Summary
Background
Androgen deprivation therapy (ADT) often worsens fatigue in patients with prostate cancer, producing symptoms similar to Chronic Fatigue Syndrome (CFS). Comparing expression (mRNA) of many fatigue-related genes in patients with ADT-treated prostate cancer versus with CFS versus healthy controls, and correlating mRNA with fatigue severity may clarify the differing pathways underlying fatigue in these conditions.
Methods
Quantitative real-time PCR was performed on leukocytes from 30 fatigued, ADT-treated prostate cancer patients (PCF), 39 patients with CFS and 22 controls aged 40–79, together with ratings of fatigue and pain severity. 46 genes from these pathways were included: 1) adrenergic/monoamine/neuropeptides, 2) immune, 3) metabolite-detecting, 4) mitochondrial/energy, 5) transcription factors.
Results
PCF patients showed higher expression than controls or CFS of 2 immune transcription genes (NR3C1 and TLR4), chemokine CXCR4, and mitochondrial gene SOD2. They showed lower expression of 2 vasodilation-related genes (ADRB2 and VIPR2), 2 cytokines (TNF and LTA), and 2 metabolite-detecting receptors (ASIC3 and P2RX7). CFS patients showed higher P2RX7 and lower HSPA2 versus controls and PCF. Correlations with fatigue severity were similar in PCF and CFS for only DBI, the GABA-A receptor modulator (r=−0.50, p<0.005 and r=−0.34, p<0.05). Purinergic P2RY1 was correlated only with PCF fatigue and pain severity (r= +0.43 and +0.59, p=0.025 and p=0.001).
Conclusions
PCF patients differed from controls and CFS in mean expression of 10 genes from all 5 pathways. Correlations with fatigue severity implicated DBI for both patient groups and P2RY1 for PCF only. These pathways may provide new targets for interventions to reduce fatigue.
Keywords: prostate cancer, Chronic Fatigue Syndrome, fatigue, qPCR, gene expression purinergic, GABA-receptor modulator, cytokine
With 98% of prostate cancer patients surviving for 5 years or more after diagnosis and initial treatment, it is increasingly important to determine the physiological pathways underlying symptoms that affect quality of life (QOL) in these survivors, and to improve medical management of these symptoms. One symptom with a powerful impact on QOL is cancer-related fatigue (CRF), defined by the National Comprehensive Cancer Network as a persistent subjective sense of tiredness that interferes with daily functioning, is not proportional to activities, is not fully relieved by rest, and results in a chronic state of exhaustion. CRF is reported to seriously affect QOL in over 40% of prostate cancer patients treated with androgen deprivation therapy (ADT) (Escalante and Manzullo, 2009; Storer et al., 2012). ADT with leuprolide and related drugs is the current treatment of choice to improve survival for metastatic prostate cancer patients, who often use ADT for many months or years. ADT is also used, with lower doses and shorter treatment periods, for localized prostate cancer.
It is clear that the initiating causes of fatigue in ADT-treated prostate cancer patients are primarily the cancer itself and the treatments for that cancer; however, the specific fatigue-related neurological, energy metabolism and immune pathways that are functionally altered by these causal factors and may enhance daily fatigue in ADT-treated prostate cancer have not been established. Jager et al. (2008) has proposed that proinflammatory changes in immune function, anemia, and altered activity of the hypothalamic-pituitary-adrenal (HPA) and serotonergic systems may individually or interactively contribute to this excess fatigue. Similarly, Ryan et al. (2007) hypothesized that: “In any individual, the etiology of CRF probably involves the dysregulation of several physiological and biochemical systems… 5-HT neurotransmitter dysregulation, vagal afferent activation, alterations in muscle and ATP metabolism, hypothalamic-pituitary- adrenal axis dysfunction, circadian rhythm disruption, and cytokine dysregulation.” If these specific dysregulated pathways are identified, targeted treatments to alleviate this CRF may be developed.
One approach that has been used to examine pathways associated with fatigue in breast cancer and during interferon-alpha treatment in chronic hepatitis, as well as in other disorders such as chronic fatigue syndrome (CFS) and multiple sclerosis (MS), is to examine peripheral blood cell gene expression (mRNA) of multiple fatigue-related genes ( Kerr, 2008; Light et al., 2009; Bower et al., 2011a; Bower et al., 2011b; Felger et al., 2012; Light et al., 2012; White et al., 2012). This method is efficient by allowing many physiological targets to be examined from a single blood sample, and the mRNA reflects both genetic (inherited) and environmental influences. Because environmental influences vary across individuals and over time in the same individual, the latter is either a strength or a vulnerability depending upon whether stability is a significant concern. For CFS, studies attempting to use such gene expression as a stable and reproducible diagnostic biomarker have been unable to replicate a consistent profile of differences from controls, due in part to the heterogeneity of the syndrome and to month-to-month variations in status (Kerr, 2008; Galbraith et al., 2011; Frampton et al., 2011). The less challenging objective in the current study was to use leukocyte gene expression to identify potentially dysregulated pathways linked to pathological fatigue in ADT-treated prostate cancer or CFS. For this objective, the same pattern of differential effects need not be present in all or even the majority of these patients but possibly only in subgroups whose levels influence group means. Of specific interest in this regard are the patients whose current fatigue level is more severe.
Many fatigue-related symptoms reported by ADT-treated prostate cancer patients overlap with those reported by patients with CFS, including earlier fatigue onset and decreased performance during exercise, low energy, disturbed sleep, muscle weakness, and increased “mental fog”, and immune-related symptoms like feelings of unwellness (Jager, et al., 2008; Saylor et al., 2009; Storer et al., 2012). Other symptoms seem specific to the syndrome of CFS, including post-exertional malaise, widespread muscle and joint pain with no evidence of injury, and orthostatic intolerance (Fukuda et al., 1994; Saiki et al., 2008; White et al., 2012). Thus, it is plausible that debilitating daily fatigue in these two chronic conditions may involve both shared and different physiological pathways.
In the present study, we used 46 genes representing 5 general pathways: 1) adrenergic, monoamine and peptides; 2) immune response and inflammation; 3) sensory ion channel receptors responding to adenosine triphosphate (ATP) and other metabolites of muscle activity; 4) mitochondrial and other genes involved in lipid/energy metabolism; and 5) transcription and growth factors (see Table 1). Among these are genes that were over-expressed during our prior studies of post-exertional fatigue in CFS: both sensory (ASIC3, P2XR4, TRPV1) and adrenergic receptors (ADRA2A, ADRA2C, ADRB1, ADRB2) and cytokines IL6, IL10, TNF (formerly TNFα) and LTA (formerly TNFβ). Because of the importance of ATP in muscle activity, we included other purinergic ion-channel ATP-receptors, P2XR1 and P2XR7, and G-protein coupled purinergic receptors P2RY1 and P2RY2, as well as another subtype of the acid-sensing family, ASIC1, and another transient vanilloid receptor, TRPV4. Based on prior research (Saiki et al., 2008; Hsiao et al., 2013),we also included genes with important roles in mitochondrial function and energy/lipid/heat metabolism (ATP5E, COX5B, DBI, HSPA2, NDUFS5, and SOD2) and regulators of transcription and cell proliferation (APP, CREB1, CXCR4, PPARA, SIRT1,STAT5A, TLR4, VEGFA, VIPR2, NR3C1, NRG1, and NFKB1). A key member of the energy, immune and transcription subgroups, the diazepam binding inhibitor (DBI, the GABA-type A receptor modulator) is involved in lipid metabolism, is able to displace benzodiazepines on the GABA receptor and thus influences mood regulation, and controls transcription of adrenal steroids influencing immune function including cortisol. TLR4, NR3C1 and NFKB1 have major roles in regulating transcription of cytokines and other inflammatory pathways that have been hypothesized to contribute to sensations of illness and malaise, altered sleep and eating patterns and other components of CRF (Barsevick et al., 2010; Gupta et al., 2011; Bower et al., 2011a).
Table 1.
Genes by general pathway and class
| Pathways | Classes | Genes1 |
|---|---|---|
| Adrenergic, monoamines, peptides (inc. 4 vasodilatory) | Adrenergic, serotonin, dopamine, oxytocin, vasoactive peptide, enzymes/peptidases | ADRA2A, ADRA2C, ADRB1, ADRB2, COMT, DRD4, HTR1D, OXT, OXTR, PSMA4, SULT1A1, VIPR2 |
| Immune response and Inflammation | Cytokines, antigen receptors, chemokine, cytotoxic | CXCR4, DBI, GZMA, IL10, IL6, LTA, NFKB1, NR3C1, TNF, TLR4 |
| Sensory ion channels (ATP- and metabolite sensing) | Purinergic ion channel and G-protein Acid sensing Transient vanilloid receptor Potassium | P2RX1, P2RX4, P2RX7, P2RY1 and 2 ASIC1, ASIC3 TRPV1, TRPV4 HCN2 |
| Mitochondria and lipid/energy metabolism | Electron Transport Chain + others | ATP5E, COX5B, DBI, NDUFS5, SOD2, HSPA2 |
| Transcription and growth factors | Transcriptional, matrix associated | APP, CREB1, DBI, NFKB1, NR3C1, NR3C2, NRG1, PPARA, SIRT1, SPARC, STAT5A, TLR4, VEGFA |
ABI primers used for all genes are identified in Supplemental Table 1.
