taken from both antrum and body of the patients diagnosed as gastritis, gastric ulcer, or gastric carcinoma at Chung-Ang University Yongsan Hospital in Korea between 2005 and 2006. ATCC 700392 was used as reference strains. The H. pylori strains were cultured on blood agar plate at 37℃ for 3 to 5 days under microaerobic conditions (5% O2, 10% CO2, 85% N2).
2. Susceptibility test
The minimum inhibitory concentrations (MIC) of amoxicillin (Sigma Chemical Co., St Lousis, MO, USA) was determined using the serial 2-fold agar dilution Method as described previously.12 The H. pylori strains were subcultured on Mueller-Hinton agar supplemented with 5% defibrinated sheep blood for 72 hours. A total of 1 mL of bacterial suspension adjusted to McFaland No.2 (6×108 colony-forming units/mL) was inoculated to each well of metallic plate and then each amount of suspension was spotted on each antibiotic-containing agar dilution plate. The MIC of the strains was determined after 72 hours of incubation. H. pylori ATCC 43504 was used for quality control. The resistance breakpoints for amoxicillin was defined as >0.5 µg/mL.
3. Polymerase chain reaction and DNA sequence analysis
To determine the amino acid substitution of amoxicillin-resistant strains, full-length pbp1 gene was amplified by polymerase chain reaction (PCR) and then DNA sequence was analyzed. For the precise sequence analysis, it was segmented by six PCR products. The used PCR primers of each segment were shown in Table 1. DNA was extracted from bacteria on blood agar plate using a genomic DNA extraction Kit (Intron Co., Seoul, Korea). The primers used in this study were designed with the DNA sequence analysis of H. pylori ATCC 700392.13 PCR was performed using Ex Taq polymerase (TaKaRa Biomedicals, Kyoto, Japan). Purified PCR products were sequenced directly using the Big Dye terminator sequencing kits and ABI PRISM 3730XL Analyzer (Applied Biosystems, Foster City, CA, USA). We repeated above procedure three times to minimize the probability of error in the DNA sequencing process.
Table 1.
Primers Used in This Study
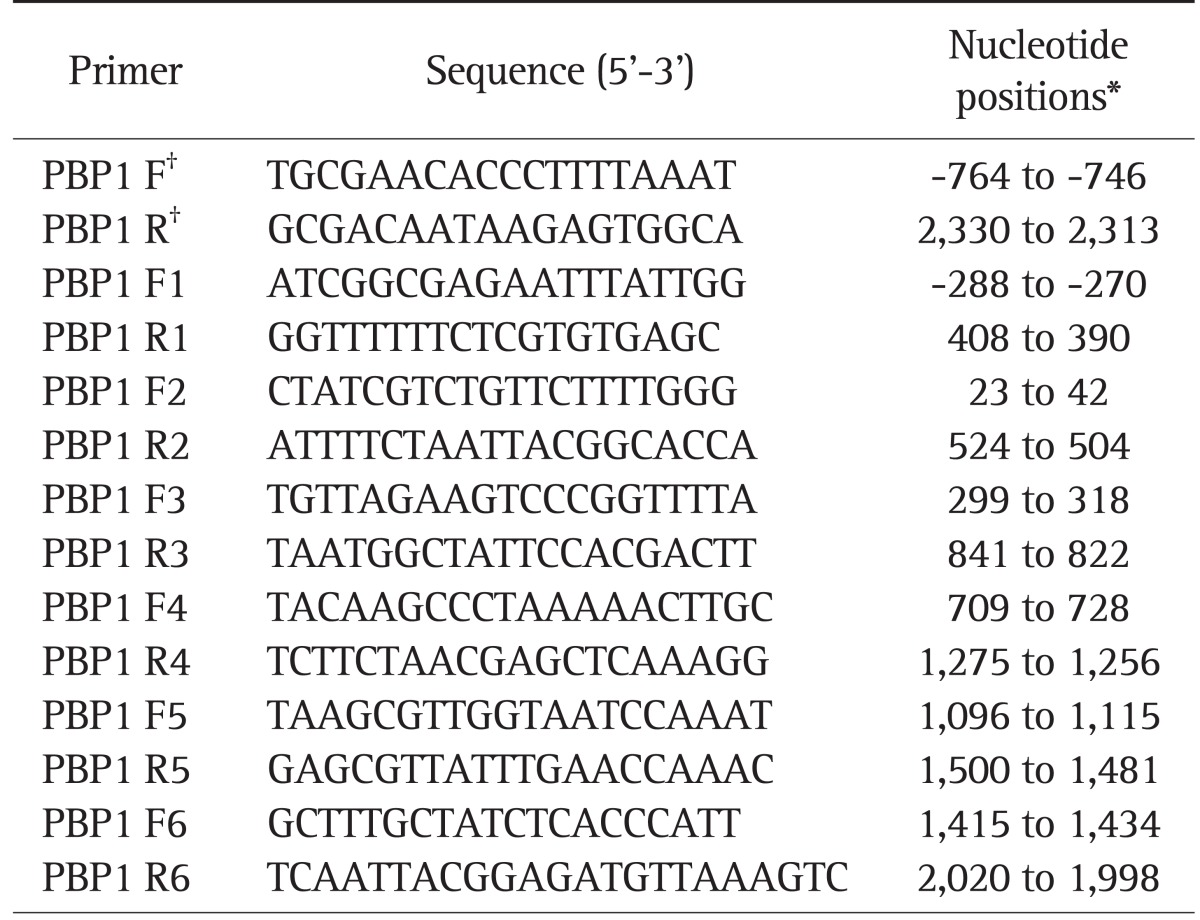
