Abstract
The Y chromosomes of 549 individuals from Siberia and the Americas were analyzed for 12 biallelic markers, which defined 15 haplogroups. The addition of four microsatellite markers increased the number of haplotypes to 111. The major Native American founding lineage, haplogroup M3, accounted for 66% of male Y chromosomes and was defined by the biallelic markers M89, M9, M45, and M3. The founder haplotype also harbored the microsatellite alleles DYS19 (10 repeats), DYS388 (11 repeats), DYS390 (11 repeats), and DYS391 (10 repeats). In Siberia, the M3 haplogroup was confined to the Chukotka peninsula, adjacent to Alaska. The second major group of Native American Y chromosomes, haplogroup M45, accounted for about one-quarter of male lineages. M45 was subdivided by the biallelic marker M173 and by the four microsatellite loci alleles into two major subdivisions: M45a, which is found throughout the Americas, and M45b, which incorporates the M173 variant and is concentrated in North and Central America. In Siberia, M45a haplotypes, including the direct ancestor of haplogroup M3, are concentrated in Middle Siberia, whereas M45b haplotypes are found in the Lower Amur River and Sea of Okhotsk regions of eastern Siberia. Among the remaining 5% of Native American Y chromosomes is haplogroup RPS4Y-T, found in North America. In Siberia, this haplogroup, along with haplogroup M45b, is concentrated in the Lower Amur River/Sea of Okhotsk region. These data suggest that Native American male lineages were derived from two major Siberian migrations. The first migration originated in southern Middle Siberia with the founding haplotype M45a (10-11-11-10). In Beringia, this gave rise to the predominant Native American lineage, M3 (10-11-11-10), which crossed into the New World. A later migration came from the Lower Amur/Sea of Okhkotsk region, bringing haplogroup RPS4Y-T and subhaplogroup M45b, with its associated M173 variant. This migration event contributed to the modern genetic pool of the Na-Dene and Amerinds of North and Central America.
Introduction
Questions about the timing and geographic origins of the migrations that led to the peopling of the Americas have been examined through use of a wide array of approaches. Greenberg et al. (1986) used linguistic, dental, and genetic evidence to propose a tripartite migration model through which the Amerinds (of North, Central, and South America), Na-Dene (of northwestern North America) and Eskimo-Aleuts (of the subarctic) emerged. Studies of maternally inherited mtDNA variation (e.g., Schurr et al. 1990; Torroni et al. 1992, 1993a, 1993b, 1994; Horai et al. 1993; Shields et al. 1993; Forster et al. 1996; Merriwether et al. 1996) have generally supported the tripartite model of Native American origins, although the number of migrations that gave rise to the Amerinds, as well as the Amerinds' interrelationship with the Na-Dene and Eskimo-Aleuts, has been subject to differences in interpretation.
More recently, studies of variation in the paternally inherited Y chromosome have complemented mtDNA analyses (Jobling and Tyler-Smith 1995). Initial studies of the repetitive DNA DYS1 system (Torroni et al. 1994) and alphoid repeats (Pena et al. 1995; Santos et al. 1996) suggested the existence of a major founding Y-chromosome haplotype for Amerinds. This conclusion was verified by the discovery of a Y-chromosome single-nucleotide polymorphism (SNP), a C→T transition at the DYS199 locus now referred to as “marker M3” (Underhill et al. 1996). M3 has been shown to delineate a major Native American founder haplotype (Bianchi et al. 1997, 1998; Lell et al. 1997).
Microsatellite variation has been used to further characterize Amerindian Y chromosomes, revealing a second founding haplotype in addition to M3 (Ruiz-Linares et al. 1999). A third Native American haplogroup, defined by the RPS4Y C→T SNP, has been identified in northern Amerind and Na-Dene–speaking populations (Bergen et al. 1999). This haplogroup has not been detected in South American natives, with the exception of two Wayus from Colombia (Karafet et al. 1999).
The search for the ancestors of the Native American Y chromosomes in Siberia and Asia has revealed that the M3 lineage is found only on the Chukotka peninsula of far northeastern Siberia, among the Chukchi and the Siberian Eskimos (Karafet et al. 1997; Lell et al. 1997). The most recent ancestors of the M3 lineage have been traced to central southern Siberia (Karafet et al. 1999; Santos et al. 1999).
The Siberian RPS4Y-T haplogroup has been located in the Lake Baikal region, east of M3 and its progenitors. This has been interpreted as indicating that these Y-chromosome lineages came to the Americas in distinct migrations (Karafet et al. 1999). Finally, Y chromosomes harboring the Tat polymorphism (haplogroup Tat-C) were found dispersed between native populations of central Asia and northern Europe (Santos et al. 1999), supporting a relatively recent link between these populations (Zerjal et al. 1997).
To further clarify the Siberian origins of Native American migrations into the New World, we have surveyed the Y-chromosome SNP and microsatellite variation in a large sample of geographically dispersed native populations of Siberia and the Americas. This analysis has revealed that two major male migrations peopled the Americas: one starting from southern Middle Siberia, giving rise to haplogroups M3 and M45a in North, Central, and South America, and a second migration from eastern Siberia, which brought Y-chromosome lineages RPS4Y-T and M45b to the Na-Dene and Amerinds of North and Central America.
Samples and Methods
Population Samples
All of the samples analyzed (fig. 1) in the present study were originally collected as whole blood. Most of the population samples presented here are subsets of previously studied population sets: southern Mexico Native Americans (Torroni et al. 1994); Navajos (Torroni et al. 1992); Seminoles (Huoponen et al. 1997); Siberian Eskimos and Chukchi (Starikovskaya et al. 1998); Koryaks and Itel’mens (Schurr et al. 1999); Yenisey Evenks, Udegeys, and Nivkhs (Torroni et al. 1993b); and Kets (Sukernik et al. 1996).
Figure 1.
Map of the approximate locations of the native Siberian and Native American populations analyzed in the present study
The samples representing southern Middle Siberia include the Turkic-speaking Tofalars and Tuvans, as well as the Mongolic-speaking Buryats. The samples from the Russian Far East were the Tungusic-speaking Ulchi/Nania, Negidals, and Okhotsk Evenks. These population aggregates of the Altaic linguistic family are located on opposite sides of the southern Siberian belt. The remnants of nonadmixed Tofalars, a small tribe of hunters and reindeer breeders occupying the Taiga area on the northern slopes of the eastern Sayan mountain range (Levin and Potapov 1964), were sampled in the villages of Alygdzher, Nerkha, and Upper Gutara, all of which are in the Nizhneudinsk administrative district of the Irkutsk Region.