Note that NFKB1, NR3C1 and TLR4 are listed as both Immune and Transcription genes, while DBI is listed as Immune, Energy metabolism and Transcription.
Also of special interest were fatigue-relevant genes previously associated with metastatic tumors. These included chemokine receptor CXCR4, a gene that has been associated with prognosis and metastases in prostate cancer (Furusato et al., 2010; Jung et al., 2011; Kasina and Macoska, 2012), and superoxide dismutase-2 (SOD2), an intracellular mitochondrial anti-oxidant enzyme associated with increased prostate cancer risk in Caucasians (Kang et al., 2007). Two other genes in our profile, vascular endothelial growth factor (VEGF) and vasoactive intestinal peptide receptor (VIPR2), are linked to cellular proliferation and prostate cancer progression, and VIPR2 activity influences vasodilation and blood flow in brain and other tissues (Collado et al., 2005; Nagakawa et al., 2005; Wang et al., 2011). Several other targeted genes also influence vasodilation, including adrenergic receptor types alpha 2a (ADRA2A) and 2c (ADRA2C) and beta-2 (ADRB2), the oxytocin prepropeptide (OXT) and its receptor (OXTR). Of these vasoactive genes, prior findings of decreased expression in ADT-treated prostate cancer cells led us to focus on the ADRB2 (Ramberg et al., 2008). If ADT also leads to decreases in ADRB2 receptors in the vasculature, this could enhance fatigue by decreasing blood flow to muscles and brain during physical and mental activity.
1. Overview of Current Research
Our two primary aims were: 1) to determine whether ADT-treated prostate cancer patients with daily fatigue (PCF group) showed differences in mean mRNA levels from non-fatigued controls and from CFS patients for 46 fatigue-relevant genes, since these differences may indicate functional dysregulation in pathways that are involved in fatigue; 2) to determine which mRNAs from our 46 fatigue-relevant genes are related to current fatigue severity, using both simple correlation and stepwise regression approaches. Quantitative real-time polymerase chain reaction (QPCR) was used to assess mRNA levels of these 46 genes on leukocytes. Mean mRNA levels were compared in 30 PCF patients, 39 CFS patients and 22 controls. Although all subjects were over age 40 to restrict the age range to that typical of patients with prostate cancer, we did not attempt to match the PCF and CFS patient groups for fatigue severity or gender. PCF patients are of course all males and CFS patients are predominantly females, with a gender ratio of 6:1 or higher (Yunus, 2002; Capelli et al., 2010). Instead our approach was to recruit and test representative samples of patients within each group, so that our findings would readily generalize to the PCF and CFS populations over age 40. We also included individuals with fatigue symptoms ranging from milder to more severe in order to support our second aim, which was to examine whether greater fatigue severity associated with greater up- or down-regulation of any of these genes. Intercorrelations of mRNA levels for different genes were also examined using cluster analysis, to assess whether increases and decreases in expression of the genes under study changed together in a coordinated fashion. Such intercorrelation clusters may reveal the ways in which modifying one or two key fatigue-related genes could influence many others and thus have broad effects.
2. Methods
2.1. Participants
Participants included 91 subjects including 30 with prostate cancer and increased daily fatigue (PCF), 39 with Chronic Fatigue Syndrome (CFS), and 22 healthy non-fatigued controls. All provided written informed consent, and study procedures were approved by the University of Utah Institutional Review Board. As is typical for older adults in Utah, the sample was 97% white, with 3% Hispanics (1 in each group). Inclusion criteria were as follows. To allow matching of the age range to our PCF group, all participants were aged 40–79 years. All PCF and CFS patients were recruited from the practices of our collaborating specialist physicians (NA and LB), who confirmed their primary diagnoses. Both PCF and CFS patients were required, during recent clinic visits, to have reported 2 or more of the following 6 symptoms linked to increased daily fatigue burden affecting QOL: easy fatigability, reduced muscle strength, sleep or mood disturbances, need for daytime rest, increased memory or concentration problems, and decreased ability to exercise and be active (range 2–6, mean= 4.8). All healthy controls were required to report no more than 1 of these symptoms (mean= 0.4). All PCF patients were required to be at least 3 months post-surgery, and to be on stable maintenance medical management of their condition that included recent treatment with the ADT agent, leuprolide. The CFS patients were required to meet the Fukuda et al diagnostic criteria for Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (CFS), consisting of a cluster of symptoms lasting at least 6 months that includes profound, remitting/relapsing fatigue that impairs functioning, for which no specific cause can be determined and which is not relieved by rest or recovery, as well as at least 4 of 8 additional specific syndromic symptoms (Fukuda et al., 1994). Healthy non-fatigued controls were recruited from the community surrounding the medical center. Exclusionary criteria included the following. For all participants, any neurocognitive or other health problem that might limit ability to provide informed consent or to complete our questionnaires. For the controls, an additional exclusion was any chronic health problem other than medically-controlled hypertension or depression, which are common in an older adult sample. Recall that female gender was not exclusionary; the large majority of patients with CFS are female, and to exclude females would greatly limit the meaningfulness of our findings for CFS (Yunus, 2002; Capelli et al., 2010). Instead we managed possible gender differences by statistical approaches described hereafter.
Also, of the 30 PCF patients, 27 (90%) had Stage IV metastatic tumors compared to only 3 with Stage I-III localized tumors. This is in part because maintenance leuprolide use, which was required for inclusion in the PCF group, is more common in metastatic prostate cancer. Aside from ADT, other features of their treatment varied, with 13 patients having had prostatectomy, 16 receiving chemotherapy and 12 treated with radiation therapy (including palliative radiation). The most common prior or current medical treatments other than leuprolide were zoledronic acid (Zometa; n=19) as treatment for hypercalcemia of malignancy, and bicalutamide (Casodex; n=15) as a second anti-androgen treatment. Due to use of leuprolide and other anti-antiandrogens, testosterone levels of all the PCF patients approached zero (mean + SEM = 3.96 + 0.72 ng/dl); although testosterone was not determined for the other groups, the normal range in men aged 40–79 is 300–890 ng/dl.
2.2. Procedure
Ratings of Chronic Daily Fatigue, Current Fatigue and Pain, and Depression
Participants were first interviewed about their medical history, gave permission to reconfirm this information in their medical records, and also completed questionnaires rating chronic daily fatigue, current fatigue and pain severity, and current depressive symptoms. To assess the effects of daily fatigue on QOL, all subjects completed the Rand Medical Outcomes Study Short Form-36 (MOS-36); our focus was on the Energy/Fatigue subscale of the MOS-36, which rates how much of the time the patient recalls feeling worn out, tired or lacking in energy over the past month. In addition to completing the MOS-36 to assess their daily fatigue status over the past month, subjects also provided numerical ratings of their current level of physical fatigue and overall body pain, using a 0–100 scale (where 0=no fatigue or no pain and 100=the most extreme fatigue or pain imaginable). To assess current depressive symptoms, participants completed the Quick Inventory of Depressive Symptomatology-Self Report 16 (QIDS); scores of 6 or higher on the QIDS are indicative of depression (Furukawa, 2010). QIDS items 1–4 address sleep difficulties, including problems falling asleep, staying asleep, and needing excessive night and daytime sleep. These items were used to identify how many patients in each group had sleep problems.
2.3. mRNA extraction and analysis
Following completion of medical history and questionnaires, a single blood sample was taken, the white blood cells (WBCs) manually removed and rapidly processed as described below. All blood processing and analyses were performed by personnel blinded to the subject’s group. Blood was collected in EDTA tubes, and a timer started when the draw was complete. Within 10 minutes after blood collection, the blood was centrifuged at 3200 rpm (1315 × g- Clay Adams Compact II Centrifuge) for 12 minutes, plasma removed, and the WBC layer carefully collected and placed in RLT+β-ME (Qiagen, Valencia, CA), then quickly frozen using a methanol-dry ice slurry, and stored at −80° C. RNA was extracted using RNeasy kits (Qiagen, Valencia, CA), according to manufacturer’s directions, and treated with RNase-free DNase-I (Qiagen, Valencia, CA) to prevent genomic contamination. Immediately following extraction, RNA was retrotranscribed to a cDNA library using the ABI High Capacity cDNA Archive Kit (Applied Biosystems, Inc., Foster City, CA). The cDNA samples were stored at −2° C until analysis. RNA integrity was assessed with a Bioanalyzer, and consistently found to have values greater than 9. The mean QPCR cycle counts for the reference gene, TF2B, were virtually identical in all groups, averaging 23.7+0.2 (SEM) for control subjects, 23.7+0.2 for PCF and 23.6+0.2 for CFS. These values and variations indicate consistent, high quality integrity and sufficient amounts of RNA for accurate analysis of the amount of mRNA in these experiments.
