Abstract
Single-stranded DNA-binding proteins (SSBs) bind single-stranded DNA (ssDNA) and participate in all genetic processes involving ssDNA, such as replication, recombination, and repair. Here we applied atomic force microscopy to directly image SSB–DNA complexes under various conditions. We used the hybrid DNA construct methodology in which the ssDNA segment is conjugated to the DNA duplex. The duplex part of the construct plays the role of a marker, allowing unambiguous identification of specific and nonspecific SSB–DNA complexes. We designed hybrid DNA substrates with 5′- and 3′-ssDNA termini to clarify the role of ssDNA polarity on SSB loading. The hybrid substrates, in which two duplexes are connected with ssDNA, were the models for gapped DNA substrates. We demonstrated that Escherichia coli SSB binds to ssDNA ends and internal ssDNA regions with the same efficiency. However, the specific recognition by ssDNA requires the presence of Mg2+ cations or a high ionic strength. In the absence of Mg2+ cations and under low-salt conditions, the protein is capable of binding DNA duplexes. In addition, the number of interprotein interactions increases, resulting in the formation of clusters on double-stranded DNA. This finding suggests that the protein adopts different conformations depending on ionic strength, and specific recognition of ssDNA by SSB requires a high ionic strength or the presence of Mg2+ cations.
Single-stranded DNA-binding proteins (SSBs) have a high affinity for single-stranded DNA (ssDNA) and participate in all genetic processes involving ssDNA as an accessory protein.1–7 SSBs play a role in separating the DNA strands during replication and prevent the ssDNA from re-forming as a double helix. During homologous recombination, SSBs are involved in the formation of RecA–DNA filaments, regulating the process of loading RecA onto ssDNA.6 The interaction of SSB with other recombination mediator proteins, such as RecO, facilitates the annealing step via the SSB-induced conformational change of SSB–ssDNA filaments.8
The Escherichia coli SSB is one of the most widely studied members of the SSB family. The monomer of the E. coli SSB contains 177 amino acids and exists as a stable homotetramer in solution.7 Each SSB monomer can bind ssDNA, but the tetramer, containing four ssDNA binding sites, appears to be the functional form of the protein.9 According to ref 7, SSB has two distinct binding modes depending on the salt concentration. At low monovalent salt concentrations (<10 mM NaCl), SSB binds ssDNA cooperatively with only two subunits of the SSB tetramer interacting with ssDNA. This mode is termed the (SSB)35 mode. At high ionic strengths (>200 mM NaCl), the (SSB)65 binding mode, all four subunits of SSB interact with ssDNA in a noncooperative manner. The crystallographic data for the SSB–DNA complexes were used to build the models for both binding modes.10 According to these models, in (SSB)65 mode, 65 nucleotides of ssDNA wrap around all monomers of the SSB tetramer, but ssDNA interacts with only two SSB monomers of the tetramer in the (SSB)35 mode. It was hypothesized that the (SSB)35 mode is utilized during DNA replication, whereas the (SSB)65 binding mode is associated with DNA recombination and repair processes.6 Both processes have different types of DNA substrates. DNA replication begins with duplex unwinding, exposing the single strands for SSB loading. The looped and gapped structures are involved in the recombination and repair processes, and SSB binding stabilizes these transient DNA structures. The difference in the position of ssDNA, either at the end of dsDNA (tail-DNA) or between the two dsDNA flanks (gap-DNA), can be a factor for selecting between (SSB)35 and (SSB)65 modes,6 and testing this assumption is one of the goals of this paper. To accomplish this goal and perform the nanoscale study of complexes of SSB with ssDNA under different salt conditions, we employed atomic force microscopy (AFM).
AFM and electron microscopy (EM) are widely used for the nanoscale characterization of SSB–DNA complexes; of particular note, in the pioneering work of Chrysogelos et al.,11 EM was used to assess SSB complexes with fd phage ssDNA as a substrate. The SSB–DNA complexes on these images appeared as beaded loops with approximately 170 nucleotides per loop. In ref 12, EM also was applied to characterize complexes of SSB with ssDNA, but a shorter DNA substrate was used. The cross-linking of the protein to DNA with glutaraldehyde was used in both publications to stabilize SSB–DNA complexes. The advantage of AFM compared with EM is a gentler sample preparation procedure, allowing one to prepare protein–DNA complexes without the cross-linking step. We developed a mica surface functionalization procedure that allowed us to image various protein–DNA complexes over a wide range of ionic strengths and pH values (reviewed in refs 13–15). Moreover, the samples can be imaged in aqueous solutions, so the dynamics of protein–DNA complexes can be imaged using the regular tapping mode of AFM.14,16 Recently, these studies were extended with the use of high-speed AFM (HS-AFM), capable of imaging dynamics of DNA, proteins, and protein–DNA complexes on the millisecond time scale.15,17 AFM images of SSB–DNA complexes were published in ref 18. A circular DNA substrate (M13 ssDNA) was used, and images at high and low ionic strengths were obtained.18 The results were in agreement with the ability of SSB to bind to ssDNA under different ionic conditions by different modes.
Here we used AFM and a set of specially designed DNA substrates to test the effect of tail-DNA or gap-DNA on SSB binding. We followed our recent paper19 in which the hybrid DNA approach was proposed. The hybrid DNA construct consists of a relatively long DNA duplex, to which ssDNA of the desired length is attached. The duplex part of the construct plays the role of a marker, allowing unambiguous identification of nonspecific SSB–DNA complexes. We designed hybrid DNA substrates with 5′- and 3′-ssDNA termini to clarify the role of ssDNA polarity in SSB loading. The hybrid substrates, in which two duplexes are connected with ssDNA, were the models for gapped DNA substrates. The data showed that in the presence of Mg2+ cations, SSB binds very specifically to ssDNA only, with no detectable binding to the duplex segments of the hybrid DNA. Using the set of constructs, we demonstrated that the efficiency of binding of SSB under these conditions to any of the constructs does not depend on the construct type. However, the SSB binding pattern changes at low ionic strengths. In addition to binding to ssDNA segments of the construct, SSB binds to double-stranded DNA forming large associates. These are novel findings, and their implications for SSB–ssDNA interactions are discussed.
MATERIALS AND METHODS
Assembly of Tail-DNA Substrates
The tail-DNA substrate was assembled as described in ref 19 using a DNA adaptor approach and is schematically shown in Figure 1, and the details for the assembly process are shown in Figure S1A of the Supporting Information. Briefly, a restriction DNA fragment (approximately 200–400 bp) with sticky ends was generated. A synthetic oligonucleotide of the required length was annealed with the adaptor to make a sticky end complementary to the end of the DNA restriction fragment. In the experiments, the length of the ssDNA varied between 27 nucleotides (nt) and 69 nt. Annealing was performed in 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, and 100 mM NaCl, with a 1:1 mixture of synthetic oligonucleotides and an adaptor. The annealed product was mixed with the dsDNA restriction fragment in a 5:1 ratio and ligated overnight at 16 °C. The ligation product, hybrid DNA, was separated from the nonligated DNA in a 1.5% agarose gel and purified using a standard Qiagen kit, as described in ref 19. The purified product was resuspended in 10 mM Tris-HCl (pH 7.5) and 1 mM EDTA. With the use of the adaptor approach, the yield of the ligated product was approximately 60%. Direct ligation of the long oligonucleotide to the DNA restriction fragment did not produce the tailed substrate with a reasonable yield. The appropriate design of the adaptors and restriction DNA fragments allowed us to make 3′- and 5′-tail-DNA substrates of different lengths.
