Abstract
Background
Observation of gene expression changes implying gene regulations using a repetitive experiment in time course has become more and more important. However, there is no effective method which can handle such kind of data. For instance, in a clinical/biological progression like inflammatory response or cancer formation, a great number of differentially expressed genes at different time points could be identified through a large-scale microarray approach. For each repetitive experiment with different samples, converting the microarray datasets into transactional databases with significant singleton genes at each time point would allow sequential patterns implying gene regulations to be identified. Although traditional sequential pattern mining methods have been successfully proposed and widely used in different interesting topics, like mining customer purchasing sequences from a transactional database, to our knowledge, the methods are not suitable for such biological dataset because every transaction in the converted database may contain too many items/genes.
Results
In this paper, we propose a new algorithm called CTGR-Span (Cross-Timepoint Gene Regulation Sequential pattern) to efficiently mine CTGR-SPs (Cross-Timepoint Gene Regulation Sequential Patterns) even on larger datasets where traditional algorithms are infeasible. The CTGR-Span includes several biologically designed parameters based on the characteristics of gene regulation. We perform an optimal parameter tuning process using a GO enrichment analysis to yield CTGR-SPs more meaningful biologically. The proposed method was evaluated with two publicly available human time course microarray datasets and it was shown that it outperformed the traditional methods in terms of execution efficiency. After evaluating with previous literature, the resulting patterns also strongly correlated with the experimental backgrounds of the datasets used in this study.
Conclusions
We propose an efficient CTGR-Span to mine several biologically meaningful CTGR-SPs. We postulate that the biologist can benefit from our new algorithm since the patterns implying gene regulations could provide further insights into the mechanisms of novel gene regulations during a biological or clinical progression. The Java source code, program tutorial and other related materials used in this program are available at http://websystem.csie.ncku.edu.tw/CTGR-Span.rar.
Background
Over the past decade, a great number of studies on time course issue have become increasingly important since most clinical/biological events, such as infection-related chronic/acute inflammatory responses [1-3], drug treatment-related experiments [4], cell cycle-arrest [5] or other important issues [6], require a period of time in which aberrant alterations in gene expression would lead to different outcomes. Therefore, through performing a consecutive monitoring of massive gene expressions and discovering their regulations during clinical/biological manifestations, the hidden layer of biological mechanisms could be unveiled. However, to our knowledge, these is no effective method can handle this issue although the high-throughput microarray is a powerful tool and has been widely utilized to efficiently detect differentially expressed genes among a group of patients in a time course experiment [3,4]. These authors only focused on how to identify differentially expressed genes varied with time but actually we did not know whether these genes are associated with each other or not. Their results did not show the valuable information.
Sequential pattern mining is one of the most important topics in the field of data mining, especially for the database systems. The fundamental meaning of a sequential pattern refers to a set of singleton frequent items/differentially expressed genes that are followed by another set of items/differentially expressed genes in the time-stamp ordered transaction. Therefore, once the potential gene regulations occurred in a period of time, it could be identified by mining such sequential patterns from a dataset-converted database. Referring to previous studies, several parental algorithms using different computational designs, such as AprioriAll [7], SPADE [8] and PrefixSpan [9], have been successfully proposed and used for different databases to discover their own sequential patterns. The apriori-like (level-wise) GSP [10] and pattern-growth-based Prefix-growth [11] as well as DELISP [12] are evolutionarily designed incorporating with many constraints such as the size of gap among the sequence-involved singleton items, or a time interval within which items are observed as belonging to the same transaction even if they originate from different transactions. Besides, any possible subpatterns derived from each parental sequential pattern also satisfy the user-set constraint values. This property is called downward closure [7-12]. Therefore, any possible subpatterns of each sequential pattern, particularly for the longer ones, need to be generated during the decomposing process that is time-consuming and space-exhausting. Once both shorter and longer sequential patterns have the same occurrence times across all transactions in the database, i.e., closed sequential patterns, the shorter ones will be eliminated from the final resulting patterns. For this purpose, some newer algorithms like incorporating with constraints, CTSP [13], and without constraints, CloSpan [14], were then designed to tackle this problem. In addition to these traditional algorithms, an increasing number of extended methods have also been performed on some interesting topics. For example, an algorithm called WSpan [15] could be used to determine weighted sequential patterns from a transactional database, and the MAGIIC [16] was designed to discover the structure motifs from protein sequences. However, to the best of our knowledge, all of the aforementioned methods are not suitable for the widely used microarray data, as a large-scale DNA microarray-based platform normally consists over tens of thousands of probes/genes, e.g., over 45,000 probes/genes in rice and over 20,000 probes/genes in human arrays. A set of differentially expressed genes (significant singleton gene items) on a single array could be individually considered as a single transaction. In that way, each transaction (each time point contained gene items) may contain too many significant singleton gene items after converting the numeric datasets into the format (discrete) of transactional databases [17]. This is called a long transaction issue. However, to date, there exists no method which can efficiently handle such kind of issue. Actually, a lot of items would frequently occur at most time points. They are similar to the housekeeping genes, which are very insensible to an extracellular stimulus; instead, they play critical roles as maintenance genes in the basic cellular functions [18]. Moreover, mining sequential patterns containing too many such items may increase the difficulty in interpreting the resulting gene regulations. The performance of the preceding sequential pattern mining methods would also be limited to these simultaneous items.
In this paper, we propose a new algorithm called CTGR-Span (Cross-Timepoint Gene Regulation Sequential pattern) with some biologically designed parameters to solve the issue mentioned above by mining CTGR-SPs (Cross-Timepoint Gene Regulation Sequential Patterns). The CTGR-Span ensures that all of the resulting patterns imply gene regulations, which take place across different time points during the course of biological observations. The method is an extended and improved version of our previous paper [19] presented in the 2012 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). The most important changes include: first, we designed a new optimal parameter tuning procedure for the proposed algorithm to ideally determine suitable conditions in pattern mining. The procedure has a merit that there is no need to additionally compute the standard deviation of time intervals in a time course dataset. Based on this design, then we compared our method with two representative sequential pattern mining algorithms, namely GSP and PrefixSpan, in execution efficiency and effectiveness. The resulting patterns were validated using a manual literature survey and an automatic Gene Ontology enrichment analysis [20]. Finally, more explanations for the proposed algorithm have also been added to this paper like i) providing complete examples for readily understanding both our proposed algorithm and the new parameter tuning procedure, and ii) performing more experimental results on the two publicly available human disease-related time course microarray datasets [3,4].
The rest of this paper is organized as follows. The proposed method and materials for analysis are described in Methods. In Results and Discussion, we give the experimental results of the proposed method on two time course gene expression datasets. Concluding remarks are given in Conclusions.
Methods
In this section, we introduce how to efficiently discover CTGR-SPs (Cross-Timepoint Gene Regulation Sequential Patterns) from a time course microarray dataset through 3 main parts: i) an introduction to the experimental background of 2 input microarray datasets, ii) how to convert a numeric dataset into a transactional database, and iii) the kernel of the CTGR-Span (Cross-Timepoint Gene Regulation Sequential pattern) and its required biologically designed arguments.
Input microarray datasets
We tested this paper presenting method using the same input datasets as our previous works [19]. In brief, 2 time course gene expression microarray datasets (GSE6377 [3] and GSE11342 [4]) were downloaded from the GEO database. In GSE6377, McDunn et al. attempted to detect 8,793 transcriptional changes in 11 ventilator-associated pneumonia patients' leukocytes across 10 time points. For the other GSE11342, Taylor et al. monitored 22,283 gene expression changes in peripheral blood monocytes of 20 hepatitis C virus infected patients across the first 10 weeks right after treating with the Peg-interferon alfa-2b plus ribavirin.