The cDNA libraries were analyzed using the ABI quantitative, real-time QPCR system on the ABI Prism 7900 Sequence Detection System (SDS) 2.4.1 (Applied Biosystems, Inc., Foster City, CA), using ABI TaqMan Master Mix (Applied Biosystems, Inc., Foster City, CA). Master Mix/primer plus primer/probe solutions and template solutions were separately loaded onto 96 well pre-plates. Then 384 well plates were robotically loaded and mixed from the 96 well plates. Each targeted gene was examined in duplicate, with TF2B standards being run in quadruplicate. Additional control samples containing no template were also run. Primer probes for the 46 genes were obtained from TaqMan Gene Expression Assays (Applied Biosystems, Inc., Foster City, CA) and are listed in Supplemental Table S1. QPCR data was processed using the ABI’s SDS program with count values for genes computed in the curve log-linear using a standard 0.2 threshold. Gene expression amounts relative to TF2B were determined using the dCT method, the count difference of each of the candidate genes from the reference gene TF2B. The efficiency of TF2B is close to 100% when the count (CT) values are less than 30 (with threshold set at 0.2). We considered as acceptable only TF2B values less than 26 to ensure that our efficiencies were high, and all samples with CT>25 were re-run using backup samples obtained concurrently with the original samples. TF2B was selected from an array of 16 references genes (including 18-S, GAP-DH, Beta-actin and PSMB6) as the most stable and having the lowest standard deviation in both controls and patients.
2.4. Statistical Analysis
Despite similar age ranges in all groups, the CFS patients and controls had lower mean ages than the PCF group (see Table 2), and included both genders. To control for possible age or gender effects contributing to any group differences, mRNA data were first analyzed using 3-group ANCOVAs with age and gender as covariates. Only when we obtained a significant effect of Group after adjusting for age- or gender-related effects did we then examine comparisons among means to determine whether the PCF group differed from CFS or Controls or both groups. To further rule out any possible gender effects, a second set of mean comparisons was performed using only data from same-gender subgroups (e.g., male controls and male CFS patients vs. the PCF patients, and female controls vs. female CFS patients). When mRNA data were found to be significantly skewed (non-normal distributions), analyses were repeated after log transformation.
Table 2.
Patient Characteristics: Ratios, Percent, Means and Standard Errors [in brackets]
| Group | Gender M/F |
Age | MOS Energy 0–100 |
Fatigue 0–100 |
Pain 0–100 |
Depression # and % |
Sleep problems # and % |
Regular exercise # and % |
|---|---|---|---|---|---|---|---|---|
| PCF | 30/0 | 65.1 [1.4] |
32.7[5.3] | 30.3 [3.9] | 18.3 [4.5] |
13 (43%) | 27 (90%) | 4 (13%) |
| CFS | 10/29 | 56.0 [1.2] |
8.3[4.5] | 71.3 [2.8] | [51.4] [4.0] |
20 (51%) | 36 (92%) | 4 (10%) |
| CON | 10/12 |
52.7 [1.7] |
72.4[6.6] | 2.9 [1.2] |
2.5 [0.8] |
0 | 6(27%) | 16(73%) |
For Gender ratio and Age, PCF>CFS and CON (p<0.01); thus, to control for these potential confounds, gender and age are used as covariates in data analyses, and mean comparisons in all subjects are repeated in males-only and females-only subgroups.
For Energy, Fatigue and Pain measures, all 3 groups differ from each other (p<0.001); thus, analyses correlating fatigue and pain severity with gene expression levels are performed within the 2 patient groups separately.
For proportions of the subgroups with depression (defined as total QIDS score >5), sleep problems (defined as score>1 on QIDS sleep items 1–4), and regular exercise (defined as brisk walking or more vigorous exercise for 20 min or longer at least twice weekly), PCF and CFS did not differ from each other, but both patient groups differed from CON (p<0.01).
Pearson correlations between current fatigue and pain severity ratings and mRNA levels for specific genes were also calculated separately for the PCF and the CFS groups, and stepwise regression was then used to identify the genes that could be combined to yield the strongest predictive models. Cluster analyses based on the associations between mRNA levels of different genes were performed within the 2 patient groups, to indicate whether increased or decreased expression of specific genes might, in fact, be linked to a larger integrated multi-gene pattern.
Unlike whole genome microarray, which involves thousands of genes, most of which have no a priori justification for expecting to obtain group differences or associations with fatigue, this study examined a much smaller set of 46 genes, each from pathways shown to modulate fatigue, pain, muscle performance, blood flow, energy metabolism or mood. Thus, the need to control for false positive findings with multiple comparisons was in fact well balanced against the potential error of false negatives in this first investigation to compare expression of these genes in CFS and PCF groups. Nevertheless, to address the increased risk of false positive findings with multiple comparisons, alpha level was set at p=0.025 rather than the conventional p=0.05 for the initial ANCOVAs comparing the 3 groups, and for the associations in the Cluster analysis. For all other analyses, p=0.05 was used.
3. Results
3.1. Fatigue and Pain Severity, Depression, Sleep and Exercise in the 3 Groups
For the Energy/Fatigue subscale of the MOS-36 (Table 2), CFS patients had consistently very low energy levels (range 0.0−45.8, mean= 8.3), and controls had consistently high energy levels (range 62.6−100.0, mean=72.4). Scores of PCF patients showed no overlap with controls, but did overlap with the CFS group, although their group mean was significantly higher than CFS (range 4.2–58.3, mean= 32.7, p<0.001).
Current ratings of fatigue severity and pain severity (Table 2) were also significantly higher in the CFS than the PCF patients (71.3 vs. 51.4 and 30.3 vs. 18.3, p<0.001). These ratings for current fatigue and pain were highly correlated for both the PCF and the CFS groups (r= +0.62 and +0.64, p<0.001, respectively), indicating that fatigue and pain are not fully independent symptoms.
For depression, 13 of 30 PCF patients (43%) and 20 of 39 CFS patients (51%) scored in the range of mild to moderate depression on the QIDS (6 or higher), proportions which were not significantly different. Both patient groups differed significantly from the CON group, where no subject scored above 5 on the QIDS (p<0.01; Table 2). Sleep problems were reported by more of the PCF and CFS patients compared to controls (90% and 92% vs. 27%, p<0.01), while more controls reported exercising regularly compared to the PCF and CFS patients (73% vs. 13% and 10%, p<0.01; Table 2).
3.2. Mean Group Differences in Gene Expression
For 10 genes, after adjustment for gender and age differences between groups, the PCF group differed from both the CFS and Controls (Table 3). PCF patients showed significantly higher mRNA than CFS and controls for 4 genes: CXCR4, NR3C1, SOD2 and TLR4. For 6 genes, the PCF group showed lower mRNA than both other groups: ADRB2, TNF, LTA, ASIC3, P2RX7, and VIPR2. In contrast, the CFS group differed from controls and the PCF group for only 2 genes (see Table 3). The CFS patients had higher P2RX7 and lower HSPA2 mRNA compared to both the PCF and control groups. Note that even for P2RX7, where both the PCF and CFS groups differed from controls, the direction of difference was the opposite. There was no gene where both the PCF and CFS groups differed from controls in the same direction. These outcomes were unchanged when repeated after log transformation for those 5 genes where raw data had a significant skew (see second p-values in Table 3 showing results after transformation). Also, when these mean comparisons were repeated using same-gender subgroups to verify that this factor was not influencing the outcomes, all group differences remained significant except P2RX7, which then was still marginally significant for the PCF vs. CFS difference (p<0.067) but no longer significant for the PCF vs. CON difference (p>0.40). Thus, we concluded that all our analyses reconfirmed that our group differences were not due to gender effects.
Table 3. Genes Showing Significant Differences in Patient Groups vs. Controls.
Mean mRNA levels relative to control gene (± SEM) after adjustment for age and sex differences where one or both were significant covariates.