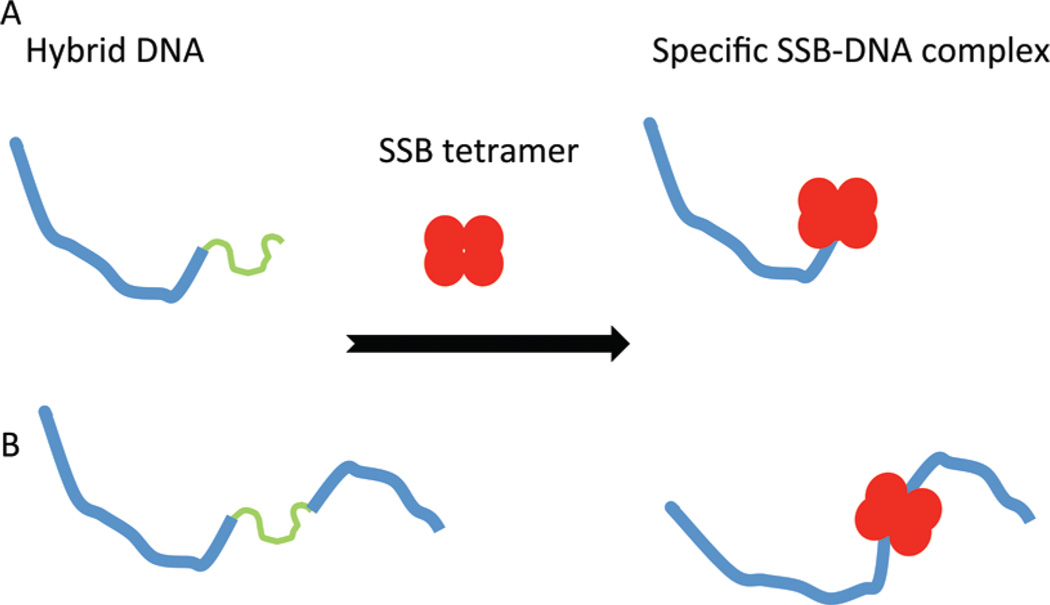
Figure 1.
Schematic presentation of the formation of the complex between the SSB tetramer and (A) the tail-DNA substrate and (B) the gap-DNA substrate. DNA duplexes are shown as blue lines; ssDNA is depicted as green lines, and SSB is shown as red spheres.
Assembly of Gap-DNA Substrates
To make a gap-DNA substrate, we mixed 3′-tail-DNA attached to the 231 bp dsDNA with the 5′-tail-DNA attached to the 441 bp dsDNA in 1:1 ratio and annealed them with a 5-fold excess of the bridging oligonucleotide. A schematic presentation of this design is shown Figure S1B of the Supporting Information. Briefly, one-half of the bridge oligo is complementary to the 5′-tail-DNA sequence, and another half is complementary to the 3′-tail-DNA. The ligation reaction was performed overnight at 16 °C. The ligation mixture was transferred into 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, and 100 mM NaCl, heated to 70 °C, and immediately put into ice, and the product was separated from the unligated products in a 1.5% agarose gel, followed by purification with a Qiagen kit. The final gap-DNA substrate has ssDNA located between the 231 and 441 bp dsDNA flanks.
Preparation of the SSB–Hybrid DNA Complex
Commercially available E. coli SSB purchased from Affymetrix (Santa Clara, CA) was used in the experiments. The complexes with DNA were prepared in a 7:1 protein:DNA ratio in different buffers. Our standard conditions refer to the buffer containing 10 mM Tris-HCl (pH7.4), 50 mM NaCl, 10 mM Mg2+, and 1 mM DTT. For experiments in low-salt solutions, we used the buffer containing 10 mM Tris-HCl (pH 7.4), 20 mM NaCl, and 1 mM DTT. The high-salt buffer contained 10 mM Tris-HCl (pH 7.4), 200 mM NaCl, and 1 mM DTT. Complexes were incubated at room temperature for 10 min followed by purification with a Montage UFC spin column, as described in ref 19.
Sample Preparation and AFM Imaging of the Dry Sample
For AFM sample preparation, the 1-(3-aminopropyl)-silatrane (APS)-functionalized mica (APS-mica) was used as described in ref 16. Five microliters of the sample was deposited on APS-mica for 2 min, rinsed with deionized water, and dried with Ar gas. Images were acquired in tapping mode in air using the Multimode Nanoscope IV system from Bruker (Santa Barbara, CA). Olympus silicon probes (Asylum Research, Santa Barbara, CA) with a 42 N/m spring constant and resonance frequencies between 310 and 340 Hz were used.
Data Analysis
For each SSB–hybrid DNA complex, we measured the length of the double-stranded part of the DNA and the height and diameter of the protein using Femtoscan Online (Advanced technologies Center, Moscow, Russia) as described in refs 19 and 20. The protein volumes were calculated as described in ref 20. The data for measurements of protein volumes were summarized into histograms using Origin version 6.0 (Originlab, Northampton, MA). Each data set contained the measurements for at least 100 complexes.
Sample Preparation for High-Speed AFM (HS-AFM)
The mica (1.5 mm × 1.5 mm) was glued to a glass cylinder attached to the AFM scanner, cleaved, and treated with an APS solution as described above. A drop of the SSB–hybrid DNA complex (2 µL), prepared as described in the section on preparation of SSB–hybrid DNA complexes, was deposited on the mica surface for 5 min. The scanner with the sample was immersed in 200 µL of a buffer solution. All images were taken in the buffer without drying the sample.
HS-AFM Imaging
The HS-AFM images were acquired by using the HS-AFM instrument developed in the Ando group (Kanazawa University, Kanazawa, Japan, and distributed by RIBM Co. Tsukuba, Japan). The data were acquired by operating the instrument in tapping mode in liquid. Silicon nitride AFM probes (BLAC10EGS, Olympus) were etched using the electron beam deposition (EBD) procedure.21 The spring constant of the AFM probes was between 0.1 and 0.2 N/m, with the resonance frequency between 400 and 1000 kHz in water. Continued scanning over the selected area (200 nm × 200 nm) was performed to follow the dynamics of A3G–DNA complexes. The scan rate varied between 720 and 990 ms/image.
RESULTS
Hybrid DNA Construct Approach
To directly visualize the complex of SSBs with ssDNA, we designed a hybrid DNA in which ssDNA was attached to a dsDNA (tail-DNA) or between two dsDNAs (gap-DNA). Schematically, each approach is shown in panel A or B of Figure 1, respectively. If the protein binds specifically to tail-DNA, the complex will attach to only one end of the hybrid DNA (Figure 1A), and specific interactions with gap-DNA will lead to the formation of a complex in which the protein is located between the two dsDNA flanks (Figure 1B).