*The nucleotide positions given are according to the pbp1 gene and start at the ATG start codon of Helicobacter pylori ATCC 700392 (GenBank accession no. AE000511); †The primers were used for the polymerase chain reaction amplification of the complete pbp1 gene.
4. Natural transformation of H. pylori
For natural transformation, 25 µL of suspension with 5 µg of H. pylori DNA was spotted onto brain heart infusion (BHI) agar plates directly. After incubated for 24 hours, cells were scraped onto the selective BHI agar plates, then were incubated for 3 to 5 days. Amoxicillin-susceptible H. pylori strain ATCC 43504 and CAUH-32 were transformed by three PCR products obtained from amplification of pbp1 genes of amoxicillin-resistant strains. One was full length of pbp1 gene. Another was N-terminal region of pbp1 gene (-288 to 841). The other was C-terminal region of pbp1 gene (709 to 2,020). Before transformation of gene product, N-terminal region of pbp1 gene was amplified with primer PBP1 F1 and PBP1 R3 and C-terminal region of pbp1 gene was amplified with primer PBP1 F4 and PBP R6. In addition, full length of pbp1 gene was also amplified with primer F and R (Table 1). In regard to two separated regions, the Val16→Ile, Val45→Ile mutations were included in one region and the Ser414→Arg, Asn562→Tyr, Thr593→Ala, Gly595→Ser, and Ala599→Thr mutations were included in the other region. Natural transformation was preceded with each divided pbp1 gene products. In this study we also tested the effect of natural transformation of whole pbp1 gene. Transformants were selected on the BHI agar plates containing 5% sheep blood.
RESULTS
1. Prevalence of amoxicillin resistant H. pylori and MIC of amoxicillin
A total of 150 H. pylori strains were isolated from 144 patients with chronic gastritis (n=75), peptic ulcer (n=66), or stomach cancer (n=3). Of them, 87 were male and 57 were female. Their mean age was 49.9±14.6 years old. Among 150 H. pylori strains, nine strains isolated from different patients showed amoxicillin resistance (6%). The MIC value of resistant strains were ranged from 1 to 4 µg/mL. Table 2 shows the MICs of H. pylori strains used in this study. Among the 12 strains, nine strains were resistant to amoxicillin and remaining three strains were susceptible to amoxicillin with MIC <0.0625 µg/L.
Table 2.