| A. Genes down-regulated in PCF Group | |||||||
|---|---|---|---|---|---|---|---|
| Gene | PCF | CFS | CON | Overall Group P |
PCF vs CFS |
PCF vs CON |
CFS vs CON |
| ADRB2** | 6.63E−01 ±8.80E−02 |
10.03E−01 ± 6.60E− 01 |
9.64E−01 ± 8.20E− 02 |
0.016 | 0.004 | 0.025 | 0.507 |
| VIPR2 | 4.17E−03 ± 2.75E−03 |
6.86E−03 ± 2.75E− 03 |
6.51E−03 ± 4.22E− 03 |
0.003 0.000 |
0.000 0.000 |
0.023 0.002 |
0.703 0.447 |
| ASIC3 | 3.94E−03 ±2.22E−03 |
6.07E−03 ± 2.44E− 03 |
5.41E−03 ± 2.02E− 03 |
0.001 | 0.000 | 0.018 | 0.291 |
| P2RX7** | 4.70E−02 ± 3.30E−02 |
7.90E−02 ± 5.00E− 03 |
6.20E−02 ± 6.00E− 03 |
0.003 | 0.009 | 0.010 | 0.007 |
| TNF** | 1.27E−01 ± 1.50E−02 |
1.90E−01 ± 1.10E− 02 |
1.78E−01 ± 1.40E− 02 |
0.015 | 0.004 | 0.026 | 0.463 |
| LTA* | 4.39E−02 ± 5.49E−03 |
6.03E−02 ± 4.20E− 03 |
6.34E−02 ± 5.07E− 03 |
0.025 | 0.036 | 0.013 | 0.633 |
| B. Genes up regulated in PCF group | |||||||
|---|---|---|---|---|---|---|---|
| Gene | PCF | CFS | CON | Overall Group P |
PCF vs CFS |
PCF vs CON |
CFS vs CON |
| CXCR4** | 2.89E+00 ± 2.11E−01 |
2.15E+00 ±1.49E−01 |
1.78E+00 ± 1.88E−01 |
0.003 | 0.013 | 0.001 | 0.116 |
| NR3C1 | 4.88E−01 ± 2.61E−01 |
3.45E−01 ± 1.84E−01 |
3.32E−01 ± 1.31E−01 |
0.008 0.032 |
0.018 0.012 |
0.010 0.05 |
0.751 0.772 |
| SOD2 | 2.28E+01 ±6.66E+00 |
1.85E+01 ±7.10E+00 |
1.66E+01 ± 6.45E+00 |
0.006 0.002 |
0.018 0.008 |
0.002 0.000 |
0.283 0.242 |
| TLR4 | 5.333E−01 ± 3.73E−01 |
3.34E−01 ± 1.11E−01 |
3.10E−01 ± 7.06E−02 |
0.000 0.000 |
0.008 0.000 |
0.003 0.000 |
0.312 0.628 |
| C. Genes up- or down-regulated in CFS group | |||||||
|---|---|---|---|---|---|---|---|
| Gene | PCF | CFS | CON | Overall Group P |
PCF vs CFS |
PCF vs CON |
CFS vs CON |
| P2RX7** | 4.70E−02 ± 3.30E−02 |
7.90E−02 ± 5.00E−03 |
6.20E−02 ± 6.00E−03 |
0.003 | 0.009 | 0.010 | 0.007 |
| HSPA2 | 1.62E−02 ± 2.20E−02 |
0.38E−02 ± 0.48E−02 |
1.46E−02 ± 2.20E−02 |
0.006 0.000 |
0.011 0.000 |
0.807 0.447 |
0.032 0.000 |
with gender only as significant covariate;
with age and gender as significant covariates, p<0.05. Expressed in scientific notation units where E+01 = ×10, E+00 = ×1, E−01 = ×0.1, E−02 = ×0.01, etc.
Where two p-values are included, this gene showed significant skew (non-normality), and the first p-value (upper) is from analyses using untransformed data and the second p-value (lower) is from log transformed data. All outcomes remained significant when repeated after log transformation.
In male-only subgroup comparisons, all differences remain significant except for P2RX7 where PCF vs. CFS p=0.067, and PCF vs. CON p=0.46. In female-only comparisons for CFS vs. CON, all differences remain significant.
3.3. Are Higher or Lower mRNA Levels Associated with Current Fatigue and Pain Severity: Correlations and Stepwise Regression Models
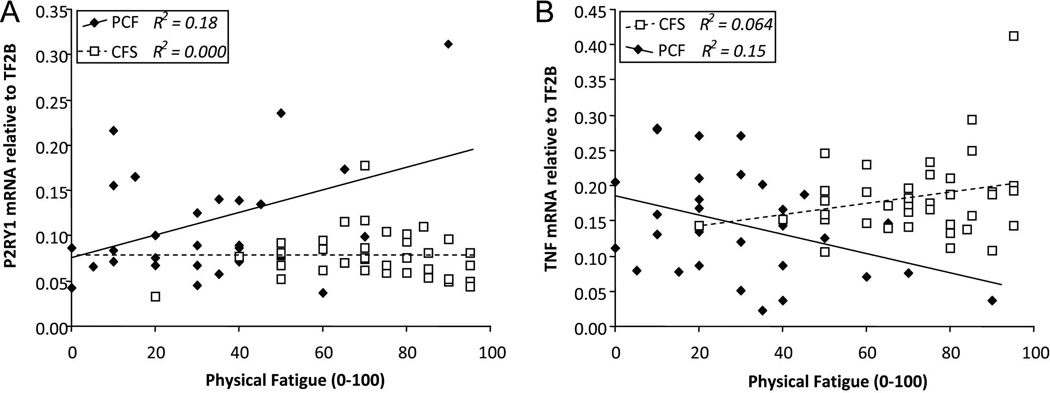
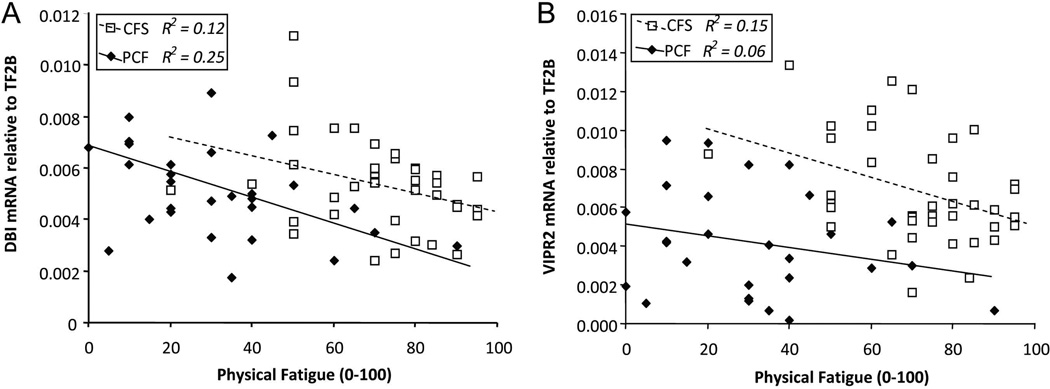
Numerical ratings of current physical fatigue and pain severity at the time of blood sampling were examined for correlations with expression of individual genes within the PCF and the CFS groups. In both the patient groups, higher current fatigue was correlated with lower expression of one gene, the GABA receptor modulator DBI (r=−0.50, p<0.005 and r=−0.34, p<0.05, respectively; see Figure 1A). Higher fatigue in CFS also was associated with lower VIPR2 (r=−0.39, p<0.025), a gene which showed decreased expression in the PCF group as a whole (see Figure 1B). Other relationships differed between the two patient groups. In the PCF group but not the CFS group, higher ratings of both fatigue and pain were correlated with higher expression of purinergic receptor P2RY1 (r=+0.43 and +0.59, p=0.025 and p=0.001, respectively; see Figure 2A for fatigue only).
Figure 1. Relationship between each patient’s rating of current fatigue severity versus: A. DBI mRNA levels, and B. VIPR2 mRNA levels.
Open squares/broken line=CFS, filled diamonds/solid line=PCF. For DBI, this relationship is significant for both patient groups (r= −0.35 and −0.50, R2= .12 and .25, p<0.05 and p<0.005, respectively). For VIPR2, this relationship is significant for CFS (r= −.39, R2= .15, p<0.05) and non-significant for PCF (r= −0.25, R2= .06, p=0.16). PCF patients have lower group mean VIPR2 levels than the CFS group (p<0.001).
Figure 2. Relationship between each patient’s rating of current fatigue severity versus: A. P2RY1 mRNA levels and B. TNF mRNA levels.
Open squares/broken line=CFS, filled diamonds/solid line=PCF. For P2YR1, this relationship is significant for PCF (r= +0.43, R2= 0.18, p<0.025) but entirely absent for CFS (r= 0.0). For TNF, this relationship is significant for PCF (r= −0.39, R2= .15, p<0.05), and non-significant for CFS (r= +0.25, R2= .06, p=0.12). Although not significant independently, in stepwise regression, higher TNF is a significant second predictor of fatigue severity for the CFS group when added to DBI (p<0.05). PCF patients have lower group mean TNF levels than the CFS group (p<0.004).
In addition to P2RY1, higher pain in the PCF group was correlated with higher expression of 3 adrenergic receptors, ADRB1, ADRA2A and ADRA2C (r=+0.48, +0.43, and +0.40, p<0.01, p<0.025 and p<0.035, respectively), and 3 mitochondrial mRNAs, ATP5E, HSPA2 and NDUFS5 (r=+0.52, +0.43, and +0.42, p<0.01, p<0.04 and p<0.05, respectively). Also, in the PCF group only, higher current fatigue was correlated with lower expression of 2 genes that were significantly decreased in the PCF group vs. CFS or controls, TNF (r=−0.39, p<0.03; see Figure 2B) and P2RX7 (r=−0.40, p<0.03), and with 3 other mRNAs, APP (r=−0.43, p<0.025), P2RX4 (r=−0.42, p<0.025) and OXT (r=−0.40, p<0.03). For the CFS group but not the PCF group, higher pain ratings were correlated with higher LTA (r=+0.37, p<0.025) and SIRT1 (r=+0.34, p<0.035). It is noteworthy that trends for the two cytokines, TNF and LTA, are opposite for the 2 patients groups, with higher gene expression linked to greater symptom severity in CFS and lower expression to greater symptom severity in PCF. These relationships were enhanced or unaltered when adjusted for age and gender differences.