The Tuvan samples were collected in Kizil, the capital of the Tuva Republic, mainly from students of Tuva University. Most of the territory of the Tuva Republic is situated in the arid steppe zone in the very center of the Asian continent, bounded by the foothills of the Sayan mountains to the north and the Mongolian arid, sandy wastes to the south (Levin and Potapov 1964). Tuva’s total population size is ∼100,000, and many individuals still pursue traditional subsistence activities similar to those of the Russian Buryats. The Russian Buryats number ∼200,000 and are regarded as the northern extension of the Mongolic-speaking ethnic groups of Mongolia and China (Levin and Potapov 1964). Buryat samples were collected in Kushun village, Nizhneudinsk District, Irkutsk Region. These samples represent the Buryats of the Sayan-Baikal upland.
Blood samples from Ulchi and Nanai individuals were collected in the villages of Old and New Bulava in the Ulchi District of the Khabarovsk Region. Samples from a geographically isolated group of Evenks were collected in several small settlements on the mainland Okhotsk Sea shore in the Tugur-Chumikan District of the Khabarovsk Region. Additionally, remnants of the Negidals, who were swamped by the expanding Evenks, were sampled in the Polina Osipenko District (Upriver Negidals) and the Nikolayevski District (Downriver Negidals) of the Khabarovsk Region. Historically, these samples represent well-defined Tungusic-speaking tribes of hunters and fishermen occupying the Lower Amur Region (Levin and Potapov 1964; Black 1988; Krauss 1988).
Many of the Central American and all of the South American samples were collected and supplied by J. V. Neel. DNA was extracted, by standard phenol/chloroform protocols, directly from whole blood, from fractionated buffy coats, or from lymphoblastoid cell lines.
Y-Chromosome Marker Analysis
Each DNA sample was typed for 16 Y-chromosome loci: 4 microsatellite loci and 12 biallelic SNPs. The four microsatellite loci (DYS19, DYS388, DYS390, and DYS391) were amplified and typed using standard primers and conditions (Deka et al. 1996; Kayser et al. 1997). Alleles were designated according to the number of repeats within the variable tri- or tetranucleotide-repeat tract at each locus. Sequence information was obtained from the literature, for DYS19 (Roewer et al. 1992), DYS390 (Forster et al. 1998), and DYS391 (Carvalho-Silva et al. 1999), and from the Human Genome Database Web site, for DYS388 (accession number G00-365-729). The variable repeat tract at each locus was confirmed by sequencing of representative alleles. Each of the four microsatellite loci were sequenced on Y chromosomes representing all haplotypes observed in our populations of southern Mexicans, Koryaks, Itelmen, and Evenks, in addition to eight European and nine African American Y chromosomes. In each case, the size of the PCR product was strictly correlated with the number of repeats in the variable tract as follows (the sequence of the repeat structure is given with the variable tract underlined): DYS19, TAA(GATA)5GTAT(TA)5GTGT(TA)6GTGTTTTA(GATA)3GGTA(GATA)nTAG (10 repeats = 186-bp PCR product); DYS388, CTCAA(ATA)nATG (10 repeats = 125-bp PCR product); DYS390, (GATA)2GAT(GATA)4GACA(GATA)n(GACA)8(GATA)2GAA (10 repeats = 212-bp PCR product); and DYS391, GCA(GATA)n(GACA)3GATA (10 repeats = 283-bp PCR product).
Our allele designations may differ from those of previous studies, which used different methods of ascertaining and defining the size of the variable-repeat numbers. For example, our DYS19 10-repeat allele (186-bp PCR product) corresponds to the 13-repeat allele of de Knijff et al. (1997). Such differences are resolvable by referring to the size of the corresponding PCR product for each allele defined by repeat number.
The 12 biallelic SNPs were analyzed using the methods of Lell et al. (1997) for M3; Underhill et al. (1997) for M9 and M17; Hammer and Horai (1995) for YAP; Jobling et al. (1996) for the DYS7C deletion; Zerjal et al. (1997) for Tat; Bergen et al. (1999) for RPS4Y 711; Su et al. (1999) for M45, M89, and M119; and Underhill et al. (2000) for M48 and M173. Selected haplotypes within the Y-chromosome phylogeny were examined for variation in nine additional SNPs (M7, M40, M50, M88, M95, M103, M110, M111, and M122); however, these proved to be essentially monophyletic and, hence, not sufficiently informative to merit further analysis.
Networks of Y-chromosome haplotypes were constructed according to the model of Deka et al. (1996) and Cooper et al. (1996). Each haplotype was connected to all other haplotypes from which it differed by one repeat unit step at a single microsatellite locus, with the networks drawn to minimize crossovers. In our networks, we defined ancestral or founder haplotypes on the basis of frequency, geographic distribution, and knowledge of ancestral states based on biallelic marker analysis.
Given the lack of a standardized nomenclature for Y-chromosome haplotypes, we have used a nomenclature system modeled after that of the mtDNA. Each major Y-chromosome lineage defined by biallelic markers is classified as a Y-chromosome haplogroup. The microsatellite-defined variants within each haplogroup further define individual haplotypes. As a shorthand for discussion, each haplogroup will be referred to by the most recently occurring marker that is common to all of the descendent haplotypes of the lineage (e.g., the haplogroup defined by the presence of the M89, M9, M45, and M3 markers will be referred to as “haplogroup M3”). Haplogroup M45 has been subdivided into subhaplogroups, M45a and M45b, with most—but not all—M45b chromosomes having the M173 variant. The presence of the M173 variant is designated by either “M173” or an asterisk (*). The individual haplotypes are further delineated by listing the number of repeats present at the microsatellites DYS19, DYS388, DYS390, and DYS391 (e.g., the Y-chromosome haplotype M45, which harbors the M173 variant and 11 repeats for each microsatellite locus, is referred to as “M45b*[11-11-11-11]”).
Results
Biallelic-Marker Distribution in the Americas
Our analysis revealed the presence of 15 distinct Y-chromosome haplogroups defined by 12 biallelic markers, in 549 individuals from Siberia and the Americas (fig. 2). The predominant Native American Y-chromosome lineage was haplogroup M3. This lineage accounted for 66% of all Native American Y chromosomes in the present study and was present at population frequencies of 27%–100% (fig. 2). The composition, distribution, and frequency of this lineage is concordant with several previous studies of Y-chromosome variation in Native American populations (Underhill et al. 1996; Bianchi et al. 1997; Karafet et al. 1997, 1999; Lell et al. 1997; Scozzari et al. 1997; Underhill et al. 2000).
Figure 2.