Minimum Inhibitory Concentrations for the Helicobacter pylori Isolates Used in This Study
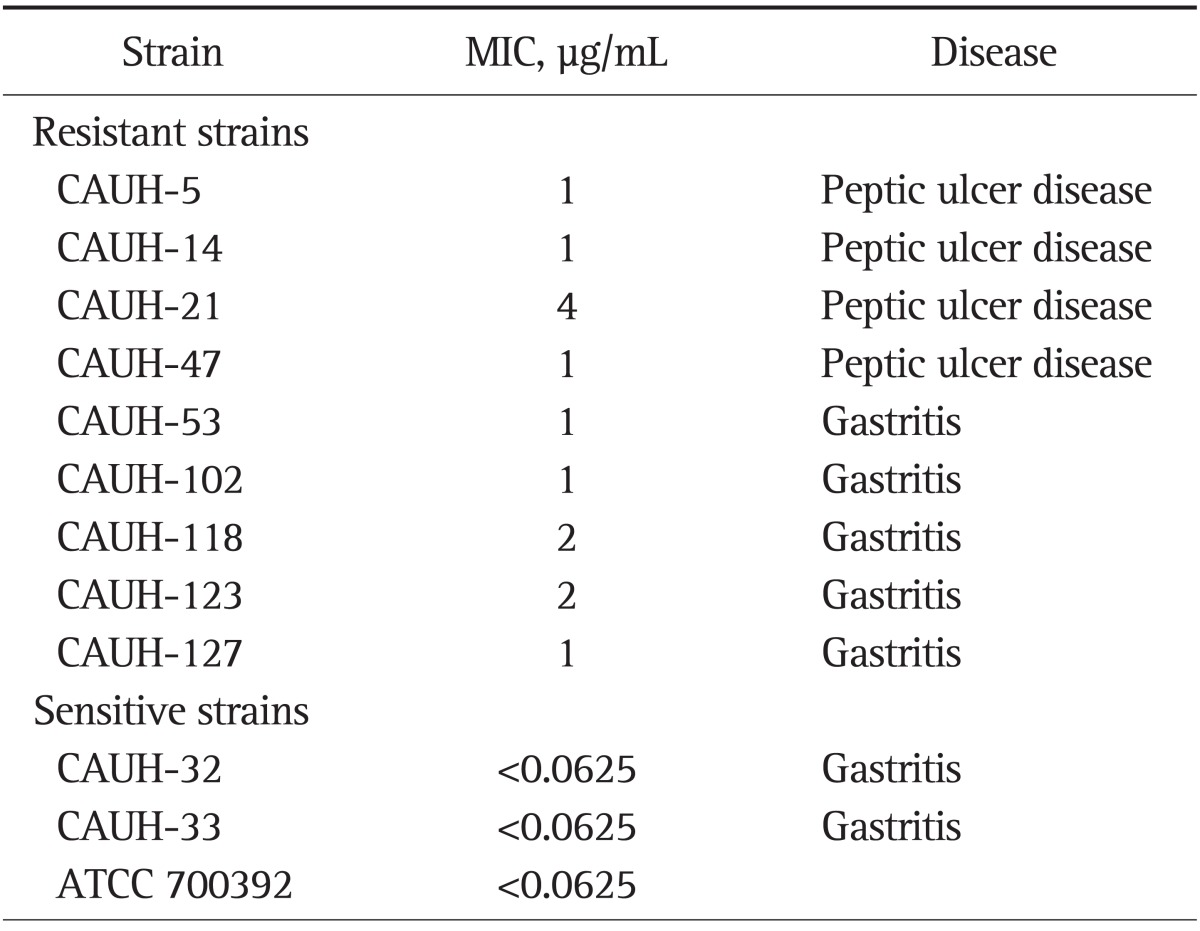
MIC, minimum inhibitory concentration.
2. Comparison of the PBP1 amino acid sequences between amoxicillin resistant and susceptible strains
The DNA sequence of full-length pbp1 gene from 12 isolated strains was determined and amino acid sequence of PBP1 was analyzed. The PCR products of each six segment were 660, 501, 542, 405, 567, and 515 bp (data not shown). To determine the amino acid substitutions specific to amoxicillin-resistant H. pylori, nine amoxicillin-resistant strains were compared with those of three amoxicillin-susceptible strains in terms of PBP1 amino acid sequences. PBP1 sequence analysis of resistant strains showed seven amino acid substitutions in Val16→Ile, Val45→Ile, Ser414→Arg, Asn562→Tyr, Thr593→Ala, Gly595→Ser, and Ala599→Thr. Intriguingly, identified amino acid substitutions were identical in all strains with repeating three times. Table 3 shows the substitutions detected in amoxicillin-resistant strains.
Table 3.