Effect of the Type of DNA Substrate on Complex Formation
This section describes the results obtained under standard conditions corresponding to room temperature in a solution containing 10 mM Tris-HCl (pH7.4), 50 mM NaCl, 10 mM Mg2+, and 1 mM DTT. Under these conditions, the SSB–DNA complex forms efficiently with all four monomers of SSB in contact with ssDNA.10,22
SSB Binding to 5′-Tail-DNA
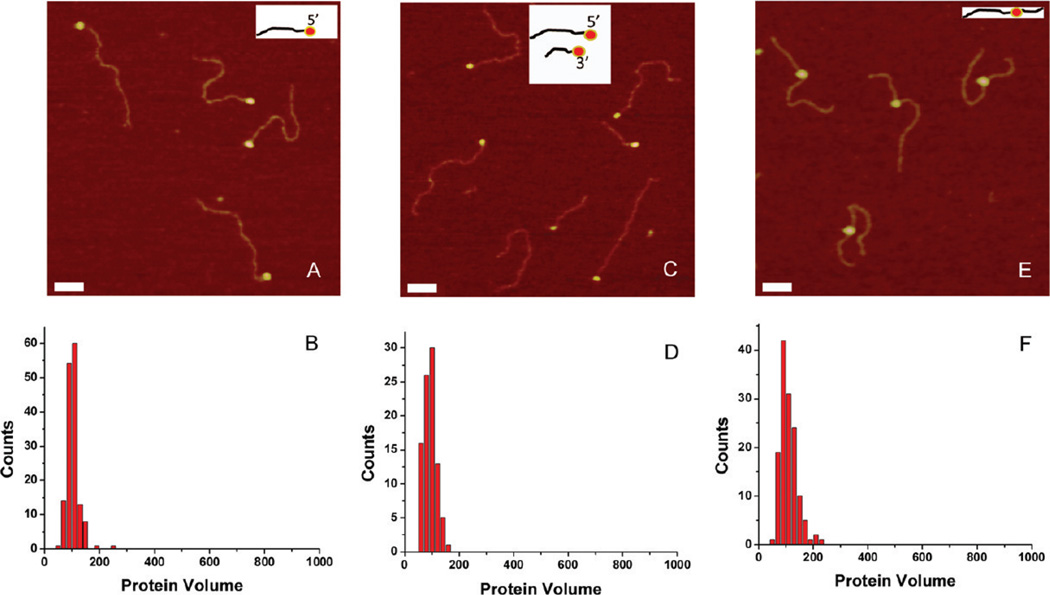
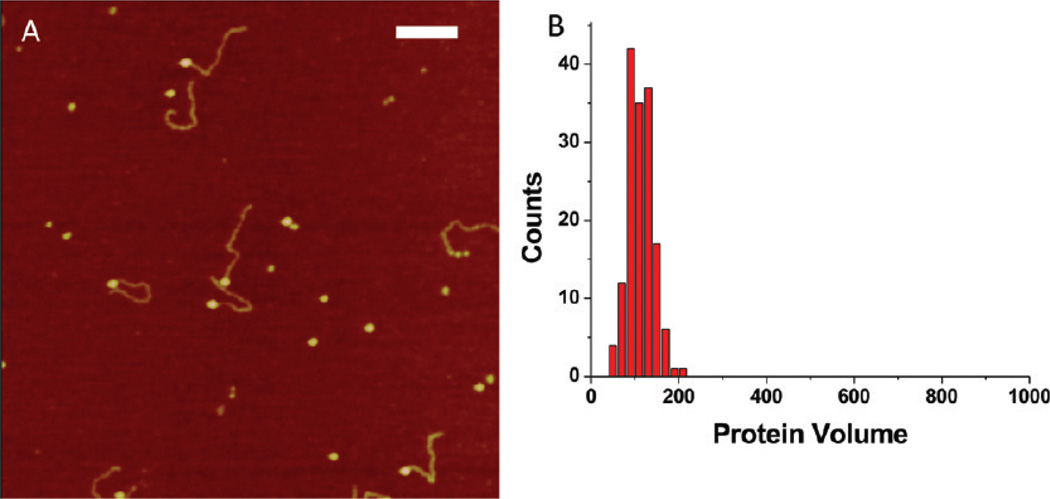
Figure 2A shows AFM images of 69-tail-DNA in complex with SSB. Only one end of the DNA substrate is occupied by the protein, which appears as a bright spherical particle (blob) on the AFM images. The tail-DNA design has one blunt end, and the other end is single-stranded. Because we are able to distinguish between blunt and ssDNA ends of the hybrid DNA,19 we conclude that SSB binds to the ssDNA end of the hybrid DNA. The yield of complexes was close to 90%, with no protein bound to either end or inside the DNA duplex, indicating a very specific binding interaction between the SSB and the ssDNA end of the DNA substrate. The blobs are uniform in size, as supported by the protein volume measurements (Figure 2B), which show a narrow histogram, with a mean value of approximately 100 nm3. These results are consistent with numerous previous data that postulate that SSB under such conditions is a homogeneous homotetramer in complex with ssDNA.7
Figure 2.
AFM data for complexes of SSB with hybrid DNA of different types. (A, C, and E) AFM images of complexes with 5′-tail-DNA, a mixture of 5′-tail-DNA and 3′-tail-DNA, and gap-DNA, respectively. The insets show the schematic presentations of all complexes. Bars are 50 nm. (B, D, and F) Histograms for the protein volumes for 5′-tail-DNA, a mixture of 5′-tail-DNA and 3′-tail-DNA, and gap-DNA, respectively.
SSB Complexes with 3′- and 5′-ssDNA Ends
To examine the effect of polarity of ssDNA ends on SSB binding, we compared the AFM data for two hybrid DNA substrates, terminated with 5′- and 3′-ssDNA ends. The dsDNA parts for both designs were different in length, allowing us to unambiguously distinguish between the two substrates when they were mixed. Figure 2C shows a typical AFM image of the SSB complexes with a 1:1 mixture of such substrates. SSB binds to the ends of long and short DNA substrate complexes, demonstrating unambiguous identification of 5′-bound and 3′-bound complexes. As seen previously (Figure 2A), the protein appears at one end of the substrate, and no nonspecific complexes are seen (Figure 2C). The yields of complexes calculated separately for each substrate were very close to each other (65% for 5′-tail-DNA and 60% for 3′-tail-DNA), suggesting that ssDNA polarity has little or no effect on SSB binding. We also measured the sizes of protein complexes for both substrates. The results for the volume measurements for 3′-tail-DNA complexes are shown in Figure 2D. The histogram is very close to the histogram shown in Figure 2B for 5′-tail-DNA, with the maximum of approximately 100 nm3. These data demonstrate that the stoichiometry of the complexes also does not depend on the polarity of ssDNA ends.
SSB Complexes with the Gap-DNA Substrate
To examine the effect of ssDNA ends on SSB loading efficiency, we designed a substrate (gap-DNA) in which 69-tail-DNA is placed between the two 231 and 441 bp DNA duplexes. AFM images for the complex of SSB with the gap-DNA are shown in Figure 2E. In these images, the protein is located between two dsDNA arms and the length measurements confirmed that SSB occupies the internal ssDNA region. There are no end-bound complexes in this case, which is again in line with the high specificity of SSB for ssDNA. The yield of the complexes (75%; N = 137) is very close to that for tailed substrates (80%; N = 150), suggesting that neither the polarity of the ssDNA substrate nor the availability of the ends is critical for the formation of the complex of SSB with ssDNA. The protein volume measurements (Figure 2F) confirm the homogeneity of the protein stoichiometry in complexes.