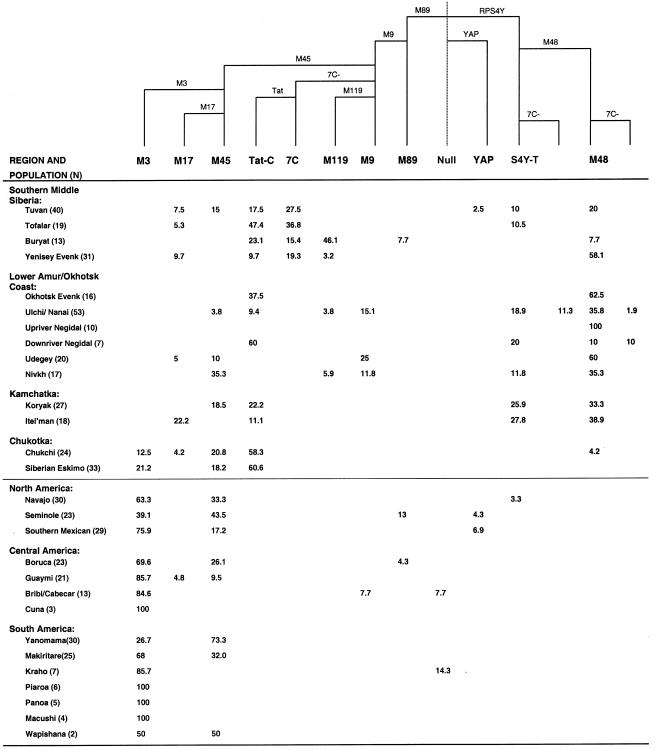
Frequency distribution of Y-chromosome haplogroups in Siberian and Native American populations
The second-most-frequent Y-chromosome lineage observed in the Americas was haplogroup M45 (fig. 2). This lineage is identical to haplogroup M3, except that it lacks the M3 variant. Haplogroup M45 was found to be widely distributed among Native American and Siberian populations. Overall, 29% of Native American Y chromosomes were defined by haplogroup M45, and its distribution explains the apparent relatedness of most non-M3 Y chromosomes in the Americas (Ruiz-Linares et al. 1999). Furthermore, haplogroup M45 could be subdivided by the M173 SNP (Underhill et al. 2000). The M45 chromosomes harboring the M173 variant were observed only in Native Americans from North and Central America, the lone exception being a Wapishana from South America.
The remaining Native American Y chromosomes were defined by a few rare haplogroups (fig. 2). One Guaymi individual from Costa Rica belonged to haplogroup M17, and a single Bribri/Cabecar from Costa Rica belonged to haplogroup M9. In addition, one Boruca individual from Costa Rica and three Seminoles from southern Florida belonged to haplogroup M89. A single Kraho from Brazil and a single Bribi/Cabecar from Costa Rica were found to have all of the biallelic markers present in their ancestral state (null haplotype). Two southern Mexican Mixes and one Seminole harbored the YAP insertion (M1), but these M1 chromosomes were probably acquired through post-Columbian gene flow from individuals of African ancestry (Huoponen et al. 1997; Lell et al. 1997). Finally, a single Navajo from New Mexico harbored a haplogroup RPS4Y-T Y chromosome (Bergen et al. 1999). This Y-chromosome lineage was previously shown to be present at significant frequencies in several northern Amerind and Na-Dene populations (Bergen et al. 1999; Karafet et al. 1999) and was postulated to represent an additional Native American founding Y-chromosome lineage (Karafet et al. 1999). Its virtual absence in Central and South American Amerind populations is confirmed in the present extensive population survey.
Biallelic-Marker Distribution in Siberia
A greater diversity of Y-chromosome types were observed in Siberia than in the Americas, a finding consistent with founder effects occurring during the peopling of the Americas (Wallace et al. 1985). Siberian Y chromosomes split into two major groups, with 57% belonging to haplogroups delineated by the ancient M89 polymorphism (macrohaplogroup M89, encompassing haplogroups M89, M9, M119, 7C, Tat-C, M45, M17, and M3) and 42% belonging to haplogroups delineated by the RPS4Y-T marker (macrohaplogroup S4Y-T) (fig. 2).
As reported elsewhere, haplogroup M3 was observed in Asia only in the far northeastern Siberian region of Chukotka (Karafet et al. 1997; Lell et al. 1997). The closely related haplogroup M45, which has been observed at high frequencies throughout Europe, was observed in populations throughout the southern belt of Siberia, as well as in the populations of the Siberian Pacific coast; however, it is virtually absent in the East Asian populations south of Siberia (Su et al. 1999; Semino et al. 2000; Underhill et al. 2000). The M45 haplotypes presumably include the most-recent ancestors of the M3 lineage. These results are consistent with previous studies that used other Y-chromosome markers, indicating that the progenitor of the M3 lineage resides in the Kets and Altayans of Middle Siberia and the Selkups of southwestern Siberia (Karafet et al. 1999; Santos et al. 1999). Haplogroup M45 could be further subdivided by the biallelic marker M173. All M173-positive M45 chromosomes were found in eastern Siberian Udegeys and Koryaks and North and Central American natives. Thus, haplogroup M45 appears to be divided into two distinct subgroups: (1) Middle Siberian, together with North, Central, and South American Amerinds, and (2) eastern Siberian, with North and Central American Na-Dene and Amerinds.
The Tat-C haplogroup was observed at significant frequencies in each of the southern Middle Siberian populations studied (fig. 2). Surprisingly, it reached its highest frequency in the Siberian Eskimos and Chukchi from the Chukotkan peninsula. The Tat-C haplogroup was absent in the Lower Amur and Sea of Okhotsk region populations that have maintained greater geographic and/or linguistic isolation (e.g., the Udegeys, Nivkhs, and Upriver Negidals) and was only detected in the populations likely to have had recent contact or shared origins with the populations of southern Middle Siberia (e.g., the Okhotsk Evenks, Ulchi/Nanai, and Downriver Negidals). Because the Tat-C polymorphism originated on a Y chromosome containing the DYS7C deletion (haplogroup 7C), which was present only in the Middle Siberian Tuvans, Buryats, Tofalars, and Yenisey Evenks, the Tat-C haplogroup probably entered the Lower Amur and eastern Siberia from southern Middle Siberia. This conclusion is consistent with the previous hypothesis that the Tat-C and 7C haplogroups arose in central Asia and migrated west to northern Europe and east to Chukotka (Zerjal et al. 1997).
Haplogroup M119 was present primarily in southern Middle Siberia, accounting for 6 of the 13 Buryat Y chromosomes. Although it is rare elsewhere in Siberia, this haplogroup occurs at significant frequencies in various ethnic Chinese populations (Su et al. 1999), indicating a northward migration of this haplogroup from East Asia. Interestingly, none of the Y chromosomes analyzed in the present study contained the M122 marker, which has been shown to represent a large proportion of northern Chinese and East Asian Y chromosomes (Su et al. 1999).