Substitutions in Penicillin-Binding Protein 1 of an Amoxicillin-Resistant Helicobacter pylori Strain
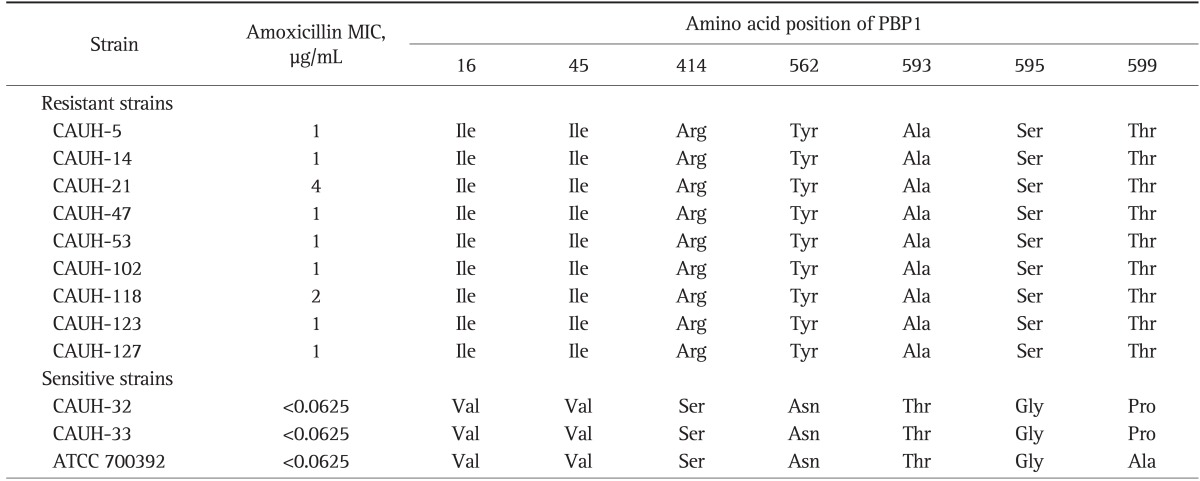
PBP1, penicillin-binding protein 1; MIC, minimum inhibitory concentration.
3. The pbp1 gene transformation of amoxicillin-resistant strains
To determine whether amoxicillin-resistance was transferable to amoxicillin-susceptible strain and to examine the change of MIC in amoxicillin-susceptible strains, amoxicillin-susceptible strains (ATCC 43504, CAUH-32) were transformed with PCR products of pbp1 gene (Table 4). The identified amino acid substitutions were existed in slightly different region. One group of five amino acid substitutions including Ser414→Arg, Asn562→Tyr, Thr593→Ala, Gly595→Ser, and Ala599→Thr was in the adjacent to penicillin-binding motif but the other two substitutions of Val16→Ile, Val45→Ile were somewhat distant from the region. Transformation was performed in the divided two segment of pbp1 gene. The amino acid substitution of Val16→Ile, Val45→Ile were included in one fragment (segment -288 to 841) and Ser414→Arg, Asn562→Tyr, Thr593→Ala, Gly595→Ser, and Ala599→Thr were contained in the other fragment (segment 709 to 2,020), respectively. The MIC range of transformants obtained by using pbp1 of amoxicillin-resistant strain which containing penicillin-binding motif were 0.25 µg/mL, which was much higher than that of the recipient strain ATCC43504. However, there was no MIC change in the transformants obtained by the other fragment of pbp1 gene. When the whole pbp1 gene of resistant strains were transferred to the susceptible strains, the MIC change was same with that of segment pbp1 gene transformation which containing the penicillin-binding motifs.
Table 4.