Converting microarray datasets into transactional databases
The sequential patterns could be mined directly from a transactional database if the data are discrete. The microarray-involved probe/gene expression values need to be discretized into singleton items within every transaction. Here we show you an example from Table 1 to 3. Table 1 shows the probe/gene expression values of 3 genes G1 to G3 over 4 time points TP1 to TP4 with a fixed interval (1 day). The experimental design is performed in 3 patients. The first time point of this example is regarded as a baseline for deriving the significant items at each time point. All of the values are then divided by the first time point. The divided values can be presented in a fold change matrix as Table 2. The absolute fold changes exceeding a fold-change threshold are further defined as the significant genes. Suppose that the threshold is set as 1.5, only the eligible significant genes can be preserved as new items as shown in Table 3. Take patient 1 for instance, up-regulated G1, down-regulated G2 and down-regulated G3 occur at the second time point that will be presented within the same parentheses (transaction). In this example, a set of 3 time-ordered transactions for each patient is called a sequence.
Table 1.
Example of time course microarray dataset
| Patient IDs | Genes | TP1 | TP2 | TP3 | TP4 |
|---|---|---|---|---|---|
| 1 | G1 | 249 | 656 | 100 | 50 |
| G2 | 333 | 100 | 777 | 989 | |
| G3 | 500 | 250 | 157 | 333 | |
| 2 | G1 | 123 | 950 | 135 | 354 |
| G2 | 222 | 987 | 592 | 80 | |
| G3 | 300 | 222 | 246 | 735 | |
| 3 | G1 | 500 | 121 | 100 | 50 |
| G2 | 400 | 777 | 520 | 60 | |
| G3 | 100 | 300 | 400 | 500 | |
TPn: gene/probe reading values at time point n.
Table 3.
Converted transactional database
| Patient IDs | Sequences |
|---|---|
| 1 | <(G1+G2-G3-)2(G1-G2+G3-)3(G1-G2+G3-)4> |
| 2 | <(G1+G2+)2(G2+)3(G1+G2-G3+)4> |
| 3 | <(G1-G2+G3+)2(G1-G3+)3(G1-G2-G3+)4> |
<>: a sequence; ()t: a transaction of time point t; G+/-: significantly up- or down-regulated gene item.
Table 2.
Fold changes of gene/probe reading values
| Patient IDs | Genes | TP1/1 | TP2/1 | TP3/1 | TP4/1 |
|---|---|---|---|---|---|
| 1 | G1 | 1.00 | 2.63 | -2.49 | -4.98 |
| G2 | 1.00 | -3.33 | 2.33 | 2.97 | |
| G3 | 1.00 | -2.00 | -3.18 | -1.50 | |
| 2 | G1 | 1.00 | 7.72 | 1.10 | 2.88 |
| G2 | 1.00 | 4.45 | 2.67 | -2.78 | |
| G3 | 1.00 | -1.35 | -1.22 | 2.45 | |
| 3 | G1 | 1.00 | -4.13 | -5.00 | -10.00 |
| G2 | 1.00 | 1.94 | 1.30 | -6.67 | |
| G3 | 1.00 | 3.00 | 4.00 | 5.00 | |
TPn/m: gene/probe reading values of time point n relative to m.
However, the content of the converted transactional databases will be affected by different threshold settings. In this study, the threshold of GSE6377 is set as 1.03 and the threshold of GSE11342 is set as 1.5, based on the same criteria used for the original datasets [3,4].
CTGR-Span: cross-timepoint gene regulation sequential pattern
Since the CTGR-Span is designed based on a pattern-growth-based manner [9] for mining CTGR-SPs, we will present the kernel procedure and meanwhile show the main differences between the traditional pattern-growth-based and our methods using a readily understood example. Finally, we present several extra biologically designed parameters toward more meaningful CTGR-SPs in biology.
Kernel procedure
The main strength of the CTGR-Span is to overcome a problem that the transactions have too many items/significant genes. According to our design, it also has several advantages: i) the items within transactions do not need to be sorted in advance, ii) the mining results will not be affected by different sorting types, iii) more meaningful sequential patterns implying gene regulations in biology can be successfully discovered relative to the traditional sequential pattern mining algorithms [7-12], and iv) massive repeated redundant patterns will not be identified. The following examples guide you how to trace the mining processes to explore the patterns from a microarray dataset-converted database. A set S of sequences containing 4 patients' transactions is shown in Table 4. Each transaction consists of several significant gene items Gn+/-. In this example, we set a minimum support (minSupp) as 50%, which means if any one of the items occur in at least 2 different individual sequences (each patient has its own sequence), we call these items as frequent items and further to generate CTGR-SPs through a prefix-projection-based manner [9] in the following steps:
Table 4.
Example of transactional database
| Patient IDs | Sequences |
|---|---|
| 1 | <(G1+)1(G2-G3+)2(G3+)3> |
| 2 | <(G1+G4-)1(G3+)2(G2-G3+)4(G5+)5> |
| 3 | <(G8-)1(G1+G2-)2(G2-G3+)3> |
| 4 | <(G7+)1(G1+G3+G6-)2(G2-G3+)3> |
<>: a sequence; ()t: a transaction of time point t; G+/-: significantly up- or down-regulated gene item.
Step 1: Find length-1 CTGR-SPs
After scanning the S, the frequent items of length-1 including <G1+>, <G2-> and <G3+> can be successfully identified since they appear over one half of the sequences. Therefore, these 3 frequent items are regarded as the lengh-1 CTGR-SPs.
Step 2: Divide search space
Each item within the set of length-1 CTGR-SPs is individually considered as a prefix to find its postfixes in which they are also frequent in the S.
Step 3: Find postfixes of CTGR-SPs
For each identified prefix, the subsets of CTGR-SPs can be identified using a depth-first search-based manner in the prefixes projected databases.
For readily understanding the above 3 steps, here we extend an example shown in Table II of our previous conference paper [19] as Table 5 and show more descriptions on the comparisons of the traditional sequential pattern-growth-based manner and our proposed CTGR-Span. First, for the proposed method, the prefixes within length-1 CTGR-SPs are shown in the left-most column. Only the subsequences prefixed with the first occurrence of the prefixes and started from the next transaction will be presented in the projected databases. As an example, the prefix <G1+> contained in the sequence <(G1+G4-)1(G3+)2(G2-G3+)4(G5+)5> of patient 2 (Table 4), only the subsequence <(G3+)2(G2-G3+)4(G5+)5> will be listed in the projected database for mining longer CTGR-SPs. According to the same principle, the sequences in S containing <G1+> are projected to form the <G1+>-projected database, which consists of 4 candidate postfixes: <(G2-G3+)2(G3+)3>, <(G3+)2(G2-G3+)4(G5+)5>, <(G2-G3+)3> and <(G2-G3+)3>. Then, by scanning <G1+>-projected database once, the length-2 CTGR-SPs having prefix <G1+> can be identified including <(G1+)(G2-)>: 4 (<(G1+)(G2-)> appears 4 times) and <(G1+)(G3+)>: 4. The CTGR-SPs longer than length-2 can be further generated from the current length-2 CTGR-SPs. After constructing their respective projected databases, the <(G1+)(G2-)>-projected database consists of two candidate postfixes: <(G3+)3> and <(G5+)5>. However, both <(G3+)> and <(G5+)> appear only once over the sequences involved in the <(G1+)(G2-)>-projected database that is lower than the minSupp (50%). Hence, the further processes for mining the <(G1+)(G2-)>-projected database will be terminated. On the other hand, recursive mining patterns from the <(G1+)(G3+)>-projected database, which contains two candidate postfixes including <(G3+)3> and <(G2-G3+)4(G5+)5>, returns one eligible postfix <G3+> to form a length-3 CTGR-SPs <(G1+)(G3+)(G3+)>. Finally, according to the same criteria, we can find the remaining CTGR-SPs prefixed with <G2-> or <G3+> by constructing their corresponding projected databases.