We next used stepwise regression to determine which of these genes in combination might yield the strongest predictive models for current fatigue and pain severity, testing separately for the PCF and CFS groups. For predicting fatigue severity, DBI together with P2RY1 yielded the best model in the PCF group (R2=.359; adjusted R2=.311, p<0.002), and DBI with TNF yielded the best model for the CFS group (R2=.341, adjusted R2=.285, p<0.002). For predicting pain severity, a model based on P2RY1, ATP5E, and NDUFS5 together accounted for most of the total pain variance in PCF patients (R2=.630; adjusted R2=.587, p<0.0001), while DBI and LTA together yielded the best model for the CFS patients (R2=.307, adjusted R2=.268, p<0.001). Thus, lower DBI expression was confirmed as a predictor of fatigue severity in both patient groups, as well as predicting pain severity in the CFS group. Higher P2RY1 contributed to prediction of fatigue severity, and together with higher expression of 2 mitochondrial genes, showed excellent prediction of pain severity in the PCF group only. Lower expression of pro-inflammatory cytokines TNF and LTA combined with lower DBI to predict fatigue and pain respectively in the CFS group.
3.4. Patterns of Up-regulation or Down-regulation of Multiple Fatigue-relevant Genes: Clusters Based on Intercorrelations of mRNA Levels
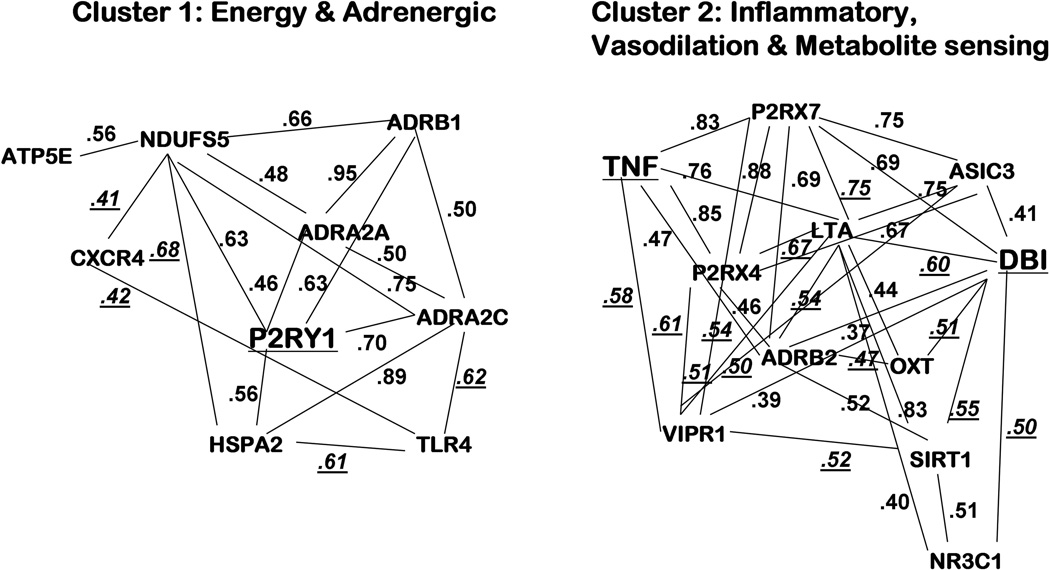
In the PCF patients, intercorrelations of mRNA levels yielded 2 clusters (see Figure 3). Cluster 1 can be termed the Energy and Adrenergic Activation Cluster. It included 9 genes: 3 adrenergic receptors (ADRB1, ADRA2a and ADRA2c), 3 mitochondrial/energy metabolism genes (HSPA2, ATP5E and NDUFS5), 1 transcription gene influencing immune function (TLR4), and the chemokine receptor (CXCR4), plus the key gene with strongest prediction of fatigue and pain severity, P2RY1 (intercorrelation r’s = +0.41-+0.95, p<0.05-p<0.0001). Cluster 2 can be termed the Inflammation and Vasodilation/Metabolite sensing Cluster. It included 2 inflammatory cytokines (TNF and LTA), 3 transcriptional genes also linked to inflammation (DBI, NR3C1 and SIRT1), 3 genes associated with vasodilation (ADBR2, VIPR2 and OXY), and 3 ion channel genes responsive to ATP and other metabolites increased by muscle activity (ASIC3, P2RX4 and P2RX7) (intercorrelation r’s = +0.37−+0.88, p<0.05-p<0.0001). Note that this second cluster included all 6 of the genes that were significantly down-regulated in the PCF group.
Figure 3. Cluster 1 (P2RY1 with energy/mitochondrial and adrenergic receptors) and Cluster 2 (DBI and TNF with inflammatory, vasodilation and metabolite sensing markers), based on intercorrelations of mRNA levels in the fatigued prostate cancer (PCF) group.
Numbers depict Pearson r values for 2 genes connected by a line (p<0.05-p<0.0001). Those r values that are underlined are significant only in the PCF group, while those that are not underlined are also significantly correlated for the CFS group. Key genes (larger type, underlined) are best predictors of fatigue severity in stepwise regression: DBI with P2RY1 for PCF group, and DBI with TNF for CFS group.
In the CFS group, intercorrelations also revealed 2 clusters, both similar to the PCF clusters. (Correlation coefficients in Figure 3 that are not underlined were significant in the CFS as well as the PCF group; r= +0.30–+0.99, p<0.05-p<0.0001). Although the same genes were included in these clusters for both the PCF and CFS groups, only 31 of the 50 correlations were significant for the CFS group (62%), suggesting that increases and decreases in expression of these genes may occur in a more tightly integrated way in the PCF versus the CFS group.
4. Discussion
4.1. Mean Group Differences in Gene Expression
Our findings reinforce the interpretation that CRF is a complex phenomenon, in part because the systems that influence fatigue are very complex. That complexity is reflected in our finding that 10 genes (consisting of 1 or more from each of the 5 pathways studied) were either over-expressed or under-expressed in the PCF group. For each of these 10 genes, the PCF group differed in the same way from the healthy controls as from the CFS group (who like the PCF group included many patients on non-study medications, and had more depression, sleep problems, and pain than controls). This suggests that our PCF group differences were not a result of these potential confounding factors. Likewise, because the PCF mean mRNA differences remained significant after use of age and gender as covariates and after restricting comparisons to single gender subgroups, we also conclude that these differences were not due to age or gender.
Of the 10 genes with altered expression in the PCF group, 5 were immune-related. The up-regulation of chemokine receptor CXCR4 was expected in view of the previous research linking this gene with prostate cancer metastases (Furusato et al., 2010; Jung et al., 2011; Kasina and Macoska, 2012), given that 90% of this PCF sample had Stage IV metastatic tumors. Similarly, TLR4 which transcriptionally regulates cytokine production was over-expressed along with another transcriptional cytokine regulator, NR3C1. However, 2 pro-inflammatory cytokines, TNF and LTA, showed reduced expression in PCF. Although these findings appear inconsistent, the lower TNF and LTA mRNAs may actually reflect over-activation of these cytokines in our PCF group, since decreased mRNA can be a marker of homeostatic secondary efforts working to correct for excessive activity.
Recently, Hsiao et al. reported both up-regulated and down-regulated expression of different mitochondrial genes in PCF patients (Hsiao et al., 2013). Of the 6 energy/mitochondrial genes in our study, SOD2 mRNA was increased in PCF, and HSPA2 mRNA was decreased in CFS. Both increased and decreased SOD2 expression has been previously reported in metastatic prostate cancer (Kang et al., 2007;Quiros et al., 2009; Sharifi et al., 2008; Thomas and Sharifi, 2012). Our finding of decreased HSPA2 in CFS also replicated prior research (Saiki et al., 2008). Although dysregulation in mitochondrial gene expression may be a result of fatigue and inactivity as well as causing fatigue, these findings suggest that pathways involved in energy functions could be targets for new therapies to reduce fatigue in both PCF and CFS.
Other genes that were down-regulated in the PCF group included 3 that we previously associated with post-exertional worsening of fatigue in CFS: ASIC3, P2RX4 and ADRB2 (Light et al., 2009; Light et al., 2012). ASIC3 and P2RX4 receptors work together as a receptor complex to detect ATP, lactate and other metabolites of muscle activity (Light et al., 2008), and ADRB2 receptors are also co-localized with them and may modulate the activity of these metabolite-detecting receptors. In our prior exercise studies, mRNA for these receptors was markedly increased at 8, 24 and 48 hours post-exercise in CFS patients, concurrent with increases in both fatigue and pain. In the present study as in those prior studies, the CFS group did not show altered expression of these receptors in the absence of an exercise challenge. In fact, only 2 genes (P2RX7 and HSPA2) were either up- or down-regulated in the CFS group using this resting baseline sample, many fewer than in the PCF group. It is acknowledged that because this sample of CFS patients were all over age 40, and included mostly peri- and postmenopausal women, these findings may not be representative of younger CFS patients, who are often studied in their 20’s and 30’s. Still, this reinforces our prior interpretation that dysregulated fatigue pathways in CFS are more clearly revealed after exercise than at rest.