Susceptibilities and Substitutions in Penicillin-Binding Protein 1 of an Amoxicillin-Resistant Helicobacter pylori Strain Transformants
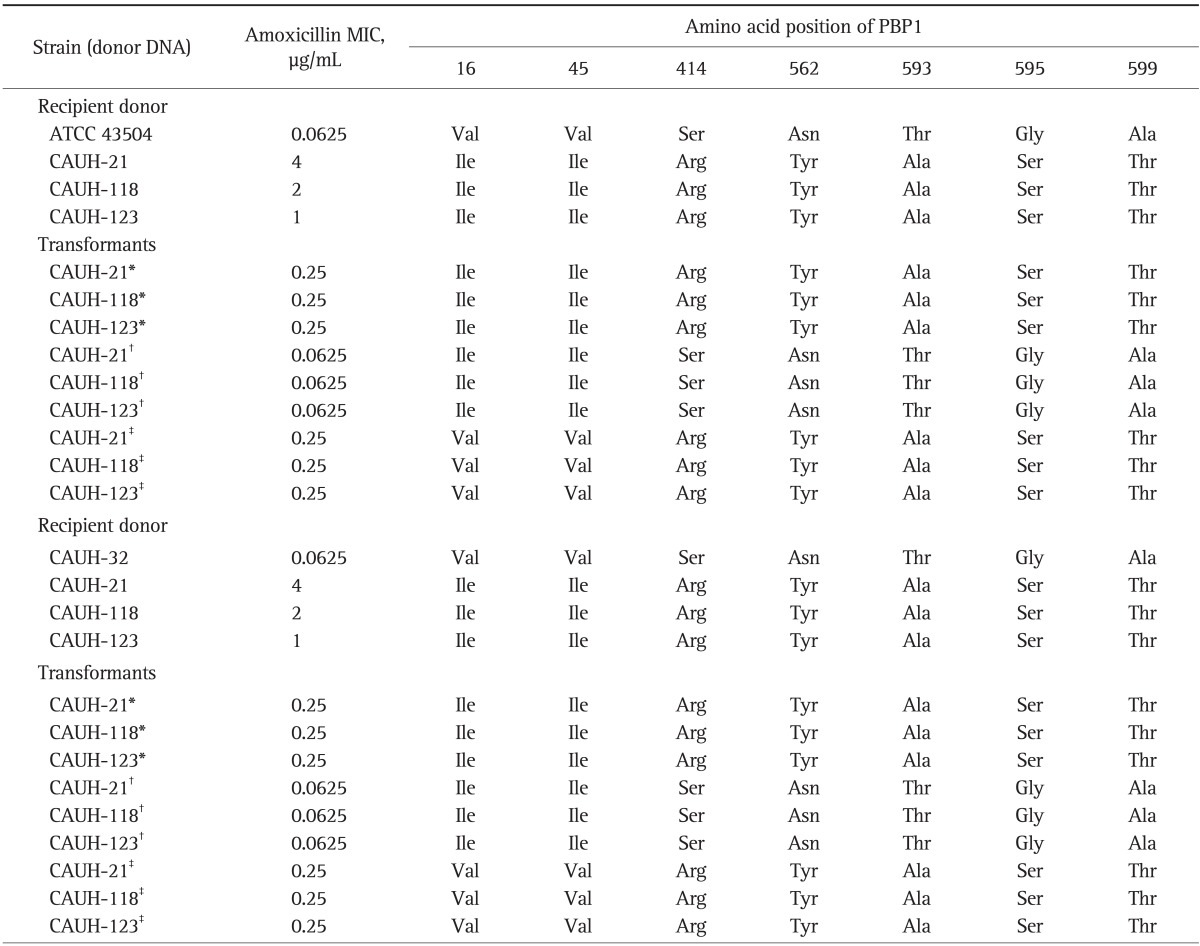
PBP1, penicillin-binding protein 1; MIC, minimum inhibitory concentration.
*The full pbp1 gene; †Region (-288 to 841); ‡Region (709 to 2,020) were used for transformation.
DISCUSSION
Prevalence of antibiotic-resistant H. pylori has been increased world wide and has been a major factor of eradication failure. Amoxicillin is a β-lactam antibiotic and constitute the primary H. pylori eradication regimen. Concerns regarding amoxicillin resistance of H. pylori are recently developed. In the past, even until the last decade it was generally considered that amoxicillin-resistance of H. pylori was absent or very rare. But currently, reports on the isolation of amoxicillin resistant H. pylori are increasing world wide. In this study we investigated the prevalence of amoxicillin-resistance H. pylori in Korea and resistance mechanism of H. pylori strains correlated with PBP1 amino acid substitution. In some regions, the prevalence rate of amoxicillin-resistant H. pylori was lower than 1%, however the world wide reports on the prevalence showed variable difference according to the geographic location.6,14-16 Such geographic difference of amoxicillin resistance is thought to be closely related with the pattern of antibiotic use of the society. Other factors including the study population might also account the difference of the reports. The prevalence rate of amoxicillin-resistance H. pylori in the present study was 6%. The previous report on the prevalence rate of amoxicillin-resistant H. pylori in Korea where the MIC breakpoint of amoxicillin-resistance H. pylori was ≥0.5 µg/mL was 5.6% in 1994 and 18.5% in 2003.17 As standardized resistance break point of amoxicillin in H. pylori has not been determined, there has been some confusion in the interpretation. When the breakpoint MIC is defined to ≥0.5 µg/mL, the prevalence was up to 10% in the present study.