Table 5.
Comparison of patterns between a traditional pattern-growth-based approach and CTGR-Span
| Prefixes | Traditional projected databases | Projected databases of CTGR-Span | Traditional sequential patterns | CTGR-SPs |
|---|---|---|---|---|
| G1+ | <(G2-G3+)2(G3+)3> <(_G4-)1(G3+)2(G2- |
<(G2-G3+)2(G3+)3> <(G3+)2(G2-G3+)4(G5+)5> |
<(G1+)(G2-)> <(G1+)(G3+)> |
<(G1+)(G2-)> <(G1+)(G3+)> |
| G3+)4(G5+)5> | <(G2-G3+)3> | <(G1+)(G2-G3+)>* | <(G1+)(G3+)(G3+)> | |
| <(_G2-)2(G2-G3+)3> | <(G2-G3+)3> | <(G1+)(G3+)(G3+)> | ||
| <(_G3+G6-)2(G2-G3+)3> | ||||
| G2- | <(_G3+)2(G3+)3> | <(G3+)3> | <(G2-)(G3+)> | <(G2-)(G3+)> |
| <(_G3+)4(G5+)5> | <(G5+)5> | <(G2-G3+)>* | ||
| <(G2-G3+)3> <(_G3+)3> |
<(G2-G3+)3> <> |
|||
| G3+ | <(G3+)3> | <(G3+)3> | <(G3+)(G3+)> | <(G3+)(G3+)> |
| <(G2-G3+)4(G5+)5> <> |
<(G2-G3+)4(G5+)5> <> |
<(G3+)(G2-)> <(G3+)(G2-G3+)>* |
<(G3+)(G2-)> | |
| <(G6-)2(G2-G3+)3> | <(G2-G3+)3> | |||
G+/-: significantly up- or down-regulated gene item; <>: a sequence; ()t: a transaction of time point t; _: indexed prefix; *: redundant patterns derived from traditional pattern-growth-based sequential pattern mining methods.
After mining all of the sequential patterns, apparently, the traditional patterns marked with an asterisk will not be discovered by our proposed method since they contain the simultaneous items at the same time point. For example, in the first row data of Table 5 one <(G1+)(G2-G3+)> out of four traditional sequential patterns contains the simultaneous item G2- and G3+, which do not imply a gene regulation in a time period but a frequent itemset. Although the pattern could be disassembled into "(G1+) → (G2-)" and "(G1+) → (G3+)", they have overlapped with the other explored sequential patterns including the traditional length-2 sequential pattern <(G1+)(G2-)> and <(G1+)(G3+)>. Therefore, a lot of redundant patterns may be identified by the traditional methods. This thorny problem can be avoided by mining CTGR-SPs. Table 5 shows the strength of the CTGR-Span and elucidates why CTGR-Span is more efficient and useful than the traditional pattern-growth-based methods.
Biological parameter designs
As stated above, we have introduced the main differences between the traditional and our proposed method. Then we intend to describe how to enrich the patterns with more meaningful in biology. In addition to the inherent parameter minSupp for mining traditional patterns, we additionally introduce 3 parameters: minimum timepoint support (minTSupp), sliding window size (SWS) and maximum time constraint (maxTC) to the CTGR-Span to mine more meaningful sequential patterns in gene regulation based on some biological characteristics. Since the fundamental definitions of these parameters have been shown in the section II, MATERIALS AND METHODS, of our previous conference paper [19], here we briefly describe their main characteristics and followed by some concrete examples.
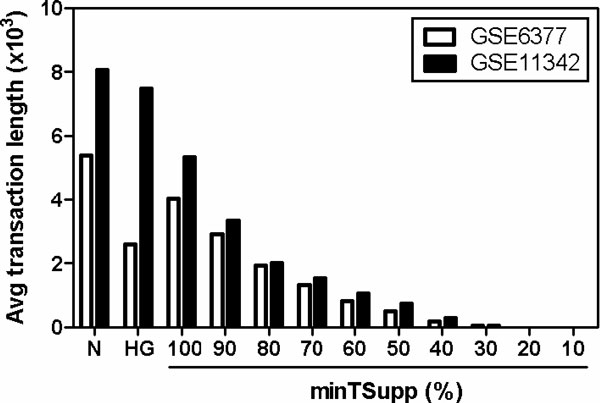
minTSupp (minimum timepoint support). After converting the input microarray datasets into the transactional datasets, thousands of items are contained in each transaction. The average lengths of the transactions of the two datasets are presented as two bars at the left-most N tick shown in Figure 1. The continuously expressed genes at all-time points may not be susceptible to the cellular responses. They may have a propensity for being housekeeping or maintenance genes [18]. In this regard, some well-studied housekeeping genes (HGs) contained in each transaction will be removed. Based on the similar concept, if the items constitutively appear in most time points, these HG-like items can also be further removed from the transactions using the proposed parameter minTSupp. The average lengths of transactions in both input datasets as the functions of varying minTSupp are shown in Figure 1.
Figure 1.
Average transaction lengths of converted transactional databases. N: converted transactional databases; HG: filter transactions of the converted transactional databases using a housekeeping gene database.
SWS (sliding window size). Mining sequential patterns implying gene regulations across fixed time points may cause the resulting patterns inadequate because the response times among a set of genes through transcription regulations are not identical. The sliding window size (SWS) parameter can flexibly allow the patterns containing items to be derived from the same/different time points. Here we show you an example extended from Table 4. Table 6 shows the projected databases of length-1 CTGR-SPs when the SWS is set as 1. Once the time intervals between the transactions contained in the length-1-projected databases and the prefixes not exceed 1 (SWS = 1), the transactions-involved items and the prefixes may actually take place at the same time point. In this case, the gene items involved in a-prime-symbol-marked transactions indicate that they occur with the prefixes at the same time point even if all of them originate from different time points.
Table 6.
Example of SWS = 1
| Prefixes | Projected databases | CTGR-SPs |
|---|---|---|
| G1+ | <(G2-G3+)2'(G3+)3> | <(G1+G2-)> |
| <(G3+)2'(G2- | <(G1+G3+)> | |
| G3+)4(G5+)5> | ||
| <(G2-G3+)3'> | ||
| <(G2-G3+)3'> | ||
| G2- | <(G3+)3'> | <(G2-G3+)> |
| <(G5+)5'> <(G2-G3+)3'> |
||
| <> | ||
| G3+ | <(G3+)3'> <(G2-G3+)4(G5+)5> |
|
| <> | <(G3+G3+)> | |
| <(G2-G3+)3'> | ||
G+/-: significantly up- or down-regulated gene item; <>: a sequence; ()t: a transaction of time point t; _: indexed prefix; *: redundant patterns derived from traditional pattern-growth-based sequential pattern mining methods.
maxTC (maximum time constraint). Normally, the cells need to react quickly to resist adverse environmental changes, massive short-term gene regulations should be more pronounced within a cellular signaling transduction. In this regard, when setting smaller values of the parameter maxTC, a pattern containing two gene items with a big time gap will not be generated. Table 7 shows the length-1-projected databases and CTGR-SPs from an extended example of Table 4 when maxTC is set as 1. The possible postfixes for generating length-2 CTGR-SPs only will be checked till the transactions marked with a prime symbol.
Table 7.