Effect of Salts on SSB–ssDNA Complexes
Complexes under Low-Salt Conditions
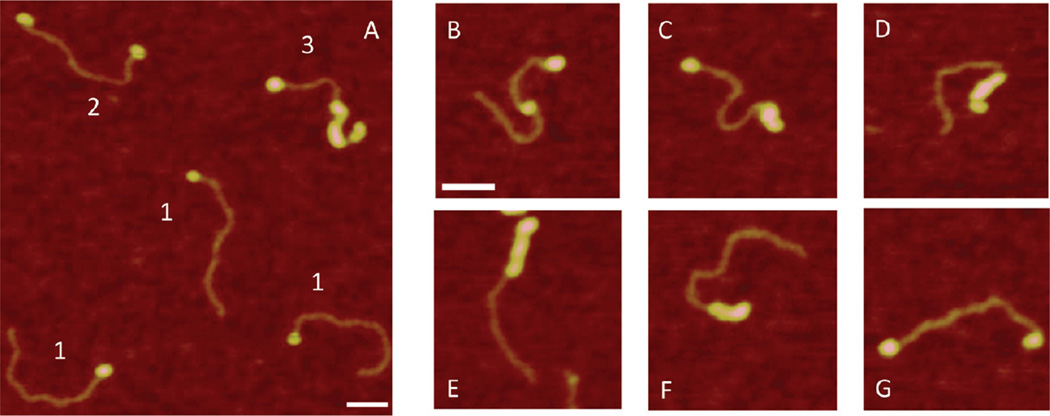
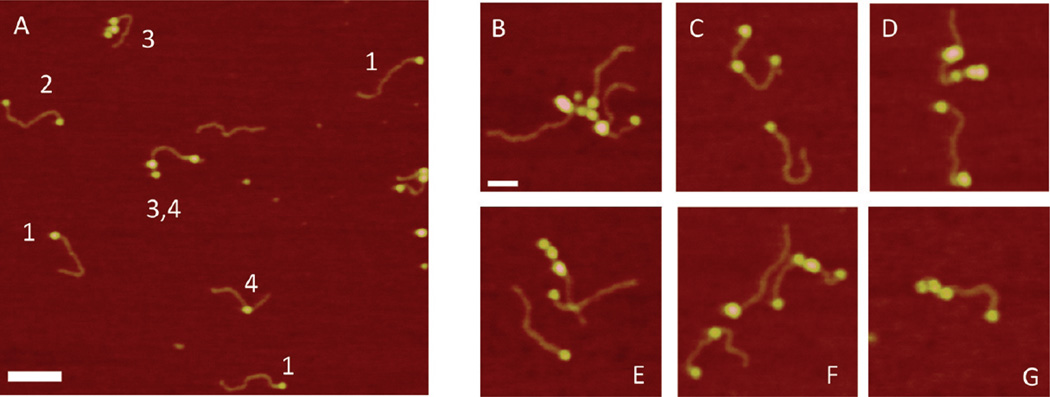
All experiments described above were conducted in the presence of Mg2+. To determine the effect of Mg2+ cations on protein binding, we imaged complexes of SSB under low-salt conditions without Mg2+ cations. According to ref 7, under these conditions, the binding mode of SSB changes, so only two monomers of the SSB tetramer are in contact with ssDNA. For experiments in the low-salt solutions, we used the buffer containing 10 mM Tris-HCl (pH 7.4), 20 mM NaCl, and 1 mM DTT. Figure 3 shows the data obtained with the 69-tail-DNA substrate. A large-scale image containing a set of complexes is shown in Figure 3A. A striking difference between this image and the previous images shown in Figure 2 is the appearance of complexes with various morphologies and their location. In addition to the end-bound complexes, indicated with 1, complexes with two blobs attached to both ends of the DNA (indicated with 2) and proteins assembled in clusters (indicated with 3) were observed. The gallery of images of individual complexes of various types is shown in Figure 3B–G. Additional AFM images for complexes of SSB with 69-tail-DNA under low-salt conditions are shown in Figure S2 of the Supporting Information. The protein blobs can appear at both ends (Figure 3C,G), inside the duplex (Figure 3B), and as clusters of blobs, which bind the dsDNA region of the substrate (Figure 3C–F). We found that the size of the protein varies dramatically. In addition to the appearance of isolated blobs at the end (Figure 3G) or inside the DNA duplex (Figure 3 B), much larger blobs and clusters appear (Figure 3C–F). These data suggest that under low-salt conditions, SSB, in addition to specifically binding to ssDNA, is capable of binding to dsDNA and forming clusters. We termed these complexes nonspecific complexes. Note that the formation of clusters does not change the contour length of the DNA duplex. This is confirmed by measuring the length of the DNA in the complexes (see Figure S2a–c of the Supporting Information). AFM analysis revealed that complexes specifically bound to one end comprised only 60% of all complexes. The breakdown for nonspecific complexes is as follows: two-end-bound, 20%; middle-bound, 11%; and clusters, 13%. Figure S3A of the Supporting Information summarizes the yield of specific and nonspecific complexes under these conditions.
Figure 3.
AFM images of complexes of 69-tail-DNA with SSB assembled under low-salt conditions. (A) Large-scale AFM image with different types of complexes. Different types of complexes are indicated with 1–3. (B–G) Gallery of AFM images of SSB–DNA complexes of various morphologies. Bars are 50 nm.
SSB–DNA Complexes under High-Salt Conditions
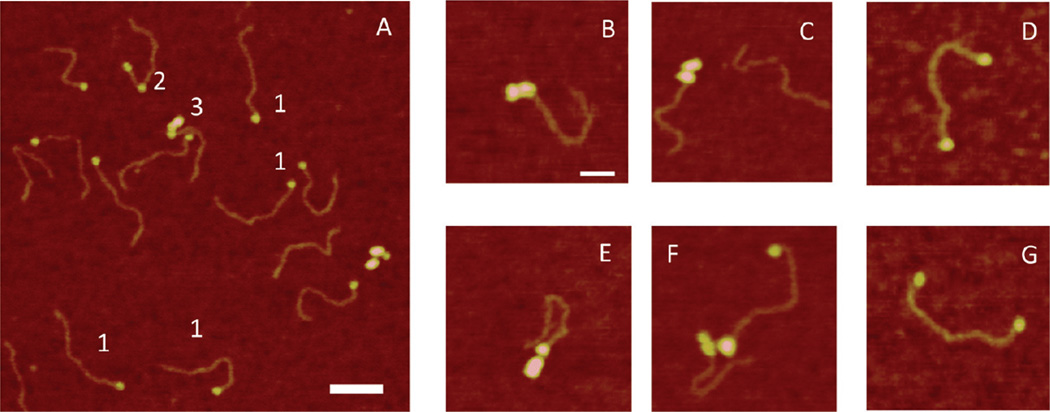
To examine the effect of elevated concentrations of NaCl on SSB–DNA interactions, we assembled the complexes in the buffer containing 10 mM Tris-HCl (pH7.4), 200 mM NaCl, and 1 mM DTT. A typical AFM image for such a sample is shown in Figure 4A, with selected zoomed images with clusters and two-end binding shown in panels B–G of Figure 4. Although the majority of complexes are the end-bound specific complexes, nonspecific complexes are seen in the image. They constitute 11% of all complexes, comprised of 7% nonspecific two-end-bound complexes and 2% of both middle-bound complexes and clusters (see the diagram in Figure S3B of the Supporting Information)
Figure 4.
AFM images of complexes of SSB formed with the hybrid 69-tail-DNA under high-salt conditions. (A) Typical large-scale image on which different morphologies of complexes are indicated with 1–3. The bar is 100 nm. (B–G) Gallery of images of SSB–DNA complexes of various morphologies. The bar is 50 nm.