Slightly less than half of the Siberian Y chromosomes belonged to macrohaplogroup S4Y-T, delineated by the RPS4Y-T marker. Previous studies have shown that RPS4Y-T is present at a high frequency in the Lake Baikal region and Mongolia (Karafet et al. 1999) but is absent in Europeans (Bergen et al. 1999). In our study, the S4Y macrohaplogroup (haplogroups RPS4Y-T and M48, with and without the DYS7C deletion) is at its highest frequency in the populations of eastern Siberia, including Kamchatka and the Lower Amur River basin (fig. 2). This lineage was almost completely absent in the Chukotkan populations in which the Native American haplogroup M3 was present. Interestingly, the single Chukchi RPS4Y-T haplotype also exhibited the M48 marker, defining an eastern Siberian subhaplogroup, M48. This suggests a more recent admixture from Southeast Asia. Derivatives of both the RPS4Y-T and M48 haplogroups have acquired DYS7C deletions. These parallel mutations generated two subbranches of these haplogroups, indicating that the DYS7C deletion should always be used in conjunction with other markers, to define paternal lineages (Jobling et al. 1996). These data indicate that macrohaplogroup S4Y chromosomes entered Siberia from the south and that the haplogroup RPS4Y-T chromosomes in the Americas could have originated from the Lower Amur River region near the Sea of Okhotsk.
Microsatellite and Network Analysis of Y-Chromosome Lineages
To further delineate Y-chromosome lineages, the biallelic haplogroups were subdivided using microsatellite markers. Analysis of 520 of our Native American and Siberian Y chromosomes through use of four microsatellite loci (DYS19, DYS388, DYS390, and DYS391) subdivided the 14 haplogroups into 111 haplotypes (table 1table 1). Networks of adjacent haplotypes were constructed for individual lineages, to aid in inferring their histories (Cooper et al. 1996) (figs. 3, 4, and 5). In these networks, the geographic localization of the haplotypes is indicated by the color of the surrounding haplotype boxes (see fig. legends).
Table 1.
Y-Chromosome Haplotypes in Native American and Siberian Populations
|
Allele at Locus |
||||||||||||||||
| Haplogroup | M89 | M9 | M45 | M173 | M3 | M17 | M119 | DYS7C | Tat | RPS4Y | M48 | YAP | DYS19 | DYS388 | DYS390 | DYS391 |
| M89 | + | 11 | 13 | 10 | 10 | |||||||||||
| M89 | + | 11 | 14 | 10 | 9 | |||||||||||
| M89 | + | 12 | 9 | 10 | 10 | |||||||||||
| M89 | + | 12 | 12 | 10 | 10 | |||||||||||
| M89 | + | 12 | 11 | 9 | 10 | |||||||||||
| M89 | + | 11 | 12 | 11 | 11 | |||||||||||
| M9 | + | + | 11 | 9 | 11 | 10 | ||||||||||
| M9 | + | + | 12 | 11 | 9 | 10 | ||||||||||
| M9 | + | + | 12 | 11 | 10 | 10 | ||||||||||
| M9 | + | + | 13 | 11 | 11 | 10 | ||||||||||
| M9 | + | + | 14 | 12 | 10 | 10 | ||||||||||
| M45 | + | + | + | 10 | 11 | 9 | 10 | |||||||||
| M45 | + | + | + | 10 | 11 | 10 | 10 | |||||||||
| M45 | + | + | + | 10 | 11 | 11 | 10 | |||||||||
| M45 | + | + | + | 10 | 11 | 11 | 11 | |||||||||
| M45 | + | + | + | 10 | 11 | 12 | 11 | |||||||||
| M45 | + | + | + | 10 | 11 | 12 | 10 | |||||||||
| M45 | + | + | + | 10 | 11 | 13 | 10 | |||||||||
| M45 | + | + | + | 10/15 | 11 | 11 | 10 | |||||||||
| M45 | + | + | + | 10 | 12 | 9 | 10 | |||||||||
| M45 | + | + | + | 10 | 12 | 9 | 11 | |||||||||
| M45 | + | + | + | 11 | 11 | 9 | 10 | |||||||||
| M45 | + | + | + | 11 | 11 | 10 | 10 | |||||||||
| M45 | + | + | + | 11 | 11 | 11 | 10 | |||||||||
| M45 | + | + | + | 11 | 11 | 11 | 9 | |||||||||
| M45 | + | + | + | 11 | 11 | 12 | 11 | |||||||||
| M45 | + | + | + | 12 | 11 | 9 | 10 | |||||||||
| M45 | + | + | + | + | 10 | 11 | 10 | 11 | ||||||||
| M45 | + | + | + | + | 11 | 11 | 11 | 11 | ||||||||
| M45 | + | + | + | + | 11 | 11 | 12 | 10 | ||||||||
| M45 | + | + | + | + | 11 | 11 | 10 | 12 | ||||||||
| M45 | + | + | + | + | 12 | 11 | 11 | 11 | ||||||||