Example of maxTC = 1
| Prefixes | Projected databases | CTGR-SPs |
|---|---|---|
| G1+ | <(G2-G3+)2'(G3+)3> | <(G1+)(G2-)> |
| <(G3+)2'(G2-G3+)4(G5+)5> | <(G1+)(G3+)> | |
| <(G2-G3+)3'> | ||
| <(G2-G3+)3'> | ||
| G2- | <(G3+)3'> | <(G2-)(G3+)> |
| <(G5+)5'> | ||
| <(G2-G3+)3'> | ||
| <> | ||
| G3+ | <(G3+)3'> | <(G3+)(G3+)> |
| <(G2-G3+)4(G5+)5> | ||
| <> | ||
| <(G2-G3+)3'> | ||
G+/-: significantly up- or down-regulated gene item; <>: a sequence; ()t: a transaction of time point t; _: indexed prefix; *: redundant patterns derived from traditional pattern-growth-based sequential pattern mining methods.
Results and discussion
In this section, we presented the experimental results of the proposed CTGR-Span of two time course gene expression datasets. Because performing the program with different parameter values would yield diverse results, all of the parameters used in this study will be tuned according to the biological backgrounds of the datasets. By introducing the tuned parameter values to the CTGR-Span, the resultant CTGR-SPs will then be evaluated with previous literature and a GO enrichment analysis to reveal their reliability in biology. Meanwhile, in terms of the performance, the execution efficacy between the traditional and our proposed methods will also be examined in this study.
Optimal parameter tuning
In addition to the inherent parameter minSupp of the traditional methods, we additionally introduced 3 parameters minTSupp, SWS and maxTC to the CTGR-Span. However, two questions might arise as to how to set these parameter values for most biologists and whether these parameters are useful for mining gene regulations. In this section, we performed an optimal parameter tuning process to obtain a general rule for setting the parameters without additionally calculating the standard deviations of the time intervals of a dataset in advance [19]. Based on the impact degree of each parameter setting to the numbers of the resulting CTGR-SPs, we examined the parameters in an order of minTSupp (Table 8 and Supplementary Table 1 to 3 in Additional file 1), minSupp (Table 8 and Supplementary Table 1 to 3 in Additional file 1), maxTC (Table 9 and 10) and SWS (Table 11 and 12). Several characteristics of the mined CTGR-SPs of two input datasets are presented in these tables. However, here arises a question as to how to judge which condition (combination of parameter values) is more suitable for further exploration - it is a trade-off that higher parameter values would allow fewer patterns to be mined, but lower parameter values would dramatically increase the number of marginal patterns. Both quantity and quality of the resultant patterns are necessary to be taken into account in this work. In the first dataset (GSE6377), McDunn et al. have proven that as the ventilator associated pneumonia (VAP) patients recovered from critical illness complicated by acute infection, the general trajectory (riboleukogram) converged, consistent with an immune attractor [3]. Eighty five genes involved in the inflammatory response were identified with consistent changes in abundance during seven days bracketing the diagnosis of VAP. For the other dataset (GSE11342), Taylor et al. identified 85 significantly up/down-regulated genes involved in the immune response from the blood monocytes of hepatitis C patients during the first 10 weeks of treatment with the Peg-interferon alfa-2b plus ribavirin in peripheral [4]. We used a Gene Ontology (GO) enrichment analysis [20] to test if the longest CTGR-SPs-involved at least two genes under the conditions are relevant to the corresponding biological manifestations (inflammatory response in GSE6377 and immune response in GSE11342). We focused on the longest CTGR-SPs containing at least two gene items because the longer patterns not only contained more significant gene items but also carried more information in a consecutive gene regulation according to the original design of the algorithm. The testing results are presented as -log(p-value) in the tables.
Table 8.
Characteristics of mined sequential patterns (minSupp = variable and minTSupp = 100%)
| GSE6377 | GSE11342 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 100% | 95% | 90% | 85% | 80% | 75% | 70% | 100% | 95% | 90% | 85% | 80% | 75% | 70% | |
| # of CTGR-SPs | 417 | 426 | 4,762 | 5,090 | 181,295 | 181,170 | 6,948,828 | 32 | 224 | 964 | 3,077 | 11,105 | 6,053 | 17,412 |
| # of longest CTGR-SPs | 81 | 81 | 59 | 59 | 176,552 | 176,552 | 208,297 | 2 | 28 | 203 | 1,717 | 4 | 283 | 4,713 |
| Maximal length of CTGR-SPs | 4 | 4 | 6 | 6 | 6 | 6 | 7 | 4 | 4 | 4 | 4 | 5 | 5 | 5 |
| # of genes in CTGR-SPs | 212 | 211 | 1,006 | 996 | 2,821 | 2,826 | 5,313 | 25 | 138 | 466 | 1,132 | 2,011 | 2,801 | 4,142 |
| # of genes in longest CTGR-SPs | 14 | 14 | 11 | 11 | 214 | 214 | 77 | 2 | 3 | 16 | 67 | 3 | 30 | 160 |
| # of gene pairs in lonest CTGR-SPs | 70 | 70 | 58 | 58 | 4,077 | 4,077 | 1,548 | 4 | 21 | 128 | 672 | 6 | 119 | 1,119 |
| -Log(p-value) | 0.34† | 0.34† | 0.00† | 0.00† | 0.55† | 0.55† | 0.29† | 0.00†† | 1.26†† | 0.26†† | 0.91†† | 0.00†† | 1.58†† | 4.11†† |
| # of GSP | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| # of PrefixSpan | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
%: minSupp value presented as percentage; †: test longest CTGR-SPs-involved genes in inflammatory response using GO enrichment analysis; ††: test longest CTGR-SPs-involved genes in immune response using GO enrichment analysis; -: no complete patterns.
Table 9.
Characteristics of mined sequential patterns in GSE6377 (maxTC = variable, minSupp = 95% and minTSupp = 100%)
| 2d | 3d | 4d | 5d | 6d | 7d | 8d | 9d | ≥ 10d | |
|---|---|---|---|---|---|---|---|---|---|
| # of CTGR-SPs | 157 | 157 | 166 | 166 | 180 | 180 | 298 | 306 | 426 |
| # of longest CTGR-SPs | 157 | 157 | 9 | 9 | 17 | 17 | 58 | 58 | 81 |
| Maximal length of CTGR-SPs | 1 | 1 | 3 | 3 | 4 | 4 | 4 | 4 | 4 |
| # of genes in CTGR-SPs | 157 | 157 | 169 | 169 | 179 | 179 | 201 | 202 | 211 |
| # of genes in longest CTGR-SPs | 0 | 0 | 7 | 7 | 10 | 10 | 12 | 12 | 14 |
| # of gene pairs in lonest CTGR-SPs | 0 | 0 | 11 | 11 | 27 | 27 | 50 | 50 | 70 |
| -Log(p-value)† | - | - | 0 | 0 | 0 | 0 | 0 | 0 | 0.34 |
d: # of days of SWS; †: test longest CTGR-SPs-involved genes in inflammatory response using GO enrichment analysis; -: no p-values.
Table 10.
Characteristics of mined sequential patterns in GSE11342 (maxTC = variable, minSupp = 95% and minTSupp = 100%)
| 28d | 31d | 34d | 37d | 40d | 43d | 46d | 49d | 52d | 55d | 58d | 61d | 64d | ≥ 67d | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # of CTGR-SPs | 112 | 112 | 120 | 126 | 157 | 165 | 160 | 163 | 163 | 161 | 194 | 194 | 220 | 242 |
| # of longest CTGR-SPs | 112 | 112 | 8 | 14 | 45 | 2 | 2 | 2 | 2 | 2 | 28 | 28 | 28 | 28 |
| Maximal length of CTGR-SPs | 1 | 1 | 3 | 3 | 3 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| # of genes in CTGR-SPs | 112 | 112 | 119 | 123 | 132 | 132 | 132 | 132 | 132 | 132 | 136 | 135 | 136 | 140 |
| # of genes in longest CTGR-SPs | 0 | 0 | 4 | 6 | 14 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | 3 |
| # of gene pairs in lonest CTGR-SPs | 0 | 0 | 7 | 11 | 42 | 4 | 4 | 4 | 4 | 4 | 21 | 21 | 21 | 21 |
| -Log(p-value)†† | - | - | 1.02 | 0.74 | 0.40 | 0 | 0 | 0 | 0 | 0 | 1.31 | 1.31 | 1.31 | 1.31 |
d: # of days of SWS; ††: test longest CTGR-SPs-involved genes in immune response using GO enrichment analysis; -: no p-values.