Gram-negative bacilli commonly become resistant to β-lactam agent. The general mechanism of β-lactam resistance of gram-negative bacilli is mostly caused by a production of β-lactamase and in some species, a mutations of PBPs decrease the binding affinity of β-lactams to PBPs.18 The resistance mechanism of H. pylori is explained by mutation of PBPs, unlike other gram-negative bacilli, β-lactamase has not been found in H. pylori.6,19 The PBPs are enzyme for formation of peptidoglycan layer of bacterial wall and has its enzymatic activity in the C-terminal region.20 It has been known that PBPs have some well conserved residues which involved in penicillin binding and the mutations which occurred in or around the regions were related with antibiotic resistance. Contribution of PBP mutation in the development of β-lactam resistance of gram-negative bacilli had been investigated in the Streptococcus pneumoniae, Hemophilus influenaze, and Proteus mirabilis.21-23 In these species the amino acid substitution in close to the penicillin-binding motifs (SXXK, SXN, and KTG) of PBPs was associated with development of β-lactam resistance. Regarding H. pylori, amino acid substitution in penicillin-binding motifs SAIK (368 to 371), SLN (433 to 435), and KTG (555 to 557) was known to be related with development of β-lactam resistance.9,11,17 Although there have been quite a few reports which elucidate the amoxicillin-resistance mechanism of H. pylori correlated with the role of PBP1, this study showed seven amino acid substitutions of PBP1 from the nine amoxicillin-resistant strains extracted from Korean patients. The identified each amino acid substitutions were confirmed with repeated amino acid sequence analysis. In the transformation of PBP1 genome of amoxicillin-resistant strains which were preceded in two separated segment of PBP1, only the one including penicillin-binding motifs caused moderate increase of amoxicillin MIC. When the whole PBP1 genome was transformed into the susceptible strains, the increase of amoxicillin MIC was same with that of transformants in which penicillin-binding motif containing fragment was transferred. The result of our study confirmed the previous reports that multiple amino acid substitutions in or close to the PBPs of PBP1 are responsible for amoxicillin-resistance H. pylori.10 In our experiment, we found seven amino acid substitutions and these point mutations were all transferred to susceptible strains. Of the amino acid substitutions, Asn562→Tyr mutation which located near the KTG (555 to 557) motif was identically found in all amoxicillin resistant strains and the mutation was also commonly reported in other strains.11,19,24 The substitution of Ser414→Arg which occurred in the adjacent to SLN (433 to 435) motif was also identically found in all resistance strains. The contribution to resistance phenotype of Ser414→Arg mutation was previously explained by Gerrits et al.11 The fact that the locations of described amino acid substitutions are adjacent to penicillin-binding motifs suggests that the mutations might be a main mechanism of amoxicillin resistance. We also found other mutation of Val16→Ile, Val45→Ile, Thr593→Ala, Gly595→Ser, and Ala599→Thr. The mutations of Thr593→Ala, Gly595→Ser were also found in previously reported resistant isolates, however, these mutations were not specific or played a role of amoxicillin resistance in the previous report.10 In the experiment of natural transformation of divided pbp1 gene, the mutation of Val16→Ile and Val45→Ile which is remote from the penicillin-binding motif, showed no relevance to amoxicillin resistance. To the recent, the influence of Thr593→Ala, Ala599→Thr mutations are not sufficiently revealed. About these mutations, further investigation of site-directed mutagenesis might be needed to define the role in the amoxicillin resistance. In the present experiment, acquisition of amoxicillin resistance of susceptible strains by natural transformation of pbp1 gene was restrictive. The MIC of amoxicillin-resistance isolates were ranged 1 to 4 µg/mL and the transformant showed only moderate increase of MIC from <0.0625 to 0.25 µg/mL. This finding is somewhat coherent with previous studies which tested natural transformation of pbp1 gene into amoxicillin-susceptible strains. The MIC changes of transformants of the previous studies were moderate and we also could not obtain highly resistant strains.10,24,25 The natural transformation studies of pbp1 gene of amoxicillin-resistant strains confirm that multiple amino acid substitutions are associated with amoxicillin resistance of H. pylori and such amino acid substitution of pbp1 gene confer some part of resistant phenotype. The whole resistance mechanism is more complex and multifactorial and is known to be interacting each other. As it turned out, the factors PBP2, PBP3, and some porin proteins are also involved in the resistance mechanism.26,27 However, the exact mechanism of amoxicillin-resistant H. pylori and its effect to eradication failure is not fully revealed so far.