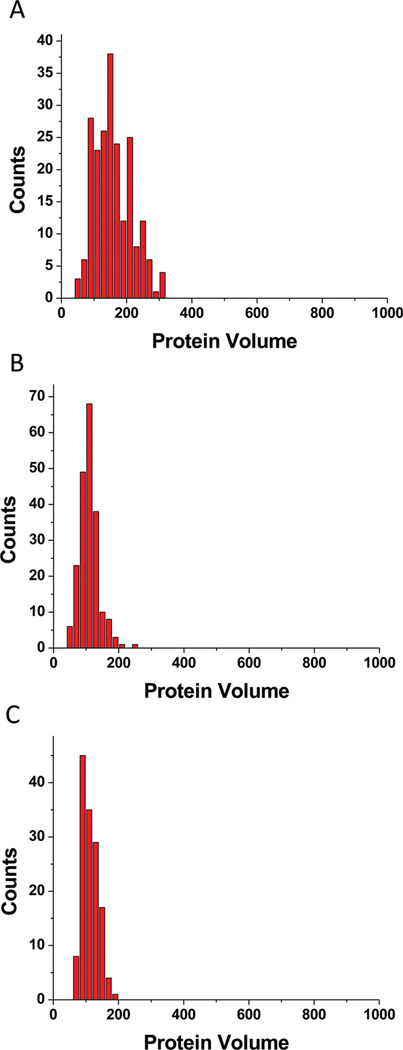
We utilized the ability of AFM to provide the molecular mass of proteins in protein–DNA complexes (e.g., refs 14, 19, 20, and 23 and references therein) to compare the stoichiometries of SSB bound to ssDNA under different ionic conditions. In this analysis, we measured volumes of the protein for the end-bound specific complexes only. The results of the volume measurements for low and high ionic strengths are presented as histograms in panels A and B of Figure 5, respectively. The distribution of protein volume for the data obtained at low ionic strengths (Figure 5A) is considerably broader than those obtained at high ionic strengths (Figure 5B). The larger size of SSB assembled on ssDNA at low ionic strengths is consistent with earlier observations of SSB clustering under these conditions.24 Although the distribution of the protein volume under the high-ionic strength condition is close to the distribution for specific binding under standard conditions, some nonspecific binding and clusters are still observed (Figure 4A–G). Such nonspecific binding was completely eliminated when 10 mM Mg2+ cations were added; only specific complexes were formed. The protein volume histogram (Figure 5C) is narrow and practically identical to that for standard conditions (Figure 2). These results directly demonstrate the critical role of Mg2+ cations in the formation of specific complexes of SSB with ssDNA, regardless of the ionic strength.
Figure 5.
Histograms of the volumes of the end-bound complexes only formed under low-salt conditions (A), high-salt conditions (B), and low-salt conditions with Mg2+ (C).
Effect of ssDNA Length on the Morphology of SSB Complexes
To understand how the size of ssDNA contributes to the properties of SSB–DNA complexes, we imaged the complexes of SSB with ssDNA with as few as 27 nucleotides. Figure 6A shows a typical AFM image of the complex assembled under standard conditions [10 mM Tris-HCl (pH 7.4), 50 mM NaCl, 10 mM Mg2+ cations, and 1 mM DTT]. Only specific complexes appear. The volume measurements produced a narrow distribution (Figure 6B) that is very similar to those obtained for the 69-tail-DNA substrate, with the maximum corresponding to the volume of a SSB tetramer (Figure 2).
Figure 6.
AFM data for complexes of SSB with the 27-tail-DNA substrate prepared under standard conditions (TE buffer, 50 mM NaCl and 10 mM Mg2+). (A) Typical AFM image of the complex. The bar is 100 nm. (B) Histogram of the complex volumes.
The data for low-ionic strength conditions for this short ssDNA substrate are shown in Figure 7A. As shown in the results for the longer substrate (Figure 3), nonspecific complexes are formed. A gallery of such images is shown in Figure 7B–G. The yield of nonspecific complexes is 35%, including 12% of the two-end-bound complexes, 17% of the middle-bound complexes, and 6% of the clusters (see the graph for the yield of the complexes in Figure S4A of the Supporting Information). The data for the high-salt conditions demonstrate the increase in the specificity of binding of SSB to ssDNA, although nonspecific complexes appear with a 10% yield with the following breakdown for different morphologies: two-end-bound, 7%; middle-bound, 3%; clusters, 1% (see Figure S4B of the Supporting Information).
Figure 7.
AFM data for complexes of SSB with the 27-tail-DNA substrate prepared under low-salt conditions. (A) Typical AFM image in which various types of complexes are numbered. The bar is 100 nm. (B–G) Gallery of selected AFM images of various complexes. The bar is 50 nm.
The data for the volume measurements obtained for the set of AFM images acquired at low and high ionic strengths are shown in Figure S5 of the Supporting Information. The measurements were performed for the specific end-bound complexes only. The histograms are rather narrow, although the distributions are skewed toward large volumes, indicating the formation of large complexes. Such large complexes are not observed for the complex assembled in the presence of Mg2+ cations (Figure 6B).
Additional evidence of the ability of SSB to bind to double-stranded DNA under low-salt conditions was provided by experiments performed with DNA duplexes containing no single-stranded regions. The data, shown in Figure S6A–F of the Supporting Information, demonstrate that the SSB blobs appear in various locations on the DNA duplex, and the size of the blobs varies. Images illustrate end-binding complexes, middle-binding complexes, and clusters of SSB. No complexes are formed if Mg2+ cations are added to the buffer (see Figure S7A,B of the Supporting Information).
To evaluate how the ionic conditions contribute to protein–protein interactions, we measured the volume of the protein in the absence of DNA under different ionic conditions. The results of the volume measurements are shown in Figure S8A–C of the Supporting Information. According to these data, the size distribution for SSB is narrow in the presence of Mg2+ cations or under high-salt conditions (panels A and B of Figure S8, respectively), but the distribution is very wide for low salt concentrations (Figure S8C), suggesting that the protein tends to aggregate under such conditions.
Dynamics of SSB–DNA Complexes Assessed with HS-AFM
In this section, we describe the results of direct imaging of the dynamics of SSB–DNA complexes obtained with the use of HS-AFM. This emerging AFM instrumentation provides high-resolution images of the sample in a fully hydrated state, allowing one to visualize the nanoscale dynamics of the system on the millisecond time scale (reviewed in refs 15 and 21). We used 69-tail-DNA and gap-DNA substrates of different sizes and performed imaging of SSB–DNA complexes under standard conditions (Tris-HCl, 50 mM NaCl, and 10 mM Mg2+). Images were recorded with a data acquisition rate of >1 s/frame (720–990 ms). Figures below show selected frames from a full set of data, comprising dozens of such frames that are assembled as movie files in the Supporting Information.