Table 11.
Characteristics of mined sequential patterns in GSE6377 (SWS = variable, maxTC = ∞ days, minSupp = 95% and minTSupp = 100%)
| 0d | 1d | 2d | 3d | 4d | 5d | 6d | 7d | 8d | 9d | ≥ 10d | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| # of CTGR-SPs | 352 | 419 | 203 | 203 | 169 | 169 | 201 | 189 | 279 | 354 | 423 |
| # of longest CTGR-SPs | 81 | 81 | 46 | 46 | 3 | 3 | 201 | 189 | 279 | 354 | 423 |
| Maximal length of CTGR-SPs | 4 | 4 | 3 | 3 | 2 | 2 | 1 | 1 | 1 | 1 | 1 |
| # of genes in CTGR-SPs | 206 | 212 | 178 | 178 | 174 | 174 | 187 | 183 | 197 | 209 | 213 |
| # of genes in longest CTGR-SPs | 14 | 14 | 11 | 11 | 2 | 2 | 11 | 9 | 15 | 20 | 21 |
| # of gene pairs in lonest CTGR-SPs | 70 | 70 | 33 | 33 | 5 | 5 | 0 | 0 | 0 | 0 | 0 |
| -Log(p-value)† | 0.37 | 0.37 | 0.44 | 0.44 | 0.44 | 0.44 | - | - | - | - | - |
d: # of days of SWS; †: test longest CTGR-SPs-involved genes in inflammatory response using GO enrichment analysis; -: no p-values.
Table 12.
Characteristics of mined sequential patterns in GSE11342 (SWS = variable, maxTC = ∞ days, minSupp = 95% and minTSupp = 100%)
| 0d | 3d | 6d | 9d | 12d | 15d | 18d | 21d | 24d | 27d | 30d | 33d | 36d | 39d | 42d | 45d | 48d | 51d | 54d | 57d | 60d | 63d | ≥ 66d | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # of CTGR-SPs | 214 | 211 | 221 | 194 | 154 | 135 | 131 | 127 | 125 | 128 | 125 | 127 | 136 | 157 | 157 | 163 | 163 | 163 | 163 | 187 | 190 | 198 | 217 |
| # of longest CTGR-SPs | 28 | 25 | 25 | 82 | 37 | 17 | 17 | 14 | 10 | 13 | 13 | 7 | 10 | 157 | 157 | 163 | 163 | 163 | 163 | 187 | 190 | 198 | 217 |
| Maximal length of CTGR-SPs | 4 | 4 | 4 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| # of genes in CTGR-SPs | 136 | 134 | 136 | 134 | 127 | 124 | 123 | 121 | 120 | 121 | 119 | 121 | 125 | 132 | 132 | 132 | 132 | 132 | 132 | 136 | 136 | 136 | 136 |
| # of genes in longest CTGR-SPs | 3 | 3 | 3 | 15 | 10 | 9 | 9 | 9 | 7 | 8 | 5 | 5 | 4 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 |
| # of gene pairs in lonest CTGR-SPs | 21 | 19 | 19 | 59 | 26 | 16 | 16 | 14 | 10 | 12 | 10 | 10 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| -Log(p-value)†† | 1.26 | 1.37 | 1.37 | 0.70 | 0.00 | 0.00 | 0.00 | 0.00 | 0.86 | 0.86 | 0.65 | 0.53 | 0.40 | - | - | - | - | - | - | - | - | - | - |
d: # of days of SWS; ††: test longest CTGR-SPs-involved genes in immune response using GO enrichment analysis; -: no p-values.
First of all, if the same significant gene items occur too frequent during a time period, they may be similar to the HGs. Then, the significant patterns should occur as frequently as possible in a group of patients. For these two reasons, we tested both minTSupp and minSupp from 70% to 100% as shown in Table 8 and Supplementary Table 1 to 3 in Additional file 1. Apparently, the longest CTGR-SPs revealed no biologically significant when minTSupp was set as 70% or 80% regardless of the values of minSupp. Although the minTSupp was set as 90%, the common values of minSupp suitable for these two input datasets were 85%, 80%, 75% and 70%. Unfortunately, the number of genes involved in the CTGR-SPs was too high (over 250 patterns). It might be difficult for most biologists to work with the high number. In spite of these limitations, we could still successfully obtain a suitable common condition for the two datasets when minTSupp and minSupp were set as 100% and 95%, respectively.
Once the values of minTSupp and minSupp have been decided, we subsequently tested all possible values of maxTC in both two datasets as shown in Table 9 (GSE6377) and Table 10 (GSE11342). The maxTC was set from the beginning as largest time interval, 2 days (21-19) in GSE6377 and 28 days (70-42) in GSE11342, to the end as the values which included most transactions bracketing the maximal time interval, 10 days (21-11) in GSE6377 and 67 days (70-3) in GSE11342. For each dataset, the maxTC would be increased with the first minimum time interval, 1 day (1-0) in GSE6377 and 3 days (3-0) in GSE11342, to ensure any possible conditions would be tested. Apparently, according to the same criteria mentioned in the above paragraph, there was a suitable common condition for the two datasets when the values of maxTC were set as ∞ days.
Finally, we fixed the previous three parameter values and tested the SWS as shown in Table 11 (GSE6377) and Table 12 (GSE11342). The values of SWS in both datasets were set from the beginning as 0 to the end as the values which included most transactions bracketing the maximal time interval, 10 days in GSE3677 and 66 days in GSE11342. The values of SWS were also increased with a fixed interval. Then, we could successfully observe a suitable common condition when the value of SWS was set as 3 days. These tables also demonstrate that these suitable common conditions were neither the rule number nor rule length dependent. Incorporating with the domain knowledge of biology to the parameter designs might had a great benefit on discovering the CTGR-SPs with potential gene regulations. Therefore, these optimal parameter values could be certainly considered as the default settings to most biologists even if they have no any experiences before.
High performance of CTGR-Span
In this section, we compared the performance of our proposed CTGR-Span and the traditional sequential pattern mining algorithms such as the GSP and PrefixSpan in terms of execution efficiency. For achieving a fair comparison, we performed the GSP, PrefixSpan and CTGR-Span with same parameter settings on both input datasets. The resultant patterns and execution times are presented in Table 8 and Table 13 respectively. However, the traditional algorithms did not allow complete patterns (indicated with "-" in Table 8) to be identified in 2 weeks. Meanwhile, their patterns already have produced tens of millions of patterns. It might be complicated for biologist for find further usage of such massive patters. In contrast, our proposed CTGR-Span only needed to take several hours in a worst case that the minSupp was set as 70%. (Table 13). These results clearly showed the efficiency of CTGR-Span.
Table 13.
Execution times (hr) of mined sequential patterns (minSupp = variable and minTSupp = 100%)
| GSE6377 | GSE11342 | |||||||||||||
| 100% | 95% | 90% | 85% | 80% | 75% | 70% | 100% | 95% | 90% | 85% | 80% | 75% | 70% | |
| GSP | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| PrefixSpan | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CTGR-Span | 0 | 0 | 0.03 | 0.03 | 1.65 | 1.65 | 220.88 | 0 | 0 | 0 | 0 | 0.05 | 0.23 | 0.93 |
%: minSupp value presented as percentage; -: over 2 weeks.