| M3 | + | + | + | + | 9 | 11 | 8 | 10 | ||||||||
| M3 | + | + | + | + | 9 | 11 | 10 | 10 | ||||||||
| M3 | + | + | + | + | 9 | 11 | 11 | 9 | ||||||||
| M3 | + | + | + | + | 10 | 11 | 8 | 10 | ||||||||
| M3 | + | + | + | + | 10 | 11 | 9 | 10 | ||||||||
| M3 | + | + | + | + | 10 | 11 | 10 | 10 | ||||||||
| M3 | + | + | + | + | 10 | 11 | 10 | 11 | ||||||||
| M3 | + | + | + | + | 10 | 11 | 11 | 10 | ||||||||
| M3 | + | + | + | + | 10 | 11 | 11 | 9 | ||||||||
| M3 | + | + | + | + | 10 | 11 | 11 | 11 | ||||||||
| M3 | + | + | + | + | 10 | 11 | 12 | 9 | ||||||||
| M3 | + | + | + | + | 10 | 11 | 12 | 10 | ||||||||
| M3 | + | + | + | + | 10 | 11 | 14 | 10 | ||||||||
| M3 | + | + | + | + | 10 | 12 | 11 | 10 | ||||||||
| M3 | + | + | + | + | 10 | 12 | 11 | 11 | ||||||||
| M3 | + | + | + | + | 10 | 12 | 12 | 10 | ||||||||
| M3 | + | + | + | + | 10 | 13 | 11 | 10 | ||||||||
| M3 | + | + | + | + | 11 | 11 | 8 | 10 | ||||||||
| M3 | + | + | + | + | 11 | 11 | 11 | 10 | ||||||||
| M3 | + | + | + | + | 11 | 11 | 12 | 10 | ||||||||
| M3 | + | + | + | + | 11 | 11 | 12 | 11 | ||||||||
| M3 | + | + | + | + | 11 | 12 | 11 | 10 | ||||||||
| M17 | + | + | + | + | 11 | 11 | 12 | 11 | ||||||||
| M17 | + | + | + | + | 13 | 9 | 12 | 10 | ||||||||
| M17 | + | + | + | + | 13 | 11 | 10 | 11 | ||||||||
| M17 | + | + | + | + | 13 | 11 | 12 | 10 | ||||||||
| M17 | + | + | + | + | 13 | 11 | 12 | 11 | ||||||||
| M17 | + | + | + | + | 13 | 11 | 13 | 11 | ||||||||
| M17 | + | + | + | + | 14 | 11 | 12 | 10 | ||||||||
| M119 | + | + | + | 12 | 11 | 10 | 10 | |||||||||
| M119 | + | + | + | 12 | 12 | 10 | 10 | |||||||||
| 7C | + | + | + | 11 | 11 | 10 | 10 | |||||||||
| 7C | + | + | + | 12 | 11 | 10 | 11 | |||||||||
| Tat-C | + | + | + | + | 11 | 11 | 10 | 10 | ||||||||
| Tat-C | + | + | + | + | 11 | 11 | 10 | 11 | ||||||||
| Tat-C | + | + | + | + | 11 | 11 | 10 | 12 | ||||||||
| Tat-C | + | + | + | + | 11 | 11 | 11 | 11 | ||||||||
| Tat-C | + | + | + | + | 11 | 12 | 10 | 11 | ||||||||
| Tat-C | + | + | + | + | 12 | 11 | 10 | 10 | ||||||||
| Tat-C | + | + | + | + | 12 | 11 | 10 | 11 | ||||||||
| S4Y-T | + | 10 | 13 | 11 | 9 | |||||||||||
| S4Y-T | + | 11 | 11 | 11 | 10 | |||||||||||
| S4Y-T | + | 11 | 13 | 11 | 9 | |||||||||||
| S4Y-T | + | 12 | 11 | 10 | 10 | |||||||||||
| S4Y-T | + | 12 | 12 | 10 | 9 | |||||||||||
| S4Y-T | + | 12 | 12 | 12 | 9 | |||||||||||
| S4Y-T | + | 12 | 13 | 9 | 9 | |||||||||||
| S4Y-T | + | 12 | 13 | 10 | 9 | |||||||||||
| S4Y-T | + | 12 | 13 | 11 | 9 | |||||||||||
| S4Y-T | + | 12 | 14 | 11 | 10 | |||||||||||
| S4Y-T | + | 12 | 14 | 12 | 10 | |||||||||||
| S4Y-T | + | 13 | 12 | 9 | 10 | |||||||||||
| S4Y-T | + | 13 | 12 | 10 | 10 | |||||||||||
| S4Y-T | + | 13 | 13 | 10 | 9 | |||||||||||
| S4Y-T | + | 13 | 13 | 11 | 9 | |||||||||||
| S4Y-T | + | 14 | 12 | 12 | 9 | |||||||||||
| S4Y-T | + | 14 | 13 | 10 | 9 | |||||||||||
| S4Y-T | + | 14 | 13 | 11 | 9 | |||||||||||
| S4Y-T | + | 14 | 13 | 12 | 9 | |||||||||||
| S4Y-T | + | 15 | 13 | 10 | 9 | |||||||||||
| S4Y-T/7C | + | + | 12 | 10 | 11 | 10 | ||||||||||
| S4Y-T/7C | + | + | 12 | 12 | 11 | 10 | ||||||||||
| M48 | + | + | 11 | 12 | 11 | 9 | ||||||||||
| M48 | + | + | 12 | 12 | 11 | 9 | ||||||||||
| M48 | + | + | 12 | 12 | 12 | 9 | ||||||||||
| M48 | + | + | 12 | 13 | 12 | 9 | ||||||||||
| M48 | + | + | 13 | 12 | 11 | 9 | ||||||||||
| M48 | + | + | 13 | 12 | 12 | 9 | ||||||||||
| M48 | + | + | 13/14 | 11 | 11 | 9 | ||||||||||
| M48 | + | + | 14 | 11 | 11 | 9 | ||||||||||
| M48 | + | + | 14 | 12 | 10 | 9 | ||||||||||
| M48 | + | + | 14 | 12 | 11 | 9 | ||||||||||
| M48/7C | + | + | + | 13 | 12 | 11 | 9 | |||||||||
| YAP | + | 10 | 11 | 10 | 10 | |||||||||||
| YAP | + | 11 | 11 | 11 | 9 | |||||||||||
| YAP | + | 10 | 11 | 11 | 11 | |||||||||||
| YAP | + | 12 | 11 | 12 | 10 | |||||||||||
| Null | 10 | 11 | 11 | 10 | ||||||||||||
| Null | 13 | 11 | 11 | 10 | ||||||||||||
Table 1.