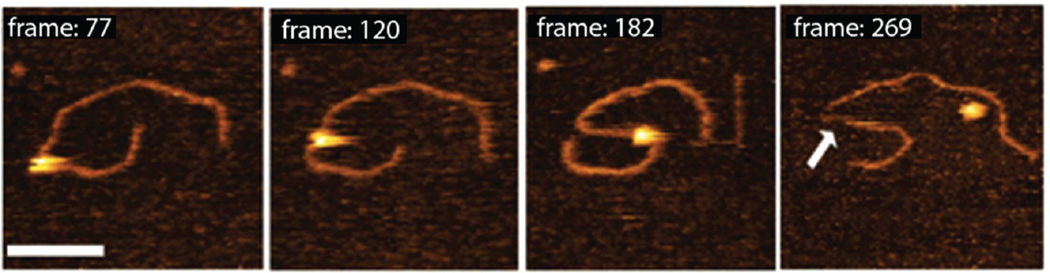
The set of a few frames for the dynamics of the complex formed with a gap substrate is shown in Figure 8, and the movie file can be viewed as Supporting Information (Movie 1). The first image in Figure 8 (frame 77) shows SSB located between two DNA duplexes. The protein remains bound to the gap regardless of DNA segmental mobility (frame 120). A dramatic change in the overall DNA shape (frame 182) did not change the protein position; it remained bound to the same location on the substrate. Eventually, the protein dissociates, as is demonstrated by the last frame in the movie, exposing the single-stranded gap between two dsDNA arms, as shown by the arrow in Figure 8 (frame 269). Note that under these conditions, SSB dissociates from the substrate in one step. The analysis of these data confirmed that SSB binds ssDNA as a tetramer and remains a tetramer after dissociation from the DNA. The slightly smaller size of the free SSB (frame 269) is explained by the contribution of ssDNA to the volume of the complex.
Figure 8.
High-speed AFM data with a few selected frames illustrating the dynamics of complexes of SSB with 69-gap-DNA, prepared under standard conditions. Each frame is 990 ms. The bar is 50 nm. The arrow points to the exposed ssDNA region appearing after protein dissociation.
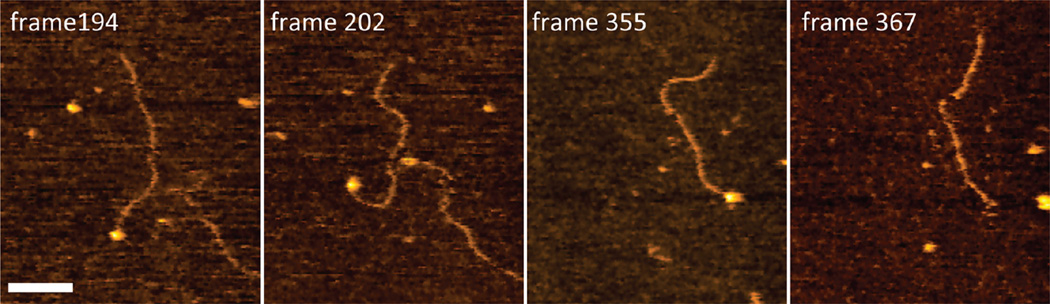
The dynamics of SSB bound to tail-DNA is demonstrated by the set of images in Figure 9 (see Movie 2 of the Supporting Information). In this figure, one end-bound complex is seen in frame 194. Frames 202 and 355 demonstrate the stable attachment of SSB to the DNA end. The complex remains stable for almost 10 s before it fully dissociates, as can be seen in frame 367. Analysis of the protein volume again confirms that SSB has a tetrameric structure and dissociates from the ssDNA as a tetramer. In the majority of cases under these conditions, SSB dissociates from DNA in a one-step fashion, although a few cases show two-step dissociation (Movie 3 of the Supporting Information). In this movie, SSB is initially bound to the end of the DNA as a tetramer, and then at some point, it splits into two dimers, with one dimer remaining attached to the DNA for a rather long time. The free dimer moves around the DNA and finally comes close to the DNA and forms a tetramer of SSB again. The tetramer of SSB then splits again into two dimers, and finally, after a while, the last dimer also dissociates from the DNA.
Figure 9.
High-speed AFM data with a few selected frames illustrating the dynamics of complexes of SSB with 69-tail-DNA prepared under standard conditions. Each frame is 720 ms. The bar is 50 nm.
DISCUSSION
In this study, we have used AFM to examine the role of cations in the structural properties of SSB in complex with ssDNA using specially designed hybrid DNA. The primary property of SSB is binding to single-stranded DNA; therefore, the main goal was to elucidate the extent to which environmental conditions affect this property of the protein. As a DNA-binding protein, SSB may also bind double-stranded DNA. We termed such complexes nonspecific ones, and our experiments allowed us to clarify this property of SSB, as well. We elaborate on each issue below.
Specificity of SSB Binding
The use of hybrid DNA substrates allowed us to directly discriminate and identify specific binding of SSB from nonspecific binding. Highly specific binding is illustrated by images in Figure 2; this specificity requires Mg2+ cations. In the presence of Mg2+, regardless of ionic strength, SSB binds exclusively to single-stranded regions of the DNA substrate. Moreover, the protein retains its tetrameric stoichiometry as evidenced by the volume measurements. The specificity is partially lost in the absence of Mg2+ cations, even under high-salt conditions. However, under low-salt conditions, the specificity of binding decreases substantially. Under these conditions, the protein binds to blunt ends of the DNA and inside of the duplex regions (Figure 3). Importantly, the morphology of the protein bound to DNA specifically in low-salt solutions changes dramatically. The individual blobs are much larger than the blobs corresponding to specific binding in the presence of Mg2+ cations. Volume measurements of the end-bound single blobs suggest an octameric morphology of SSB, and this finding is in line with the model for the low-salt (SSB)35 binding mode, in which two SSB tetramers bind to one ssDNA target.25 Moreover, SSB can form clusters in which several adjacent large blobs occupy the DNA duplex, and this side-by-side morphology is similar to that observed with AFM for SSB assembled on long ssDNA under low-salt conditions.18 The 10-fold increase in ionic strength increases SSB specificity, although approximately 10% of nonspecific complexes still can be found (Figure 4 and Figure S3B of the Supporting Information).
Overall, these data suggest that the specificity of binding of SSB to ssDNA strongly depends on ionic conditions. The high-salt conditions are close to physiological conditions and provide specificity for SSB binding, but the stringency of the interaction is provided by Mg2+ cations.