P2RX7 was not included in our prior research, but it is also activated by ATP, and like P2RX4, it has a role in modulation of nociception, particularly for inflammatory pain states (Toulme et al., 2010). In this study, the expression of P2RX7 was decreased in the PCF group. This is in contrast to the CFS patients, who showed increased expression of P2RX7. Since decreased mRNA may reflect a secondary response to increased levels of specific receptors, future studies examining protein levels of this purinergic receptor will be needed to verify whether both patient groups show similar or different alterations in P2RX7 function.
In addition to its synaptic interactions with the metabolite-sensing receptors, the adrenergic receptor ADRB2 influences vasodilation, and hypertrophy of muscle fibers and its supportive vasculature. Previously, Ramberg and colleagues (2008) found that ADRB2 mRNA was up-regulated in prostate carcinoma cells, and ADT led to reduced expression of those receptors on the malignant cells. We had hypothesized that ADT would have this down-regulation effect on ADRB2 receptors on leukocytes as well, and our findings supported this hypothesis, suggesting a generalized decrease in these receptors throughout the body. Such decreases in ADRB2 could contribute directly to the muscle mass decreases, decreased exercise performance and sensations of weakness that many patients on ADT experience. For patients who are able to maintain muscle mass, decreased ADRB2 could influence fatigue by impairing the local regulation of blood flow that occurs during physical or even mental activity.
4.2. mRNA Levels Correlated with Fatigue or Pain Severity and With Expression of Other Fatigue-related Genes
For the PCF group, the stepwise regression indicated that fatigue severity was best predicted by 2 genes that were not altered in this patient group as a whole: lower DBI and higher P2RY1. In CFS patients, it was best predicted by lower DBI and higher TNF. Named the diazepam binding inhibitor (but more commonly known as the GABA receptor modulator), DBI is a gene involved in energy regulation via lipid metabolism, modulation of mood via the GABA-A receptor, and transcription of adrenal steroids including testosterone and corticosterone (thereby affecting immune function). DBI can thus directly affect fatigue through all the general pathways that we examined except metabolite-detecting sensory ion channels, and it may influence that last pathway indirectly since our cluster analysis showed DBI to be strongly correlated with P2RX7 and ASIC3. In our PCF sample, lower expression of both purinergic receptors P2RX7 and P2RX4, like lower DBI, was correlated with more severe current fatigue. Dysregulation of DBI mRNA was previously reported in a sample of CFS patients (Saiki et al., 2008), but to our knowledge, ours is the first study linking it to CRF. Regarding functions of P2RY1, activation of this G-protein coupled metabotropic receptor increases mobilization of intracellular calcium ions, and influences signaling by thermal polymodal receptors, microglial activity and inflammation-related pain sensitivity (Tsuda et al., 2010). Recently, P2RY1 mRNA was shown to be increased in the dorsal root ganglion and dorsal horn in a rat model of cancer-induced bone pain, and application of a P2RY1 antagonist reduced nociceptive behavior (Chen et al., 2012). These observations reinforce the interpretation that some dysregulated pathways, such as DBI, are shared by fatigued patients with both disorders, but others, such as P2RY1, appear to be specific to prostate cancer fatigue. Higher TNF was associated with greater fatigue and pain in the CFS but not the PCF group, who had lower overall TNF and LTA levels than controls or CFS. Previous studies attempting to correlate fatigue severity with higher circulating levels of multiple pro-inflammatory cytokines in cancer patients have yielded both positive and negative findings (Wang, et al., 2010; Bower et al., 2011a; Bower et al., 2011b; Wang, et al., 2012; de Raaf et al., 2012; Cameron et al., 2012). TNF was unrelated to CRF severity in a meta-analysis of 18 correlational studies (Schubert et al., 2007).
Fatigue severity was highly correlated with pain severity in both the PCF and the CFS groups (r= +0.62 and +0.64), which may partly explain why DBI was a predictor of both symptoms for CFS, and P2RY1 was a predictor of both symptoms for PCF patients. Prior studies have noted that pain, fatigue and depression are part of a co-occurring symptom cluster in cancer survivors, and patients with those symptoms frequently show signs of dysregulated adrenergic, HPA-axis and immune functions (Thornton et al., 2010; Bower, 2012; Jaremka et al., 2013; Wang et al., 2012; Wood and Weymann, 2013). Our findings generally support this pattern. As shown in our intercorrelation clusters for the PCF group (Figure 3), DBI and glucocorticoid receptor NR3C1 (both involved in transcription of immune genes) correlated with cytokines TNF and LTA, and with vasodilatory genes and ion channel receptors responsive to ATP and other metabolites of muscle activity. In the other intercorrelated cluster, P2RY1 expression correlated with adrenergic receptors ADR2A, ADR2C, and ADRB1 as well as mitochondrial genes ATP5E, NDUFS5 and HSPA2. Greater pain severity in the PCF group was also correlated with expression of these adrenergic and mitochondrial genes, as well as P2RY1. These intercorrelation clusters indicate that fatigue and pain in PCF (and to a lesser extent, in CFS) involve multiple pathways that are relatively tightly integrated. Thus, by influencing individual components, it may be possible to affect several or all of the cluster components, and thus have a wider impact on the patient’s symptoms. Pain and sympathetic nervous system activity have been directly associated in many studies, and both alpha-adrenergic agonists and beta-adrenergic antagonists have analgesic as well anti-inflammatory effects (Light et al., 2009; Light and Vierck, 2009). Because of its primary role in energy regulation, mitochondrial dysfunction has a more obvious link to fatigue and decreased activity than to pain (Norheim et al., 2011; Saiki et al., 2008); however, Reichling and Levine (2009) have described ways in which mitochondrial dysfunction can influence sensory nerve fibers and lead to hyperactivity in primary afferent nociceptors.
Another pathway with evidence of dyregulation in both patient groups was VIPR2, which was under-expressed in the CFS patients reporting higher fatigue severity, and down-regulated in PCF patients as a group. This gene has roles regulating vasodilation and hypoxia within the central nervous system as well as the periphery, which could influence both physical and mental fatigue, and also influences nociception and immune function (Staines et al., 2009). Brenu et al. (2011) previously reported an increase in expression of this gene in a younger sample of CFS patients. Our findings in this CFS patient group aged 40–71 may differ in direction from prior observations, but they are consistent in identifying this pathway as dysregulated in CFS, and suggest that this dysregulation may contribute to fatigue in prostate cancer patients as well.
More severe fatigue in PCF patients was also associated with lower expression of APP, OXT, and TNF. The APP gene influences multiple pathways, including cell adhesion, neuronal development, synaptogenesis and neurite outgrowth, transcriptional regulation, and apoptosis (Zheng and Koo, 2011). In our lab, lower plasma oxytocin has been associated with decreased vasodilation during stressors (Grewen and Light, 2011), and OXT adds a third pathway to the lower VIPR2 and ADRB2 already described as vasodilatory factors that were down-regulated in PCF patients. ADRB2, OXT and APP expression were not related to fatigue or pain severity in the CFS group, again indicating that not all fatigue-related pathways are shared by both disorders.
In our PCF sample, we assume that the initiating causes of the patients’ fatigue are the cancer itself and the treatments for this condition, particularly the long-term treatment with ADT. Based on the encouraging findings of the present initial study suggesting that the maintenance of this fatigue may be influenced by changes in multiple neural and immune pathways, future investigations may be designed to test prostate cancer patients before and at multiple time points after starting ADT. Comparing ADT-treated patients with fatigued prostate cancer patients who have not been maintained on ADT would also provide valuable information on whether the patterns we observed relate solely to use of this treatment modality. This would permit examination of whether some of our observed increases or decreases in gene expression are due to the cancer itself rather than the combined effects of cancer plus ADT. Such studies would also permit assessment of within-subject increases and decreases in fatigue, and whether directional changes in gene expression co-occur with these fatigue variations. This type of repeated assessment is valuable in studying both cancer-related fatigue and CFS, which are both chronic conditions where symptoms and their severity change over time. These additional studies would help in laying the foundation for defining new targets for interventions to help manage fatigue in these disabling chronic disorders.
4.3. Study Limitations
One limitation to the present initial study is that our protocol was limited to leukocyte gene expression, not in brain, muscle or other tissues, and did not include determination of protein levels associated with any of the genes under study. Increased or decreased mRNA does not necessarily indicate that proteins/receptors for that specific gene are changed, and future studies should confirm that mRNA and protein differences are the same. Another limitation is that we did not examine whether our findings were due to changes in specific leukocyte sub-populations. Morse and McNeel (2010) have previously reported that although ADT does not lead to significant changes in numbers of circulating lymphocytes (T- and B-cells), one small T-cell subset did increase: the CD4+ RTE group. Because these represent such a small fraction of the total leukocytes, it is unlikely that this increase could explain our findings.