Evaluation with literature
After performing the optimal parameter tuning process, we set the parameter SWS = 3 days, maxTC = ∞ days, minSupp = 95% and minTSupp = 100% for the further exploration of CTGR-SPs in biology. As stated in the section of optimal parameter tuning, the evaluation criteria for GO enrichment analysis were based on the experimental backgrounds of those two datasets to preliminarily test which condition with longest CTGR-SPs-involved genes is much related to the inflammatory response caused by the ventilator-associated pneumonia (GSE6377) and the immune response after drug treatments in hepatitis C patients (GSE11342). In this section, we attempted to further address whether these patterns contain potential genes/regulations which have not been reported in previous literature yet. We scrutinized and evaluated the longest CTGR-SPs derived genes from the two input datasets using a manual literature survey. Table 14 and Table 15 show the evaluation results of GSE6377 and GSE11342, respectively. If the patterns contain same items, they will be presented as a single item from left (prefix) to right. For example, in the top-4 data rows of Table 14, there are 4 CAV1+-prefixed CTGR-SPs: <(CAV1+)(GNG7+)(EIF2D+)>, <(CAV1+)(GNG7+)(FTSJ2+)>, <(CAV1+)(GNG7+)(NR2E1-)> and <(CAV+)(GNG7+)(TMOD3-)>. The CAV1+ and GNG7+ can be individually grouped and presented as a single item in the table.
Table 14.
Longest CTGR-SPs of GSE6377 (SWS = 3 days, maxTC = ∞ days, minSupp = 95% and minTSupp = 100%)
| I1 | I2 | I3 | Supports |
|---|---|---|---|
| CAV1+ [21] | GNG7+ | EIF2D+ [24] | 100% (11/11) |
| FTSJ2+ | 100% (11/11) | ||
| NR2E1- [22] | 100% (11/11) | ||
| TMOD3- [25] | 100% (11/11) | ||
| CCL20- [26] | KIF4A+ [27] | FTSJ2+ | 100% (11/11) |
| TMOD3- [25] | 100% (11/11) | ||
| CSF3R- [28] | GNG7+ | CHST7+ | 100% (11/11) |
| EIF2D+ [24] | 100% (11/11) | ||
| FTSJ2+ | 100% (11/11) | ||
| NR2E1- [22] | 100% (11/11) | ||
| TMOD3- [25] | 100% (11/11) | ||
| KIF4A+ [27] | FTSJ2+ | 100% (11/11) | |
| NR2E1- [22] | 100% (11/11) | ||
| TMOD3- [25] | 100% (11/11) | ||
| DGKQ+ [29] | GNG7+ | FTSJ2+ | 100% (11/11) |
| NUDT4+ [30] | CDC25A+ [31] | NR2E1- [22] | 100% (11/11) |
| GNG7+ | NR2E1- [22] | 100% (11/11) | |
| KIF4A+ [27] | EIF2D+ [24] | 100% (11/11) | |
| FTSJ2+ | 100% (11/11) | ||
| NR2E1- [22] | 100% (11/11) | ||
| SOAT1- [32] | 100% (11/11) | ||
| TLR6- [33] | CORO1A+ [34] | 100% (11/11) | |
| KAT2B- [35] | 100% (11/11) | ||
| NR2E1- [22] | 100% (11/11) | ||
| PLAGL1- [22] | 100% (11/11) | ||
| NUDT4P1+ | CDC25A+ [31] | NR2E1- [22] | 100% (11/11) |
| GNG7+ | NR2E1- [22] | 100% (11/11) | |
| KIF4A+ [27] | EIF2D+ [24] | 100% (11/11) | |
| FTSJ2+ | 100% (11/11) | ||
| NR2E1- [22] | 100% (11/11) | ||
| SOAT1- [32] | 100% (11/11) | ||
| TLR6- [33] | CORO1A+ [34] | 100% (11/11) | |
| KAT2B- [35] | 100% (11/11) | ||
| NR2E1- [22] | 100% (11/11) | ||
| PLAGL1- [22] | 100% (11/11) | ||
| STX4- [36] | CDC25A+ [31] | NR2E1- [22] | 100% (11/11) |
| TMOD3- [25] | 100% (11/11) | ||
| KIF4A+ [27] | EIF2D+ [24] | 100% (11/11) | |
| FTSJ2+ | 100% (11/11) | ||
| NR2E1- [22] | 100% (11/11) | ||
| TMOD3- [25] | 100% (11/11) | ||
| TLR6- [33] | CORO1A+ [34] | 100% (11/11) | |
| KAT2B- [35] | 100% (11/11) | ||
| LSM7+ [37] | 100% (11/11) | ||
| NR2E1- [22] | 100% (11/11) | ||
| PLAGL1- [22] | 100% (11/11) | ||
[]: pneumonia-associated genes reported in previous literature; In: the nth item in a CTGR-SP; +: expressed genes; -: repressed genes.
Table 15.
Longest CTGR-SPs of GSE11342 (SWS = 3 days, maxTC = ∞ days, minSupp = 95% and minTSupp = 100%)
| L1 | L2 | L3 | L4 | Supports |
|---|---|---|---|---|
| CXCL10+ [23] | IFIT2+ [38] | ZNF710- | FECH+ [39] | 95% (19/20) |
| BPGM+ [40] | 95% (19/20) | |||
| SNCA+ [41] | 95% (19/20) | |||
| SELENBP1+ [42] | 95% (19/20) | |||
| HBZ+ | FECH+ [39] | 95% (19/20) | ||
| BPGM+ [40] | 95% (19/20) | |||
| SNCA+ [41] | 95% (19/20) | |||
| SELENBP1+ [42] | 100% (20/20) | |||
| TRIM46+ | 95% (19/20) | |||
| SELENBP1+ [42] | HBZ+ | 95% (19/20) | ||
| SELENBP1+ [42] | 95% (19/20) | |||
| PPP4R4+ | SELENBP1+ [42] | 95% (19/20) | ||
| IFIT2+ [38] | IFIT2+ [38] | ZNF710- | FECH+ [39] | 95% (19/20) |
| BPGM+ [40] | 95% (19/20) | |||
| SNCA+ [41] | 95% (19/20) | |||
| SELENBP1+ [42] | 95% (19/20) | |||
| HBZ+ | FECH+ [39] | 95% (19/20) | ||
| BPGM+ [40] | 95% (19/20) | |||
| SNCA+ [41] | 95% (19/20) | |||
| SELENBP1+ [42] | 100% (20/20) | |||
| TRIM46+ | 95% (19/20) | |||
| SELENBP1+ [42] | HBZ+ | 95% (19/20) | ||
| SELENBP1+ [42] | 95% (19/20) | |||
| PPP4R4+ | SELENBP1+ [42] | 95% (19/20) | ||
| TNFSF10+ [43] | IFIT2+ [38] | HBZ+ | SELENBP1+ [42] | 95% (19/20) |
[]: hepatitis C-associated genes reported in previous literature; In: the nth item in a CTGR-SP; +: expressed genes; -: repressed genes.
After the evaluating process, 78% (54/69 hits) in Table 14 and 73% (29/40 hits) in Table 15 of the patterns-involved genes could be successfully referred to some literature. In other words, the remaining genes might play potential roles during the time course. As stated in the previous example, it has been proven that up-regulated caveolin-1 (CAV1) would regulate NF-kappa B activation and lung inflammatory response to sepsis induced by lipopolysaccharide [21]. The upregulation of nuclear receptor subfamily 2, group E, member 1 (NR2E1) has been revealed by a microarray analysis of mice infected with influenza virus A and Streptococcus pneumonia [22]. A relation/regulation might exist between these two genes since they were strongly related to the pneumonia [21,22]. Coincidentally, in Table 15, upregulated chemokine (C-X-C motif) ligand 10 (CXCL10) has also been reported in the original paper that CXCL10 would be transiently induced early in treatment with Peg-interferon alfa-2b plus ribavirin in peripheral blood monocytes (PBMC) of hepatitis C patients [4]. It could be successfully regarded as plasma indicator for predicting the outcome of antiviral therapy in patients with hepatitis C [23]. Therefore, via this literature evaluation, we postulated that the remaining unreported genes and their relations of the identified patterns in both datasets are highly valuable to be explored in the future.