Modes of Binding of SSB to the DNA Substrates
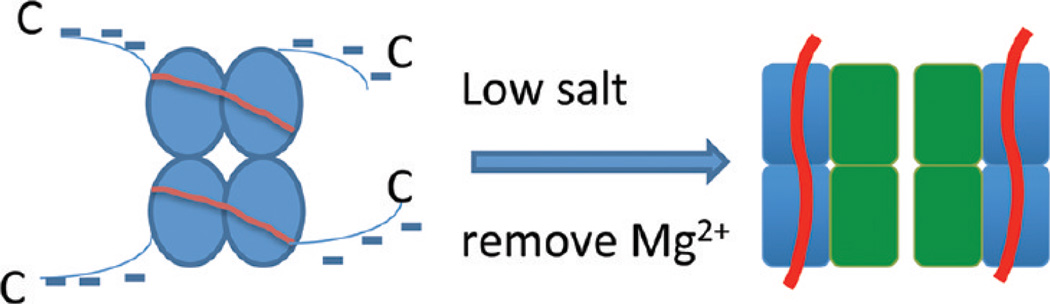
It was proposed that SSB binds to the DNA substrate by two major modes, (SSB)35 and (SSB)65 (see a review, see ref 6 and references therein). The models were built on the basis of crystallographic data obtained for the complex of the DNA binding domain of SSB with 35 nt ssDNA.10 According to this crystallographic structure, SSB forms a tetramer with two symmetrically arranged dimers, and the two 35 nt DNA substrates wrap around all four monomeric units of the tetramer. This model was called the (SSB)65 mode of SSB binding. It was postulated that in low-salt solutions, the same protein tetramer could bind a 35 nt DNA substrate [(SSB)35 mode] with only two subunits of SSB in contact with the DNA substrate.10 However, the models for the SSB–DNA complexes utilized structural data obtained for the DNA binding domain of the protein rather than for the entire protein. Although this domain (SSBc) comprises the essential N-terminal part of the protein (residues 1–135), the unstructured C-terminus of SSB plays an important role in the interaction of SSB with other proteins involved in various DNA transactions, as reviewed in ref 6. The model for the full SSB–ssDNA complex proposed in that paper suggests that the C-terminal segments of the protein are located at the periphery of the tetramer and are not involved in the interaction of the protein with DNA. An important property of C-termini is that they contain three acidic Asp residues at the end, rendering the entire C-terminus highly acidic. In such a model, schematically shown in Figure 10, electrostatic repulsion prevents the interaction between the SSB tetramers, which is in agreement with numerous data (including this work), suggesting the lack of association of SSB tetramers at high ionic strengths. Importantly, the electrostatic repulsion between the tetramers is not compensated by divalent Mg2+ cations. Moreover, in their presence, the specificity of binding of SSB to ssDNA is maximal, with no tendency for the SSB tetramer to associate. Thus, the proposed model for the tetrameric SSB with an exterior location of negatively charged C-termini explains the integrity of the tetrameric SSB.
Figure 10.
Schematic illustration of the salt-dependent conformational transition of SSB. Proteins are shown by ellipses of different shapes to illustrate the difference in the protein conformations under both conditions. At the left is the specific SSB–ssDNA complex. The protein C-termini are shown with blue lines. DNA is shown with red lines. At the right is the low-salt SSB–ssDNA complex. One dimer is shown in complex with DNA (red line). DNA is shown with red lines.
Given the fact that the negatively charged C-termini prevent the association of the tetramers at high ionic strengths, the electrostatic effect should be even stronger at lower ionic strengths. However, observations suggest otherwise. The protein associates into larger structures under low-salt conditions. Therefore, we hypothesize that SSB under low-salt conditions undergoes a structural transition in which the negative charge of the C-termini is compensated through the interaction with positively charged segments of the N-terminus of SSB. Schematically, the salt-dependent conformational transition of SSB is depicted in Figure 10. In this conformation, SSB is capable of assembling into larger oligomers; however, the protein in this conformation losses its ssDNA binding specificity. The volume measurements of the free protein demonstrate that associates as large as SSB octamers are the largest SSB associates of the free protein; therefore, it is reasonable to assume that SSB associates into dimers of tetramers. Although the assembly of SSB in large associates is observed in the absence of DNA, we cannot exclude the possibility that SSB tetramers can associate further on the DNA substrate. Because of interprotein interactions, SSB on DNA can form long side-by-side arrays that are visualized in AFM images and appear in high yields.
Thus, the conformation of SSB depends on ionic strength. Most importantly, the SSB binding specificity strongly depends on protein conformation. The specific ssDNA–SSB complexes are formed under high-salt conditions corresponding to physiological conditions, and this inherent protein property is lost in low-salt solutions.
SSB Binding to Various Types of ssDNA Substrates
The availability of hybrid DNA substrates of various types allowed us to test the hypothesis that SSB binding differs according to the cellular DNA process it is mediating. We did not find differences in the binding efficiency between gap-DNA and tail-DNA, and the SSB stoichiometry was identical for both substrates (Figure 2). The experiments were performed in the presence of Mg2+ cations in which the protein binds very specifically to ssDNA. The polarity of the ssDNA end for tail-DNA substrates also does not play an essential role. The preference for the 3′-end was described in ref 26. However, a short 30 nt substrate that is more than 2 times shorter than the ssDNA required for the formation of the (SSB)65 complex was used in that paper. There also might be the effect of the ssDNA sequence on complex formation. Note that sequences used in ref 26 and in our paper are different. Both issues are important for further understanding the mechanism of SSB–DNA interaction and require special attention. Altogether, these data suggest that the SSB binding modes during DNA repair (gap-substrate) and replication (tail-substrate) are similar and SSB binds to the substrates primarily as a tetramer.
Dynamics of SSB–DNA Complexes
The time-lapse AFM images revealed that SSB binds to the substrate strongly and remains specifically bound to the ssDNA target region during the long observation period in which the DNA undergoes extensive segmental motion. This high stability of the SSB–DNA complex reflects the high yield of complexes observed with AFM and is consistent with the analyses of thermodynamics and dissociation kinetics.27 The dissociation of the protein was also observed in the HS-AFM time-lapse experiments and occurs rapidly, between two adjacent time frames; i.e., the dissociation time is <1 s. The temperature jump experiments showed that the SSB unwrapping dynamics span between a few to ten milliseconds,27 which is in line with our estimates. Primarily, the protein dissociates from the DNA in the same morphology as it was in the complex, which according to the volume measurements, is a tetramer. However, we also observed two-step dissociation events in which one SSB dimer dissociates and another one remains bound to the same position on the DNA substrate. An example of this dissociation event is shown in Movie 3 of the Supporting Information. The stability of the dimeric SSB–ssDNA complexes is relatively high, so the complex remained stable over dozens of frames of the time-lapse experiments. These observations suggest that the SSB dimer is capable of binding ssDNA, although this is not a primary mode of SSB–DNA interaction.
The tip–sample interaction can contribute to the dynamics of protein–DNA complexes and can facilitate SSB dissociation. However, as we discussed previously,17 because of a low value of the tip oscillation amplitude (~1 nm) and a very short interval of the tip–sample contact (~100 ns), the effect of the tip on the sample dynamics in the HS-AFM experiments is negligible. Moreover, along with dissociation of SSB, we observed association events, which provides additional evidence of the gentle operation of the HS-AFM instrument.
Hybrid DNA Substrates for Other Types of SSBs
SSBs are key players in all DNA transactions, such as replication, recombination, and repair.1 Of particular note, a class of DNA-modifying enzymes, the deaminases, operate on ssDNA only.28 The mechanistic understanding of their functions is modest, which primarily is due to the lack of appropriate experimental approaches. The methodology described here fills this gap and provides the foundation for nanoscale studies of SSBs in complexes with DNA. Indeed, typical substrates for SSB are single-stranded regions at the ends of the DNA duplex or gaps within the duplexes, as exemplified by the model hybrid DNA substrates described in this work. Importantly, SSB typically works in concert with other structural proteins and enzymes, and the proposed technology can be modified for more complex systems. We applied AFM for the nanoscale studies, but the approach is fully applicable for studies of the complexes by electron microscopy.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. Bianco for critical review of the manuscript and useful suggestions and the Lyubchenko lab members for the discussion of the results and critical comments.
Funding
The work was supported by National Institutes of Health Grants 1P01GM091743-01A1 and 1 R01 GM096039-01A1, U.S. Department of Energy Grant DE-FG02-08ER64579, and a Nebraska Research Initiative grant to Y.L.L.