Y-Chromosome Haplotypes in Native American and Siberian Populations
|
Allele at Locus |
||||||||||||||||
| Haplogroup | M89 | M9 | M45 | M173 | M3 | M17 | M119 | DYS7C | Tat | RPS4Y | M48 | YAP | DYS19 | DYS388 | DYS390 | DYS391 |
| M89 | + | 11 | 13 | 10 | 10 | |||||||||||
| M89 | + | 11 | 14 | 10 | 9 | |||||||||||
| M89 | + | 12 | 9 | 10 | 10 | |||||||||||
| M89 | + | 12 | 12 | 10 | 10 | |||||||||||
| M89 | + | 12 | 11 | 9 | 10 | |||||||||||
| M89 | + | 11 | 12 | 11 | 11 | |||||||||||
| M9 | + | + | 11 | 9 | 11 | 10 | ||||||||||
| M9 | + | + | 12 | 11 | 9 | 10 | ||||||||||
| M9 | + | + | 12 | 11 | 10 | 10 | ||||||||||
| M9 | + | + | 13 | 11 | 11 | 10 | ||||||||||
| M9 | + | + | 14 | 12 | 10 | 10 | ||||||||||
| M45 | + | + | + | 10 | 11 | 9 | 10 | |||||||||
| M45 | + | + | + | 10 | 11 | 10 | 10 | |||||||||
| M45 | + | + | + | 10 | 11 | 11 | 10 | |||||||||
| M45 | + | + | + | 10 | 11 | 11 | 11 | |||||||||
| M45 | + | + | + | 10 | 11 | 12 | 11 | |||||||||
| M45 | + | + | + | 10 | 11 | 12 | 10 | |||||||||
| M45 | + | + | + | 10 | 11 | 13 | 10 | |||||||||
| M45 | + | + | + | 10/15 | 11 | 11 | 10 | |||||||||
| M45 | + | + | + | 10 | 12 | 9 | 10 | |||||||||
| M45 | + | + | + | 10 | 12 | 9 | 11 | |||||||||
| M45 | + | + | + | 11 | 11 | 9 | 10 | |||||||||
| M45 | + | + | + | 11 | 11 | 10 | 10 | |||||||||
| M45 | + | + | + | 11 | 11 | 11 | 10 | |||||||||
| M45 | + | + | + | 11 | 11 | 11 | 9 | |||||||||
| M45 | + | + | + | 11 | 11 | 12 | 11 | |||||||||
| M45 | + | + | + | 12 | 11 | 9 | 10 | |||||||||
| M45 | + | + | + | + | 10 | 11 | 10 | 11 | ||||||||
| M45 | + | + | + | + | 11 | 11 | 11 | 11 | ||||||||
| M45 | + | + | + | + | 11 | 11 | 12 | 10 | ||||||||
| M45 | + | + | + | + | 11 | 11 | 10 | 12 | ||||||||
| M45 | + | + | + | + | 12 | 11 | 11 | 11 | ||||||||
| M3 | + | + | + | + | 9 | 11 | 8 | 10 | ||||||||
| M3 | + | + | + | + | 9 | 11 | 10 | 10 | ||||||||
| M3 | + | + | + | + | 9 | 11 | 11 | 9 | ||||||||
| M3 | + | + | + | + | 10 | 11 | 8 | 10 | ||||||||
| M3 | + | + | + | + | 10 | 11 | 9 | 10 | ||||||||
| M3 | + | + | + | + | 10 | 11 | 10 | 10 | ||||||||
| M3 | + | + | + | + | 10 | 11 | 10 | 11 | ||||||||
| M3 | + | + | + | + | 10 | 11 | 11 | 10 | ||||||||
| M3 | + | + | + | + | 10 | 11 | 11 | 9 | ||||||||
| M3 | + | + | + | + | 10 | 11 | 11 | 11 | ||||||||
| M3 | + | + | + | + | 10 | 11 | 12 | 9 | ||||||||
| M3 | + | + | + | + | 10 | 11 | 12 | 10 | ||||||||
| M3 | + | + | + | + | 10 | 11 | 14 | 10 | ||||||||
| M3 | + | + | + | + | 10 | 12 | 11 | 10 | ||||||||
| M3 | + | + | + | + | 10 | 12 | 11 | 11 | ||||||||
| M3 | + | + | + | + | 10 | 12 | 12 | 10 | ||||||||
| M3 | + | + | + | + | 10 | 13 | 11 | 10 | ||||||||
| M3 | + | + | + | + | 11 | 11 | 8 | 10 | ||||||||
| M3 | + | + | + | + | 11 | 11 | 11 | 10 | ||||||||
| M3 | + | + | + | + | 11 | 11 | 12 | 10 | ||||||||
| M3 | + | + | + | + | 11 | 11 | 12 | 11 | ||||||||
| M3 | + | + | + | + | 11 | 12 | 11 | 10 | ||||||||
| M17 | + | + | + | + | 11 | 11 | 12 | 11 | ||||||||
| M17 | + | + | + | + | 13 | 9 | 12 | 10 | ||||||||
| M17 | + | + | + | + | 13 | 11 | 10 | 11 | ||||||||
| M17 | + | + | + | + | 13 | 11 | 12 | 10 | ||||||||
| (continued) | ||||||||||||||||
Table 1 (continued).
|
Allele at Locus |
||||||||||||||||
| Haplogroup | M89 | M9 | M45 | M173 | M3 | M17 | M119 | DYS7C | Tat | RPS4Y | M48 | YAP | DYS19 | DYS388 | DYS390 | DYS391 |
| M17 | + | + | + | + | 13 | 11 | 12 | 11 | ||||||||
| M17 | + | + | + | + | 13 | 11 | 13 | 11 | ||||||||
| M17 | + | + | + | + | 14 | 11 | 12 | 10 | ||||||||
| M119 | + | + | + | 12 | 11 | 10 | 10 | |||||||||
| M119 | + | + | + | 12 | 12 | 10 | 10 | |||||||||
| 7C | + | + | + | 11 | 11 | 10 | 10 | |||||||||
| 7C | + | + | + | 12 | 11 | 10 | 11 | |||||||||
| Tat-C | + | + | + | + | 11 | 11 | 10 | 10 | ||||||||
| Tat-C | + | + | + | + | 11 | 11 | 10 | 11 | ||||||||
| Tat-C | + | + | + | + | 11 | 11 | 10 | 12 | ||||||||
| Tat-C | + | + | + | + | 11 | 11 | 11 | 11 | ||||||||
| Tat-C | + | + | + | + | 11 | 12 | 10 | 11 | ||||||||
| Tat-C | + | + | + | + | 12 | 11 | 10 | 10 | ||||||||
| Tat-C | + | + | + | + | 12 | 11 | 10 | 11 | ||||||||
| S4Y-T | + | 10 | 13 | 11 | 9 | |||||||||||
| S4Y-T | + | 11 | 11 | 11 | 10 | |||||||||||
| S4Y-T | + | 11 | 13 | 11 | 9 | |||||||||||
| S4Y-T | + | 12 | 11 | 10 | 10 | |||||||||||
| S4Y-T | + | 12 | 12 | 10 | 9 | |||||||||||
| S4Y-T | + | 12 | 12 | 12 | 9 | |||||||||||
| S4Y-T | + | 12 | 13 | 9 | 9 | |||||||||||
| S4Y-T | + | 12 | 13 | 10 | 9 | |||||||||||
| S4Y-T | + | 12 | 13 | 11 | 9 | |||||||||||
| S4Y-T | + | 12 | 14 | 11 | 10 | |||||||||||
| S4Y-T | + | 12 | 14 | 12 | 10 | |||||||||||
| S4Y-T | + | 13 | 12 | 9 | 10 | |||||||||||
| S4Y-T | + | 13 | 12 | 10 | 10 | |||||||||||
| S4Y-T | + | 13 | 13 | 10 | 9 | |||||||||||
| S4Y-T | + | 13 | 13 | 11 | 9 | |||||||||||
| S4Y-T | + | 14 | 12 | 12 | 9 | |||||||||||
| S4Y-T | + | 14 | 13 | 10 | 9 | |||||||||||
| S4Y-T | + | 14 | 13 | 11 | 9 | |||||||||||
| S4Y-T | + | 14 | 13 | 12 | 9 | |||||||||||
| S4Y-T | + | 15 | 13 | 10 | 9 | |||||||||||
| S4Y-T/7C | + | + | 12 | 10 | 11 | 10 | ||||||||||
| S4Y-T/7C | + | + | 12 | 12 | 11 | 10 | ||||||||||
| M48 | + | + | 11 | 12 | 11 | 9 | ||||||||||
| M48 | + | + | 12 | 12 | 11 | 9 | ||||||||||
| M48 | + | + | 12 | 12 | 12 | 9 | ||||||||||
| M48 | + | + | 12 | 13 | 12 | 9 | ||||||||||
| M48 | + | + | 13 | 12 | 11 | 9 | ||||||||||
| M48 | + | + | 13 | 12 | 12 | 9 | ||||||||||
| M48 | + | + | 13/14 | 11 | 11 | 9 | ||||||||||
| M48 | + | + | 14 | 11 | 11 | 9 | ||||||||||
| M48 | + | + | 14 | 12 | 10 | 9 | ||||||||||
| M48 | + | + | 14 | 12 | 11 | 9 | ||||||||||
| M48/7C | + | + | + | 13 | 12 | 11 | 9 | |||||||||
| YAP | + | 10 | 11 | 10 | 10 | |||||||||||
| YAP | + | 11 | 11 | 11 | 9 | |||||||||||
| YAP | + | 10 | 11 | 11 | 11 | |||||||||||
| YAP | + | 12 | 11 | 12 | 10 | |||||||||||
| Null | 10 | 11 | 11 | 10 | ||||||||||||
| Null | 13 | 11 | 11 | 10 | ||||||||||||
Figure 3.