Conclusions
In this study, our proposed CTGR-Span overcomes the flaws of the traditional sequential pattern mining methods. Although the transactional databases converted from the large-scale time course microarray gene expression datasets have too many items/significant genes within every transaction, the gene regulations over a period of time can still be efficiently identified. The CTGR-Span runs dramatically faster than the traditional methods. In addition to the improvement of execution times, we incorporated the characteristics of gene regulation in the parameter designs and further used a GO enrichment analysis to yield the CTGR-SPs more meaningful biologically. After evaluating with previous literature, the identified patterns correlate very well with the experimental backgrounds of the two input datasets. Therefore, we postulated that our approach could provide more biological insights into the underlying mechanisms of certain biological or clinical progresses, and it also could be readily applied to other research topics of interest.
List of abbreviations
CTGR-Span: Cross-Timepoint Gene Regulation Sequential pattern; CTGR-SPs Cross-Timepoint Gene Regulation Sequential Patterns; minTSupp: minimum timepoint support; minSupp: minimum support; SWS: sliding window size; maxTC: maximum time point support; GO: gene ontology.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
CPC, YCL, YLT and VST conceived and designed the entire experiments. CPC carried out the computational studies, performed the statistical analysis and drafted the manuscript. YCL participated in the data interpretations and helped to draft the manuscript. YLT carried out the experiments. VST obtained funding and made critical study supervision. All authors read and approved the final manuscript.
Supplementary Material
Characteristics of mined sequential patterns (minSupp = 70~100% and minTSupp =70%~90%)
Contributor Information
Chun-Pei Cheng, Email: ccp0625@gmail.com.
Yu-Cheng Liu, Email: uchenliu@gmail.com.
Yi-Lin Tsai, Email: jameslovee10@gmail.com.
Vincent S Tseng, Email: tsengsm@mail.ncku.edu.tw.
Acknowledgements
We thank Lukas Horak (lukas.horak@gmail.com) for revising this paper.
Based on "CTGR-Span: Efficient mining of cross-timepoint gene regulation sequential patterns from microarray datasets", by Chun-Pei Cheng, Yi-Lin Tsai and Vincent S. Tseng which appeared in Bioinformatics and Biomedicine (BIBM), 2012 IEEE International Conference on. © 2012 IEEE [19].
Declarations
This research was funded and supported by the National Science Council of Taiwan under contract number "NSC 101-2627-B-006-003" and the National Cheng Kung University Top University Project of Ministry of Education.
This article has been published as part of BMC Bioinformatics Volume 14 Supplement 12, 2013: Selected articles from the IEEE International Conference on Bioinformatics and Biomedicine 2012: Bioinformatics. The full contents of the supplement are available online at http://www.biomedcentral.com/bmcbioinformatics/supplements/14/S12.
References
- Shie B-E, Cheng J-H, Chuang K-T, Tseng V. In: A One-Phase Method for Mining High Utility Mobile Sequential Patterns in Mobile Commerce Environments Advanced Research in Applied Artificial Intelligence. Jiang H, Ding W, Ali M, Wu X, editor. Vol. 14. Springer Berlin/Heidelberg; 2012. pp. 616–626. [Google Scholar]
- Cheng CP, Lee PF, Chin CY, Liu WC, Wu IC, Chang TT, Tseng VS. Discovering Clinical Biomarkers of Chronic Hepatitis B by Mining Mutation Hotspots. Technologies and Applications of Artificial Intelligence (TAAI), 2011 International Conference on: 11-13 Nov. 2011. 2011. pp. 51–56.
- McDunn JE, Husain KD, Polpitiya AD, Burykin A, Ruan J, Li Q, Schierding W, Lin N, Dixon D, Zhang W. et al. Plasticity of the systemic inflammatory response to acute infection during critical illness: development of the riboleukogram. PloS one. 2008;14(2):e1564. doi: 10.1371/journal.pone.0001564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MW, Tsukahara T, McClintick JN, Edenberg HJ, Kwo P. Cyclic changes in gene expression induced by Peg-interferon alfa-2b plus ribavirin in peripheral blood monocytes (PBMC) of hepatitis C patients during the first 10 weeks of treatment. Journal of translational medicine. 2008;14:66. doi: 10.1186/1479-5876-6-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizuka N, Yoshizuka-Chadani Y, Krishnan V, Zeichner SL. Human immunodeficiency virus type 1 Vpr-dependent cell cycle arrest through a mitogen-activated protein kinase signal transduction pathway. Journal of virology. 2005;14(17):11366–11381. doi: 10.1128/JVI.79.17.11366-11381.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciuto AM, Phillips CS, Orzolek LD, Hege AI, Moran TS, Dillman JF. Genomic analysis of murine pulmonary tissue following carbonyl chloride inhalation. Chemical research in toxicology. 2005;14(11):1654–1660. doi: 10.1021/tx050126f. [DOI] [PubMed] [Google Scholar]
- Agrawal R, Srikant R. Mining Sequential Patterns. Proceedings of the Eleventh International Conference on Data Engineering. 1995. pp. 3–14. 655281: IEEE Computer Society.
- Zaki MJ. SPADE: an efficient algorithm for mining frequent sequences. Machine Learning. 2001;14:31–60. doi: 10.1023/A:1007652502315. [DOI] [Google Scholar]
- Pei J, Han J, Mortazavi-asl B, Pinto H, Chen Q, Dayal U, Hsu M-c. PrefixSpan: Mining Sequential Patterns Efficiently by Prefix-Projected Pattern Growth. Proceedings of the 17th International Conference on Data Engineering. 2001. p. 215. 879716: IEEE Computer Society.
- Srikant R, Agrawal R. Mining Sequential Patterns: Generalizations and Performance Improvements. Proceedings of the 5th International Conference on Extending Database Technology: Advances in Database Technology. 1996. pp. 3–17. 650382: Springer-Verlag.
- Pei J, Han J, Wang W. Mining sequential patterns with constraints in large databases. Proceedings of the eleventh international conference on Information and knowledge management; McLean, Virginia, USA. 2002. pp. 18–25. 584799: ACM.
- Lin M-Y, Lee S-Y. Efficient mining of sequential patterns with time constraints by delimited pattern growth. Knowl Inf Syst. 2005;14(4):499–514. doi: 10.1007/s10115-004-0182-5. [DOI] [Google Scholar]
- Lin M-Y, Hsueh S-C, Chang C-W. Mining Closed Sequential Patterns with Time Constraints. J Inf Sci Eng. 2008. pp. 33–46.
- Yan X, Han J, Afshar R. CloSpan: Mining Closed Sequential Patterns in Large Datasets. 2003. pp. 166–177. In SDM.
- Yun U. A new framework for detecting weighted sequential patterns in large sequence databases. Knowl-Based Syst. 2008;14(2):110–122. doi: 10.1016/j.knosys.2007.04.002. [DOI] [Google Scholar]
- Hsu C-M, Chen C-Y, Hsu C-C, Liu B-J. Efficient discovery of structural motifs from protein sequences with combination of flexible intra- and inter-block gap constraints. Proceedings of the 10th Pacific-Asia conference on Advances in Knowledge Discovery and Data Mining; Singapore. 2006. pp. 530–539. 2097117: Springer-Verlag.