Regarding our sample, the age range and medical history of our patient samples limit the generalization of the present findings. For the PCF group, the observations may not generalize to patients with localized prostate tumors since 90% of our sample had metastatic tumors. Since our prostate cancer patients all were treated with ADT, our findings cannot distinguish the effect of the cancer itself from the effect of ADT treatment. Also, our CFS patients were aged 40 and older, and most of them had been diagnosed with this disorder many years ago. Our findings thus may not apply to younger CFS patients, or those in the earlier stages of this disorder. As in most studies of older adults, our PCF and CFS groups had other age-related health issues, including mild hypertension and clinical depression, and some were on medications to treat these problems. To restrict our sample to those patients who had no such issues would make recruitment difficult, and furthermore would make our sample non-representative of the typical patient populations of interest. Because the PCF group differed in the same direction from the CFS patients with similar health problems/medications as from the healthy control who were mostly free of such potential confounds, we conclude that these factors did not substantially influence our findings. Lastly, since we did not employ a challenge such as exercise, which previously has been very useful in revealing altered function of a number of the pathways under study in CFS patients, our findings may under-estimate the pathways that are dysregulated in both patient groups.
Summary
In sum, our gene expression profiles indicated both similarities and differences in fatigue-relevant pathways that may be dysregulated in ADT-treated prostate cancer and CFS patients, with many more mRNA differences from controls seen in the prostate cancer group. The diazepam binding inhibitor and vasoactive intestinal peptide receptor genes were linked to fatigue and pain severity in both disorders, while purinergic 2Y1 receptor, other vasodilatory pathways and energy/mitochondrial genes were implicated in the cancer group only. Future efforts to develop new interventions to reduce fatigue and improve quality of life in these patient groups could target one or more of these pathways.
Supplementary Material
Acknowledgments
We wish to acknowledge the staff at the Huntsman Cancer Institute who assisted with phlebotomy and the patient and non-patient participants who made this possible.
Funding Sources
This research was supported by NIH R01 AR060336 and by the Cancer Control and Population Sciences Pilot Grant Award Program from the Huntsman Cancer Institute, Salt Lake City, UT, USA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosures
Kathleen C. Light and Alan R. Light have non-commercialized intellectual property related to biomarkers of fatigue. All authors declare that there are no other conflicts of interest.
Contributers
KCL designed the study and wrote the first draft of the paper. NA and LB recruited and screened patients. ATW performed phlebotomy and statistical analysis. EI recruited and tested controls. TV, HA and RWH performed qPCR and other lab work. AYK helped to design the study and obtain grant support. ARL supervised all lab work, quantified and reviewed data, and helped write the paper. All authors contributed to and approved the manuscript.
References
- Barsevick A, Frost M, Zwinderman A, Hall P, Halyard M. I'm so tired:biological and genetic mechanisms of cancer-related fatigue. Qual. Life Res. 2010;19:1419–1427. doi: 10.1007/s11136-010-9757-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower JE. Fatigue, brain, behavior and immunity: Summary of the 2012 Named Series on fatigue. Brain Behav. Immun. 2012;26:1220–1223. doi: 10.1016/j.bbi.2012.08.009. [DOI] [PubMed] [Google Scholar]
- Bower JE, Ganz PA, Irwin MR, Arevalo JM, Cole SW. Fatigue and gene expression in human leukocytes: increased NF-kappaB and decreased glucocorticoid signaling in breast cancer survivors with persistent fatigue. Brain Behav. Immun. 2011a;25:147–150. doi: 10.1016/j.bbi.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower JE, Ganz PA, Irwin MR, Kwan L, Breen EC, Cole SW. Inflammation and behavioral symptoms after breast cancer treatment: do fatigue, depression, and sleep disturbance share a common underlying mechanism. J. Clin Oncol. 2011b;29:3517–3522. doi: 10.1200/JCO.2011.36.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenu EW, Van Driel ML, Staines DR, Ashton KJ, Ramos SB, Keane J, Klimas NG, Marshall-Gradisnik SM. Immunological abnormalities as potential biomarkers in Chronic Fatigue Syndrome/Myalgic Encephalomyelitis. J. Translat. Med. 2011;9:81. doi: 10.1186/1479-5876-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron BA, Bennett B, Li H, Boyle F, DeSouza P, Wilcken N, Friedlander M, Goldstein D, Lloyd AR. Post-cancer fatigue is not associated with immune activation or altered cytokine production. Ann Oncol. 2012;23(11):2890–2895. doi: 10.1093/annonc/mds108. [DOI] [PubMed] [Google Scholar]; Capelli E, Zola R, Lorusso L, Venturini L, Sardi F, Ricevuti G. Chronic fatigue syndrome/myalgic encephalomyelitis: an update. Int. J Immunopathol Pharmacol. 2010;23(4):981–989. doi: 10.1177/039463201002300402. [DOI] [PubMed] [Google Scholar]
- Chen J, Wang L, Zhang Y, Yang J. P2Y1 purinoceptor inhibition reduces extracellular signal-regulated proteinkinase 1/2 phosphorylation in spinal cord and dorsal root ganglia: implicationsfor cancer-induced bone pain. Acta Biochim. Biophys. Sin. 2012;44:367–372. doi: 10.1093/abbs/gms007. [DOI] [PubMed] [Google Scholar]
- Collado B, Carmena MJ, Sanchez-Chapado M, Ruiz-Vilaespesa A, Bajo A, Fernandez-Martinez AB, Varga JL, Schally AV, Prieto J. Expression of vasoactive intestinal peptide and functional VIP receptors in human prostate cancer: antagonistic action of a growth-hormone-releasing hormone analog. Int. J. Oncol. 2005;26:1629–1635. doi: 10.3892/ijo.26.6.1629. [DOI] [PubMed] [Google Scholar]
- de Raaf PJ, Sleijfer S, Lamers CH, Jager A, Gratama JW, van der Rijt CC. Inflammation and fatigue dimensions in advanced cancer patients and cancer survivors: an explorative study. Cancer. 2012;118(23):6005–6011. doi: 10.1002/cncr.27613. [DOI] [PubMed] [Google Scholar]
- Escalante CP, Manzullo EF. Cancer-related fatigue: the approach and treatment. J. Gen. Intern. Med. 2009;24(Suppl 2):S412–S416. doi: 10.1007/s11606-009-1056-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felger JC, Cole SW, Pace TW, Hu F, Woolwine BJ, Doho GH, Raison CL, Miller AH. Molecular signatures of peripheral blood mononuclear cells during chronic interferon-alpha treatment: relationship with depression and fatigue. Psychol Med. 2012;42:1591–1603. doi: 10.1017/S0033291711002868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton D, Kerr J, Harrison TJ, Kellam P. Assessment of a 44 gene classifier for the evaluation of chronic fatigue syndrome from peripheral blood mononuclear cells. PLoS One. 2011;6(3):e16872. doi: 10.1371/journal.pone.0016872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda K, Straus SE, Hickie I, Sharpe MC, Dobbins JG, Komaroff A. The chronic fatigue syndrome: a comprehensive approach to its definition and study. International Chronic Fatigue Syndrome Study Group. Ann. Intern Med. 1994;121:953–959. doi: 10.7326/0003-4819-121-12-199412150-00009. [DOI] [PubMed] [Google Scholar]
- Furukawa TA. Assessment of mood: guides for clinicians. J Psychosom Res. 2010 Jun;68(6):581–589. doi: 10.1016/j.jpsychores.2009.05.003. 2010. [DOI] [PubMed] [Google Scholar]
- Furusato B, Mohamed A, Uhlen M, Rhim JS. CXCR4 and cancer. Pathol Int. 2010;60:497–505. doi: 10.1111/j.1440-1827.2010.02548.x. [DOI] [PubMed] [Google Scholar]
- Galbraith S, Cameron B, Li H, Lau D, Vollmer-Conna U, Lloyd AR. Peripheral blood gene expression in post-infective fatigue syndrome following from 3 different triggering infections. J. Infect. Dis. 2011;204(10):1632–1640. doi: 10.1093/infdis/jir612. [DOI] [PubMed] [Google Scholar]
- Grewen KM, Light KC. Plasma oxytocin is related to lower cardiovascular and sympathetic reactivity to stress. Biol Psychol. 2011;87:340–349. doi: 10.1016/j.biopsycho.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SC, Kim JH, Kannappan R, Reuter S, Dougherty PM, Aggarwal BB. Role of nuclear factor kappaB-mediated inflammatory pathways in cancer-related symptoms and their regulation by nutritional agents. Exper. Biol Med. 2011;236:658–671. doi: 10.1258/ebm.2011.011028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao CP, Wang D, Kaushal A, Saligan L. Mitochondria-related geneExpression changes are associated with fatigue in patients with nonmetastatic prostate cancer receiving external beam radiation therapy. Cancer Nurs. 2013;36:189–197. doi: 10.1097/NCC.0b013e318263f514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager A, Sleijfer S, van der Rijt CC. The pathogenesis of cancer related fatigue: could increased activity of pro-inflammatory cytokines be the common denominator. Eur. J. Cancer. 2008;44(2):175–181. doi: 10.1016/j.ejca.2007.11.023. [DOI] [PubMed] [Google Scholar]