Network of haplotypes comprising haplogroups M45 and M3. Each box represents one microsatellite haplotype for the SNP haplogroup encompassed by the figure. The number of repeats for each microsatellite allele is given in the order DYS19-DYS388-DYS390-DYS391. An asterisk (*) in the lower left-hand corner of the box indicates that this haplotype harbors the M173 variant. The number under the haplotype indicates the number of individuals in the present study who harbor that haplotype. A dashed box represents an intermediate haplotype that was not observed in the present study. The colors of the haplotype boxes indicate the geographic region in which that haplotype was detected, with the color code defined in the figures.
Figure 4.

Network of haplotypes comprising haplogroups DYS7C and Tat-C. See figure 3 legend for explanation.
Figure 5.
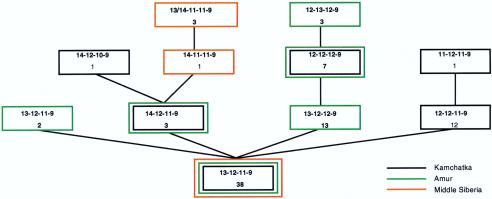
Network of haplotypes comprising the RPS4Y-T subhaplogroup M48. See figure 3 legend for explanation.
In the network of the M45 and M3 haplotypes (fig. 3), the proposed founder of the M3 lineage is the haplotype M3(10-11-11-10). This haplotype is the most frequent M3 haplotype (found in 43 individuals) and has the widest geographic distribution, appearing in all continental regions of the Americas as well as in Chukotka. Furthermore, it forms a direct link with the M45 haplogroup through the haplotype M45(10-11-11-10), the two differing by only the M3 SNP. Moreover, M45(10-11-11-10) is found from southern Middle Siberia all the way to Chukotka in the northeast. Thus, M45(10-11-11-10) migrated from southern Middle Siberia to northeastern Siberia, where the M3 marker arose.
M3(10-11-11-10) most likely arose in northeastern Siberia, rather than arising in the Americas and entering Chukotka via back migration from the Americas, as suggested elsewhere (Karafet et al. 1999). This conclusion is based on our discovery of two derivatives of M3(10-11-11-10)—M3(10-12-12-10) and M3(10-13-11-10)—that are Chukotkan specific and not found in Native Americans. This Chukotka-specific radiation of M3(10-11-11-10) indicates that M3(10-11-11-10) has resided in greater Beringia longer than it has resided in the Americas. Hence, the M3 haplogroup arose in a Beringian population ancestral to both Native Americans and northeastern Siberians.
M45(10-11-11-10) has the widest geographic distribution of all M45 haplotypes, with a range from southern Siberia to South America. Thus, this Y-chromosome haplotype appears to have been borne by an initial Native American male migration that originated in southern Middle Siberia, moved through Beringia, and colonized the Americas, giving rise to the M3(10-11-11-10) haplotype along the way.
The microsatellite data revealed a dichotomy between the M45 haplotypes of eastern Siberia and southern Middle Siberia (fig. 3). One cluster of four M45 haplotypes (subhaplogroup M45a) was characteristic of southern Middle Siberia. Of these four M45 haplotypes, two (including the progenitor of the M3 lineage) were shared with Native Americans from North, Central, and South America. A third M45a haplotype was observed only in the Americas, and the fourth, a southern Middle Siberian–specific M45 haplotype—M45(10/15-11-11-10)—was found in a single individual and differed from the ancestor of M3 only by a duplication at the DYS19 locus.
The second cluster of M45 haplotypes (subhaplogroup M45b) encompassed all of the Y chromosomes harboring the M173 variant and was found in the Lower Amur and Kamchatka Regions of eastern Siberia. These haplotypes were shared with Native Americans from North and Central America but not with those from South America, with one exception, a South American Wapishana. Hence, the M45b subhaplogroup is defined by both the M173 variant (designated by an asterisk [*]) and distinctive microsatellite alleles (fig. 3).
Thus, the M45 haplotype associations between Siberia and the Americas indicate two distinct migrations. The Middle Siberian subhaplogroup M45a was the progenitor of the initial Amerind migration to the New World, whereas the eastern Siberian subhaplogroup M45b was among the progenitors that contributed to a second migration that contributed to the North American Na-Dene and adjacent Amerind populations.
Consistent with this conclusion, the microsatellite allele frequencies of M45 haplotypes were found to differ greatly between northern and southern populations of Native Americans (table 2). The most frequent alleles at the DYS19, DYS390, and DYS391 loci differed between northern and southern Amerindian populations. In fact, microsatellite DYS390 differed between North and South America by two mutational steps. This striking shift in allele frequencies was not observed in the M3 haplotypes in North and Central America versus those in South America (table 3), where the predominant DYS19, DYS388, and DYS391 alleles were the same and where the predominant allele at the more mutable DYS390 differed by only one step. These data are consistent with a single recent origin of haplogroup M3 in Chukotka and the Americas and a with dual origin of Native American haplogroup M45 Y chromosomes, M45a and M45b, in Middle and eastern Siberia, respectively.
Table 2.