- Kim M, Shin H, Su Chung T, Joung JG, Kim JH. Extracting regulatory modules from gene expression data by sequential pattern mining. BMC Genomics. 2011;14(Suppl 3):S5. doi: 10.1186/1471-2164-12-S3-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CW, Cheng WC, Chen CR, Shu WY, Tsai ML, Huang CL, Hsu IC. Identification of human housekeeping genes and tissue-selective genes by microarray meta-analysis. PloS one. 2011;14(7):e22859. doi: 10.1371/journal.pone.0022859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C-P, Tsai Y-L, Tseng VS. CTGR-Span: Efficient mining of cross-timepoint gene regulation sequential patterns from microarray datasets. Bioinformatics and Biomedicine (BIBM), 2012 IEEE International Conference on: 4-7 October 2012. 2012. pp. 1–4. [DOI]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;14(5):P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- Garrean S, Gao XP, Brovkovych V, Shimizu J, Zhao YY, Vogel SM, Malik AB. Caveolin-1 regulates NF-kappaB activation and lung inflammatory response to sepsis induced by lipopolysaccharide. J Immunol. 2006;14(7):4853–4860. doi: 10.4049/jimmunol.177.7.4853. [DOI] [PubMed] [Google Scholar]
- Zhang H, Su YA, Hu P, Yang J, Zheng B, Wu P, Peng J, Tang Y, Zhang L. Signature patterns revealed by microarray analyses of mice infected with influenza virus A and Streptococcus pneumoniae. Microbes and infection/Institut Pasteur. 2006;14(8):2172–2185. doi: 10.1016/j.micinf.2006.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butera D, Marukian S, Iwamaye AE, Hembrador E, Chambers TJ, Di Bisceglie AM, Charles ED, Talal AH, Jacobson IM, Rice CM. et al. Plasma chemokine levels correlate with the outcome of antiviral therapy in patients with hepatitis C. Blood. 2005;14(4):1175–1182. doi: 10.1182/blood-2005-01-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr I, Sonenberg N. Host translation at the nexus of infection and immunity. Cell host & microbe. 2012;14(4):470–483. doi: 10.1016/j.chom.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong J, Zhang W, Wang X, Fan H, Lu C, Yao H. Identification of candidate susceptibility and resistance genes of mice infected with Streptococcus suis type 2. PloS one. 2012;14(2):e32150. doi: 10.1371/journal.pone.0032150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Ferrari JD, Cao Y, Ramirez MI, Jones MR, Quinton LJ, Mizgerd JP. Type I alveolar epithelial cells mount innate immune responses during pneumococcal pneumonia. J Immunol. 2012;14(5):2450–2459. doi: 10.4049/jimmunol.1200634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perini G, Tupler R. Altered gene silencing and human diseases. Clinical genetics. 2006;14(1):1–7. doi: 10.1111/j.1399-0004.2005.00540.x. [DOI] [PubMed] [Google Scholar]
- Germeshausen M, Ballmaier M, Welte K. Incidence of CSF3R mutations in severe congenital neutropenia and relevance for leukemogenesis: Results of a long-term survey. Blood. 2007;14(1):93–99. doi: 10.1182/blood-2006-02-004275. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Bielawski J, Dunn K, Yang Z. Phylogenomic analysis of natural selection pressure in Streptococcus genomes. BMC evolutionary biology. 2007;14:154. doi: 10.1186/1471-2148-7-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng BH, Liu Y, Xuei X, Liao CP, Lu D, Lasbury ME, Durant PJ, Lee CH. Microarray studies on effects of Pneumocystis carinii infection on global gene expression in alveolar macrophages. BMC microbiology. 2010;14:103. doi: 10.1186/1471-2180-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitomo T, Daigo Y, Matsuda K, Ueda K, Nakamura Y. Critical function for nuclear envelope protein TMEM209 in human pulmonary carcinogenesis. Cancer research. 2012;14(16):4110–4118. doi: 10.1158/0008-5472.CAN-12-0159. [DOI] [PubMed] [Google Scholar]
- Gantier MP. The not-so-neutral role of microRNAs in neutrophil biology. J Leukoc Biol. 2013. [DOI] [PubMed]
- Shimizu T, Kida Y, Kuwano K. A dipalmitoylated lipoprotein from Mycoplasma pneumoniae activates NF-kappa B through TLR1, TLR2, and TLR6. J Immunol. 2005;14(7):4641–4646. doi: 10.4049/jimmunol.175.7.4641. [DOI] [PubMed] [Google Scholar]
- Baguma-Nibasheka M, Angka HE, Inanlou MR, Kablar B. Microarray analysis of Myf5-/-:MyoD-/- hypoplastic mouse lungs reveals a profile of genes involved in pneumocyte differentiation. Histology and histopathology. 2007;14(5):483–495. doi: 10.14670/HH-22.483. [DOI] [PubMed] [Google Scholar]
- Rahman I, MacNee W. Role of transcription factors in inflammatory lung diseases. Thorax. 1998;14(7):601–612. doi: 10.1136/thx.53.7.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Naren AP, Gao X, Ahmmed GU, Malik AB. Protease-activated receptor-1 activation of endothelial cells induces protein kinase Calpha-dependent phosphorylation of syntaxin 4 and Munc18c: role in signaling p-selectin expression. The Journal of biological chemistry. 2005;14(5):3178–3184. doi: 10.1074/jbc.M410044200. [DOI] [PubMed] [Google Scholar]
- Swiatlo E, Brooks-Walter A, Briles DE, McDaniel LS. Oligonucleotides identify conserved and variable regions of pspA and pspA-like sequences of Streptococcus pneumoniae. Gene. 1997;14(2):279–284. doi: 10.1016/S0378-1119(96)00823-2. [DOI] [PubMed] [Google Scholar]
- Urban TJ, Thompson AJ, Bradrick SS, Fellay J, Schuppan D, Cronin KD, Hong L, McKenzie A, Patel K, Shianna KV. et al. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology. 2010;14(6):1888–1896. doi: 10.1002/hep.23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumoulin FL, von dem Bussche A, Li J, Khamzina L, Wands JR, Sauerbruch T, Spengler U. Hepatitis C virus NS2 protein inhibits gene expression from different cellular and viral promoters in hepatic and nonhepatic cell lines. Virology. 2003;14(2):260–266. doi: 10.1006/viro.2002.1701. [DOI] [PubMed] [Google Scholar]
- Ren F, Wu H, Lei Y, Zhang H, Liu R, Zhao Y, Chen X, Zeng D, Tong A, Chen L. et al. Quantitative proteomics identification of phosphoglycerate mutase 1 as a novel therapeutic target in hepatocellular carcinoma. Molecular cancer. 2010;14:81. doi: 10.1186/1476-4598-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou J, Liu P, Wang J, Zhang X. Effect of hepatitis C virus core shadow protein expressed in human hepatoma cell line on human gene expression profiles. Journal of gastroenterology and hepatology. 2006;14(12):1794–1800. doi: 10.1111/j.1440-1746.2006.04380.x. [DOI] [PubMed] [Google Scholar]
- M DIS, Volpe MG, Colonna G, Nazzaro M, Polimeno M, Scala S, Castello G, Costantini S. A possible predictive marker of progression for hepatocellular carcinoma. Oncology letters. 2011;14(6):1247–1251. doi: 10.3892/ol.2011.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann KA, Bjorkstrom NK, Veber H, Ciesek S, Riese P, Wiegand J, Hadem J, Suneetha PV, Jaroszewicz J, Wang C. et al. Interferon-alpha-induced TRAIL on natural killer cells is associated with control of hepatitis C virus infection. Gastroenterology. 2010;14(5):1885–1897. doi: 10.1053/j.gastro.2010.01.051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics of mined sequential patterns (minSupp = 70~100% and minTSupp =70%~90%)