- Jaremka LM, Fagundes CP, Glaser R, Bennett JM, Malarkey WB, Kiecolt-Glaser JK. Loneliness predicts pain, depression and fatigue: understanding the role of immune dysregulation. Psychoneuroendocrinology. 2013;38:1310–1317. doi: 10.1016/j.psyneuen.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SJ, Kim CI, Park CH, Chang HS, Kim BH, Choi MS, Jung HR. Correlation between chemokine receptor CXCR4 expression and prognostic factors in patients with prostate cancer. Korean J. Urol. 2011;52:607–611. doi: 10.4111/kju.2011.52.9.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D, Lee KM, Park SK, Berndt SI, Peters U, Reding D, Chatterjee N, Welch R, Chanock S, Huang WY, Hayes RB. Functional variant of manganese superoxide dismutase (SOD2 V16A) polymorphism is associated with prostate cancer risk in the prostate, lung, colorectal, and ovarian cancer study. Cancer Epidemiol. Biomarkers Prevent. 2007;16:1581–1586. doi: 10.1158/1055-9965.EPI-07-0160. [DOI] [PubMed] [Google Scholar]
- Kasina S, Macoska JA. The CXCL12/CXCR4 axis promotes ligand-independent activation of the androgen receptor. Mol. Cell Endocrinol. 2012;351:249–263. doi: 10.1016/j.mce.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JR. Gene profiling of patients with Chronic Fatigue Syndrome/Infectious encephalomyelitis. Curr. Rheumatol. Rep. 2008;10(6):482–491. doi: 10.1007/s11926-008-0079-5. [DOI] [PubMed] [Google Scholar]
- Light AR, Bateman L, Jo D, Hughen RW, VanHaitsma TA, White AT, Light KC. Gene expression alterations at baseline and following moderate exercise in patients with Chronic Fatigue Syndrome and Fibromyalgia Syndrome. J. Intern Med. 2012;271:64–81. doi: 10.1111/j.1365-2796.2011.02405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light AR, Hughen RW, Zhang J, Rainier J, Liu Z, Lee J. Dorsal root ganglion neurons innervating skeletal muscle respond to physiological combinations of protons, ATP, and lactate mediated by ASIC, P2X, and TRPV1. J Neurophysiol. 2008;100:1184–1201. doi: 10.1152/jn.01344.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light AR, White AT, Hughen RW, Light KC. Moderate exercise increases expression for sensory, adrenergic, and immune genes in chronic fatigue syndrome patients but not in normal subjects. J Pain. 2009;10:1099–1112. doi: 10.1016/j.jpain.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light KC, Vierck CJ. HPA axis and sympathetic influences on fibromyalgia, chronic fatigue and overlapping functional pain syndromes. In: Mayer E, Bushell MC, editors. Functional pain syndromes: Presentation and pathophysiology. Seattle WA: IASP Press; 2009. pp. 301–317. [Google Scholar]
- Morse MD, McNeel DG. Prostate cancer patients on androgen deprivation therapy develop persistent changes in adaptive immune response. Hum Immunol. 2010;7(5):496–504. doi: 10.1016/j.humimm.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagakawa O, Junicho A, Akashi T, Koizumi K, Matsuda T, Fuse H, Saiki I. Vasoactive intestinal peptide and pituitary adenylate cyclase activating polypeptide stimulate interleukin-6 production in prostate cancer cells and prostatic epithelial cells. Oncol Rep. 2005;13:1217–1221. [PubMed] [Google Scholar]
- Norheim KB, Jonsson G, Omdal R. Biological mechanisms of chronic fatigue. Rheumatology. 2011;50:1009–1018. doi: 10.1093/rheumatology/keq454. [DOI] [PubMed] [Google Scholar]
- Quiros I, Sainz RM, Hevia D, Garcia-Suarez O, Astudillo A, Rivas M, Mayo JC. Upregulation of manganese superoxide dismutase (SOD2) is a common pathway for neuroendocrine differentiation in prostate cancer cells. International J. Cancer. 2009;125:1497–1504. doi: 10.1002/ijc.24501. [DOI] [PubMed] [Google Scholar]
- Ramberg H, Eide T, Krobert KA, Levy FO, Dizeyi N, Bjartell AS, Abrahamsson PA, Tasken KA. Hormonal regulation of beta2-adrenergic receptor level in prostate cancer. The Prostate. 2008;68:1133–1142. doi: 10.1002/pros.20778. [DOI] [PubMed] [Google Scholar]
- Reichling DB, Levine JD. Critical role of nociceptor plasticity in chronic pain. Trends Neurosci. 2009;32:611–618. doi: 10.1016/j.tins.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan JL, Carroll JK, Ryan EP, Mustian KM, Fiscella K, Morrow GR. Mechanisms of cancer-related fatigue. Oncologist. 2007;12(Suppl 1):22–34. doi: 10.1634/theoncologist.12-S1-22. [DOI] [PubMed] [Google Scholar]
- Saiki T, Kawai T, Morita K, Ohta M, Saito T, Rokutan K, Ban N. Identification of marker genes for differential diagnosis of chronic fatigue syndrome. Mol. Med. 2008;14:599–607. doi: 10.2119/2007-00059.Saiki. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saylor PJ, Keating NL, Smith MR. Prostate cancer survivorship: prevention and treatment of the adverse effects of androgen deprivation therapy. J. Gen. Intern Med. 2009;24(Suppl 2):S389–S394. doi: 10.1007/s11606-009-0968-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert C, Hong S, Natarajan L, Mills PJ, Dimsdale JE. The association between fatigue and inflammatory marker levels in cancer patients: a quantitative review. Brain Behav. Immun. 2007;21(4):413–427. doi: 10.1016/j.bbi.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Sharifi N, Hurt EM, Thomas SB, Farrar WL. Effects of manganese superoxide dismutase silencing on androgen receptor function and gene regulation: implications for castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6073–6080. doi: 10.1158/1078-0432.CCR-08-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staines DR, Brenu EW, Marshall-Gradisnik S. Postulated vasoactiveneuropeptide immunopathology affecting the blood-brain/blood-spinal barrier in certain neuropsychiatric fatigue-related conditions: A role for phosphodiesterase inhibitors in treatment. Neuropsychiat. Dis Treat. 2009;5:81–89. [PMC free article] [PubMed] [Google Scholar]
- Storer TW, Miciek R, Travison TG. Muscle function, physical performance and body composition changes in men with prostate cancer undergoing androgen deprivation therapy. Asian J Androl. 2012;14:204–221. doi: 10.1038/aja.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R, Sharifi N. SOD mimetics: a novel class of androgen receptor inhibitors that suppresses castration-resistant growth of prostate cancer. Mol Cancer Ther. 2012;11:87–97. doi: 10.1158/1535-7163.MCT-11-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton LM, Andersen BL, Blakely WP. The pain, depression, and fatigue cluster in advanced breast cancer: Health Psychol. 2010;29:333–337. doi: 10.1037/a0018836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulme E, Tsuda M, Khakh BS, Inoue K. On the role of ATP-gated P2X receptors in acute, inflammatory and neuropathic pain. In: Krueger L, Light AR, editors. Translational Pain Research: From Mouse to Man. Boca Raton, FL: CRC Press; 2010. pp. 235–249. Chapter 10 Series. [PubMed] [Google Scholar]
- Tsuda M, Tozaki-Saitoh H, Inoue K. Pain and purinergic signaling. Brain Res Rev. 2010;63(1–2):222–232. doi: 10.1016/j.brainresrev.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Wang M, Chen GY, Song HT, Hong X, Yang ZY, Sui GJ. Significance of CXCR4, phosphorylated STAT3 and VEGF-A expression in resected non-small cell lung cancer. Exper. Ther Med. 2011;2:517–522. doi: 10.3892/etm.2011.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XS, Shi Q, Williams LA, Cleeland CS, Komaki RR, Mobley GM, Liao Z. Inflammatory cytokines are associated with the development of symptom burden in patients with NSCLC undergoing concurrent chemoradiation therapy. Brain Behav Immun. 2010;24:968–974. doi: 10.1016/j.bbi.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XS, Williams LA, Krishnan S, Liao Z, Liu P, Mao L, Shi Q, Mobley GM, Woodland JF, Cleeland CS. Serum sTNF-R1, IL-6, and the development of fatigue in patients with gastrointestinal cancer undergoing chemoradiation therapy. Brain Behav Immun. 2012;26:699–705. doi: 10.1016/j.bbi.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AT, Light AR, Hughen RW, VanHaitsma TA, Light KC. Differences in metabolite-detecting, adrenergic, and immune gene expression after moderate exercise in patients with chronic fatigue syndrome, patients with multiple sclerosis, and healthy controls. Psychosom Med. 2012;74:46–54. doi: 10.1097/PSY.0b013e31824152ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LJ, Weymann K. Inflammation and neural signalling: etiologic mechanisms of the cancer treatment-related symptom cluster. Curr. Opin. Support. Palliat. Care. 2013;7(1):54–59. doi: 10.1097/SPC.0b013e32835dabe3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunus MB. Gender differences in fibromyalgia and other related syndromes. J Gend. Specif Med. 2002;5(2):42–47. [PubMed] [Google Scholar]
- Zheng H, Koo EH. Biology and pathophysiology of the amyloid precursor protein. Mol. Neurodegener. 2011 Apr 28;6(1):27. doi: 10.1186/1750-1326-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.