The prevalence of amoxicillin-resistant H. pylori in Korean society has been much increased during the past few decades. Injudicious use of antibiotics in Korea has been a social issue and related antibiotic resistance of H. pylori is expected to increase furthermore. Further investigations on the nature of amoxicillin resistance and the contribution to the eradication failure of amoxicillin resistance are needed.
In conclusion, we showed that multiple amino acid substitutions of PBP1 are associated with amoxicillin resistance of H. pylori and amino acid substitution which adjacent to the penicillin-binding motif was associated with moderate MIC increase of isolate strains. Some of the amino acid substitutions of the present study were consistent with previous experiments which comprising Ser414→Arg, Asn562→Tyr. More information on the genetic mechanism of amoxicillin resistance and clinical access to the molecular diagnosis of resistance strain might provide some guidance to establish eradication regimen.
ACKNOWLEDGEMENTS
This work was supported by the Korean College of Helicobacter and Upper Gastrointestinal Research Foundation Grant. We thank Ki Sung Kim for his excellent technical help.
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.Parsonnet J, Friedman GD, Vandersteen DP, et al. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 2.Forman D, Newell DG, Fullerton F, et al. Association between infection with Helicobacter pylori and risk of gastric cancer: evidence from a prospective investigation. BMJ. 1991;302:1302–1305. doi: 10.1136/bmj.302.6788.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mégraud F. Antibiotic resistance in Helicobacter pylori infection. Br Med Bull. 1998;54:207–216. doi: 10.1093/oxfordjournals.bmb.a011671. [DOI] [PubMed] [Google Scholar]
- 4.Versalovic J, Shortridge D, Kibler K, et al. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40:477–480. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodwin A, Kersulyte D, Sisson G, Veldhuyzen van Zanten SJ, Berg DE, Hoffman PS. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol Microbiol. 1998;28:383–393. doi: 10.1046/j.1365-2958.1998.00806.x. [DOI] [PubMed] [Google Scholar]
- 6.Dore MP, Osato MS, Realdi G, Mura I, Graham DY, Sepulveda AR. Amoxycillin tolerance in Helicobacter pylori. J Antimicrob Chemother. 1999;43:47–54. doi: 10.1093/jac/43.1.47. [DOI] [PubMed] [Google Scholar]
- 7.Han SR, Bhakdi S, Maeurer MJ, Schneider T, Gehring S. Stable and unstable amoxicillin resistance in Helicobacter pylori: should antibiotic resistance testing be performed prior to eradication therapy? J Clin Microbiol. 1999;37:2740–2741. doi: 10.1128/jcm.37.8.2740-2741.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dore MP, Piana A, Carta M, et al. Amoxycillin resistance is one reason for failure of amoxycillin-omeprazole treatment of Helicobacter pylori infection. Aliment Pharmacol Ther. 1998;12:635–639. doi: 10.1046/j.1365-2036.1998.00350.x. [DOI] [PubMed] [Google Scholar]
- 9.Paul R, Postius S, Melchers K, Schäfer KP. Mutations of the Helicobacter pylori genes rdxA and pbp1 cause resistance against metronidazole and amoxicillin. Antimicrob Agents Chemother. 2001;45:962–965. doi: 10.1128/AAC.45.3.962-965.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rimbara E, Noguchi N, Kawai T, Sasatsu M. Correlation between substitutions in penicillin-binding protein 1 and amoxicillin resistance in Helicobacter pylori. Microbiol Immunol. 2007;51:939–944. doi: 10.1111/j.1348-0421.2007.tb03990.x. [DOI] [PubMed] [Google Scholar]
- 11.Gerrits MM, Schuijffel D, van Zwet AA, Kuipers EJ, Vandenbroucke-Grauls CM, Kusters JG. Alterations in penicillin-binding protein 1A confer resistance to beta-lactam antibiotics in Helicobacter pylori. Antimicrob Agents Chemother. 2002;46:2229–2233. doi: 10.1128/AAC.46.7.2229-2233.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim JJ, Reddy R, Lee M, et al. Analysis of metronidazole, clarithromycin and tetracycline resistance of Helicobacter pylori isolates from Korea. J Antimicrob Chemother. 2001;47:459–461. doi: 10.1093/jac/47.4.459. [DOI] [PubMed] [Google Scholar]