ABBREVIATIONS
- AFM
atomic force microscopy
- ssDNA
single-stranded DNA
- dsDNA
double-stranded DNA
- APS
1-(3-aminopropyl)-silatrane
- SSB
single-stranded DNA-binding protein
- nt
nucleotide
Footnotes
ASSOCIATED CONTENT
Supporting Information
Additional information for Materials and Methods, schematics explaining the design of the DNA substrate, AFM images of SSB–DNA complexes, graphs characterizing SSB–DNA complexes, and movies of animated time-lapse AFM images. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes
The authors declare no competing financial interest.
REFERENCES
- 1.Kur J, Olszewski M, Dlugolecka A, Filipkowski P. Single-stranded DNA-binding proteins (SSBs): Sources and applications in molecular biology. Acta Biochim. Pol. 2005;52:569–574. [PubMed] [Google Scholar]
- 2.Krauss G, Sindermann H, Schomburg U, Maass G. Escherichia coli single-strand deoxyribonucleic acid binding protein: Stability, specificity, and kinetics of complexes with oligonucleotides and deoxyribonucleic acid. Biochemistry. 1981;20:5346–5352. doi: 10.1021/bi00521a040. [DOI] [PubMed] [Google Scholar]
- 3.Dillingham MS, Tibbles KL, Hunter JL, Bell JC, Kowalczykowski SC, Webb MR. Fluorescent single-stranded DNA binding protein as a probe for sensitive, real-time assays of helicase activity. Biophys. J. 2008;95:3330–3339. doi: 10.1529/biophysj.108.133512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McEntee K, Weinstock GM, Lehman IR. recA protein-catalyzed strand assimilation: Stimulation by Escherichia coli single-stranded DNA-binding protein. Proc. Natl. Acad. Sci. U.S.A. 1980;77:857–861. doi: 10.1073/pnas.77.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochkarev A, Bochkareva E. From RPA to BRCA2: Lessons from single-stranded DNA binding by the OB-fold. Curr. Opin. Struct. Biol. 2004;14:36–42. doi: 10.1016/j.sbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein: Multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 8.Ryzhikov M, Koroleva O, Postnov D, Tran A, Korolev S. Mechanism of RecO recruitment to DNA by single-stranded DNA binding protein. Nucleic Acids Res. 2011;39:6305–6314. doi: 10.1093/nar/gkr199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Overman LB, Lohman TM. Linkage of pH, anion and cation effects in protein-nucleic acid equilibria Escherichia coli SSB protein-single stranded nucleic acid interactions. J. Mol. Biol. 1994;236:165–178. doi: 10.1006/jmbi.1994.1126. [DOI] [PubMed] [Google Scholar]
- 10.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 11.Chrysogelos S, Griffith J. Escherichia coli single-strand binding protein organizes single-stranded DNA in nucleosome-like units. Proc. Natl. Acad. Sci. U.S.A. 1982;79:5803–5807. doi: 10.1073/pnas.79.19.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider RJ, Wetmur JG. Kinetics of transfer of Escherichia coli single strand deoxyribonucleic acid binding protein between single-stranded deoxyribonucleic acid molecules. Biochemistry. 1982;21:608–615. doi: 10.1021/bi00533a002. [DOI] [PubMed] [Google Scholar]
- 13.Lyubchenko YL. DNA structure and dynamics: An atomic force microscopy study. Cell Biochem. Biophys. 2004;41:75–98. doi: 10.1385/CBB:41:1:075. [DOI] [PubMed] [Google Scholar]
- 14.Lyubchenko YL, Shlyakhtenko LS. AFM for analysis of structure and dynamics of DNA and protein-DNA complexes. Methods. 2009;47:206–213. doi: 10.1016/j.ymeth.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyubchenko YL, Shlyakhtenko LS, Ando T. Imaging of nucleic acids with atomic force microscopy. Methods. 2011;54:274–283. doi: 10.1016/j.ymeth.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyubchenko YL. Preparation of DNA and nucleoprotein samples for AFM imaging. Micron. 2011;42:196–206. doi: 10.1016/j.micron.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyagi A, Ando T, Lyubchenko YL. Dynamics of nucleosomes assessed with time-lapse high-speed atomic force microscopy. Biochemistry. 2011;50:7901–7908. doi: 10.1021/bi200946z. [DOI] [PubMed] [Google Scholar]
- 18.Hamon L, Pastre D, Dupaigne P, Le Breton C, Le Cam E, Pietrement O. High-resolution AFM imaging of single-stranded DNA-binding (SSB) protein-DNA complexes. Nucleic Acids Res. 2007;35:e58. doi: 10.1093/nar/gkm147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shlyakhtenko LS, Lushnikov AY, Li M, Lackey L, Harris RS, Lyubchenko YL. Atomic force microscopy studies provide direct evidence for dimerization of the HIV restriction factor APOBEC3G. J. Biol. Chem. 2011;286:3387–3395. doi: 10.1074/jbc.M110.195685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shlyakhtenko LS, Gilmore J, Kriatchko AN, Kumar S, Swanson PC, Lyubchenko YL. Molecular mechanism underlying RAG1/RAG2 synaptic complex formation. J. Biol. Chem. 2009;284:20956–20965. doi: 10.1074/jbc.M109.028977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uchihashi T, Ando T. High-speed atomic force microscopy and biomolecular processes. Methods Mol. Biol. 2011;736:285–300. doi: 10.1007/978-1-61779-105-5_18. [DOI] [PubMed] [Google Scholar]
- 22.Lohman TM, Overman LB, Datta S. Salt-dependent changes in the DNA binding co-operativity of Escherichia coli single strand binding protein. J. Mol. Biol. 1986;187:603–615. doi: 10.1016/0022-2836(86)90338-4. [DOI] [PubMed] [Google Scholar]
- 23.Shlyakhtenko LS, Lushnikov AY, Lyubchenko YL. Dynamics of nucleosomes revealed by time-lapse atomic force microscopy. Biochemistry. 2009;48:7842–7848. doi: 10.1021/bi900977t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bujalowski W, Overman LB, Lohman TM. Binding mode transitions of Escherichia coli single strand binding protein-single-stranded DNA complexes. Cation, anion, pH, and binding density effects. J. Biol. Chem. 1988;263:4629–4640. [PubMed] [Google Scholar]
- 25.Roy R, Kozlov AG, Lohman TM, Ha T. Dynamic structural rearrangements between DNA binding modes of E. coli SSB protein. J. Mol. Biol. 2007;369:1244–1257. doi: 10.1016/j.jmb.2007.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delagoutte E, Heneman-Masurel A, Baldacci G. Single-stranded DNA binding proteins unwind the newly synthesized double-stranded DNA of model miniforks. Biochemistry. 2011;50:932–944. doi: 10.1021/bi101583e. [DOI] [PubMed] [Google Scholar]
- 27.Kuznetsov SV, Kozlov AG, Lohman TM, Ansari A. Microsecond dynamics of protein-DNA interactions: Direct observation of the wrapping/unwrapping kinetics of single-stranded DNA around the E. coli SSB tetramer. J. Mol. Biol. 2006;359:55–65. doi: 10.1016/j.jmb.2006.02.070. [DOI] [PubMed] [Google Scholar]
- 28.Pham P, Calabrese P, Park SJ, Goodman MF. Analysis of a single-stranded DNA-scanning process in which activation-induced deoxycytidine deaminase (AID) deaminates C to U haphazardly and inefficiently to ensure mutational diversity. J. Biol. Chem. 2011;286:24931–24942. doi: 10.1074/jbc.M111.241208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.