- 13.Tomb JF, White O, Kerlavage AR, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 14.Graham DY. Antibiotic resistance in Helicobacter pylori: implications for therapy. Gastroenterology. 1998;115:1272–1277. doi: 10.1016/s0016-5085(98)70100-3. [DOI] [PubMed] [Google Scholar]
- 15.Poon SK, Chang CS, Su J, et al. Primary resistance to antibiotics and its clinical impact on the efficacy of Helicobacter pylori lansoprazole-based triple therapies. Aliment Pharmacol Ther. 2002;16:291–296. doi: 10.1046/j.1365-2036.2002.01184.x. [DOI] [PubMed] [Google Scholar]
- 16.Mendonca S, Ecclissato C, Sartori MS, et al. Prevalence of Helicobacter pylori resistance to metronidazole, clarithromycin, amoxicillin, tetracycline, and furazolidone in Brazil. Helicobacter. 2000;5:79–83. doi: 10.1046/j.1523-5378.2000.00011.x. [DOI] [PubMed] [Google Scholar]
- 17.Kim JM, Kim JS, Kim N, Kim SG, Jung HC, Song IS. Comparison of primary and secondary antimicrobial minimum inhibitory concentrations for Helicobacter pylori isolated from Korean patients. Int J Antimicrob Agents. 2006;28:6–13. doi: 10.1016/j.ijantimicag.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 18.Therrien C, Levesque RC. Molecular basis of antibiotic resistance and beta-lactamase inhibition by mechanism-based inactivators: perspectives and future directions. FEMS Microbiol Rev. 2000;24:251–262. doi: 10.1111/j.1574-6976.2000.tb00541.x. [DOI] [PubMed] [Google Scholar]
- 19.van Zwet AA, Vandenbroucke-Grauls CM, Thijs JC, van der Wouden EJ, Gerrits MM, Kusters JG. Stable amoxicillin resistance in Helicobacter pylori. Lancet. 1998;352:1595. doi: 10.1016/s0140-6736(98)00064-6. [DOI] [PubMed] [Google Scholar]
- 20.Goffin C, Ghuysen JM. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev. 1998;62:1079–1093. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Overweg K, Bogaert D, Sluijter M, de Groot R, Hermans PW. Molecular characteristics of penicillin-binding protein genes of penicillin-nonsusceptible Streptococcus pneumoniae isolated in the Netherlands. Microb Drug Resist. 2001;7:323–334. doi: 10.1089/10766290152773338. [DOI] [PubMed] [Google Scholar]
- 22.Ubukata K, Chiba N, Hasegawa K, Kobayashi R, Iwata S, Sunakawa K. Antibiotic susceptibility in relation to penicillin-binding protein genes and serotype distribution of Streptococcus pneumoniae strains responsible for meningitis in Japan, 1999 to 2002. Antimicrob Agents Chemother. 2004;48:1488–1494. doi: 10.1128/AAC.48.5.1488-1494.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neuwirth C, Siébor E, Duez JM, Péchinot A, Kazmierczak A. Imipenem resistance in clinical isolates of Proteus mirabilis associated with alterations in penicillin-binding proteins. J Antimicrob Chemother. 1995;36:335–342. doi: 10.1093/jac/36.2.335. [DOI] [PubMed] [Google Scholar]
- 24.Kwon DH, Dore MP, Kim JJ, et al. High-level beta-lactam resistance associated with acquired multidrug resistance in Helicobacter pylori. Antimicrob Agents Chemother. 2003;47:2169–2178. doi: 10.1128/AAC.47.7.2169-2178.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamoto T, Yoshiyama H, Nakazawa T, et al. A change in PBP1 is involved in amoxicillin resistance of clinical isolates of Helicobacter pylori. J Antimicrob Chemother. 2002;50:849–856. doi: 10.1093/jac/dkf140. [DOI] [PubMed] [Google Scholar]
- 26.Rimbara E, Noguchi N, Kawai T, Sasatsu M. Mutations in penicillin-binding proteins 1, 2 and 3 are responsible for amoxicillin resistance in Helicobacter pylori. J Antimicrob Chemother. 2008;61:995–998. doi: 10.1093/jac/dkn051. [DOI] [PubMed] [Google Scholar]
- 27.Co EM, Schiller NL. Resistance mechanisms in an in vitro-selected amoxicillin-resistant strain of Helicobacter pylori. Antimicrob Agents Chemother. 2006;50:4174–4176. doi: 10.1128/AAC.00759-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