Microsatellite Allele Frequencies on Haplogroup M45 Chromosomes from the Americas
|
Frequency ina |
||
| MicrosatelliteLocus andAllele | Central and North America (n=33) | South America (n=31) |
| DYS19: | ||
| 10 | .242 | .968 |
| 11 | .697 | .032 |
| 12 | .061 | .00 |
| DYS388: | ||
| 11 | 1.00 | .839 |
| 12 | .00 | .161 |
| DYS390: | ||
| 9 | .091 | .581 |
| 10 | .091 | .387 |
| 11 | .727 | .032 |
| 12 | .091 | .00 |
| DYS391: | ||
| 9 | .030 | .00 |
| 10 | .485 | .935 |
| 11 | .485 | .032 |
| 12 | .00 | .032 |
Most frequent allele is shown in boldface italics.
Table 3.
Microsatellite Allele Frequencies on Haplogroup M3 Chromosomes from the Americas
|
Frequency ina |
||
| MicrosatelliteLocus andAllele | Central and North America (n=98) | South America (n=47) |
| DYS19: | ||
| 9 | .020 | .064 |
| 10 | .918 | .532 |
| 11 | .061 | .404 |
| DYS388: | ||
| 11 | .990 | .915 |
| 12 | .010 | .085 |
| 13 | .00 | .00 |
| DYS390: | ||
| 8 | .00 | .149 |
| 9 | .051 | .00 |
| 10 | .163 | .234 |
| 11 | .571 | .255 |
| 12 | .214 | .340 |
| 14 | .00 | .021 |
| DYS391: | ||
| 9 | .071 | .00 |
| 10 | .827 | .936 |
| 11 | .102 | .064 |
Most frequent allele is shown in boldface italics.
The network of Tat-C and DYS7C haplotypes (fig. 4) revealed that the ancestral Tat-C haplotype (7C[11-11-10-10]) was found only in southern Middle Siberia, indicating that this Y-chromosome lineage arose in that region. Moreover, the limited microsatellite diversity and resulting compact nature of the network indicates that the Tat-C lineage arose relatively recently (Zerjal et al. 1997). The absence of the Tat-C haplogroup in the Americas, with the exception of a single Navajo (Karafet et al. 1999), along with its high frequency in both northern Europe and northeastern Siberia, indicates that the Tat-C lineage was disseminated from central Asia by both westward and eastward male migrations, the eastward migration reaching Chukotka after the Bering Land Bridge was submerged. Both the M45 and Tat-C haplogroups have been found in Europe, indicating both ancient and recent central Asian influences. However, neither of these major Middle Siberian Y-chromosome lineages appears to have been greatly influenced by the paternal gene pool of Han Chinese or other East Asian populations (Su et al. 1999).
The network of all RPS4Y-T/M48 haplotypes revealed a high degree of diversity in eastern Siberia, with lower diversity in Middle Siberia. The greatest diversity was observed in the Koryaks from Kamchatka and the Ulchi/Nanai from the Lower Amur, both of which showed six different microsatellite haplotypes. This eastern Siberian RPS4Y-T heterogeneity was further demonstrated when the subset of RPS4Y-T haplotypes defined by the M48 marker were included in a network (fig. 5). Thus, the RPS4Y-T and M48 haplogroups appear to have arisen in populations ancestral to the modern inhabitants of Kamchatka and the Lower Amur and may have only recently expanded into the Middle Siberian populations around Lake Baikal.
Discussion
The prevalence of the distinctive Y-chromosome M3 lineage in Native Americans that is further defined by a specific DYS19 allele and alphoid repeat variant initially led to the suggestion that a single patrilineal migration gave rise to all of the linguistically diverse Native American populations (Pena et al. 1995; Santos et al. 1996; Underhill et al. 1996; Bianchi et al. 1997, 1998). The observation that most of the non-M3 Y chromosomes in five Colombian populations were similar to each other suggested that there was a second Native American founder Y chromosome (Ruiz-Linares et al. 1999) but was still consistent with there having been a single migration.
A subsequent study of six Y-chromosome loci (Santos et al. 1999) suggested a single origin for most Native American Y chromosomes, traced through northeastern Siberia back to Middle Siberia, along with a possible second entry from Beringia, a scenario reminiscent of several mtDNA studies (Forster et al. 1996; Bonatto and Salzano 1997). The subsequent discovery of haplogroup RPS4Y-T in several northern Amerind and Na-Dene populations, along with its increased frequency around Lake Baikal and Mongolia, supported the theory of two independent migratory events that gave rise to present-day Native American Y chromosomes (Karafet et al. 1999)
Our results support and extend the hypothesis of at least two major male migrations from Asia to the Americas. The first migration brought haplogroup M3 Y chromosomes from Chukotka to the Americas. The founder haplotype of this lineage, M3(10-11-11-10), was derived from the southern Middle Siberian haplotype M45(10-11-11-10) during the latter’s migration through Chukotka and across the Bering Land Bridge down into North, Central, and South America. The southern Middle Siberian origin of this initial migration is further supported by the presence of M45(10-11-11-10) and of the closely related haplotype M45(10-11-10-10) in the Tuvan population. This population currently lives near the geographic center of Asia, in the region of arid steppes between Mongolia and the Sayan Mountains. Thus, the first Siberian migration into the Americas arose in southern Middle Siberia.
The Tuvans and three other populations from the Upper Yenisey region west of Lake Baikal (the Tofalars, Buryats, and Yenisey Evenks) also harbored the ancestral Tat-C haplotypes. Thus, the Tuvans contain remnants of the source of both major male expansions from central Asia into Siberia. The earlier M45 migration, which acquired the M3 variant in northeastern Siberia, moved into the Americas. The later Tat-C migration reached the northeast Siberian coast but did not enter the Americas. Both migrations also moved westward into Europe. Indeed, the dispersal of the Tat-C haplogroup most likely accounts for the clustering of Y chromosomes from certain Middle Siberian populations with those of Europeans rather than with those of other Siberians or East Asians (Santos et al. 1999).
The M45 haplogroup is divided into two subhaplogroups, M45a from Middle Siberia and M45b from eastern Siberia. These two lineages are distinguished by the M173 variant in the eastern Siberian M45b lineage as well as by different microsatellite alleles. The M45a subhaplogroup connects Middle Siberians with the North, Central, and South American Amerinds. The M45b/M173 subhaplogroup connects eastern Siberians with the North and Central American Na-Dene and surrounding Amerinds (fig. 3).
The distinctive form, frequency, and distribution of the M45b subhaplogroup confirms that two separate migrations occurred. The M173 marker is only found in the M45 Y chromosomes of the eastern Siberians and North and Central American natives and not in those of the Middle Siberians or South Americans. Furthermore, three of the distinct North and Central American M45 haplotypes (M45[*,M173][11-11-11-11] and M45*[12-11-11-11]) are only shared with the populations of the Lower Amur and the Sea of Okhotsk region (fig. 3).
This second Siberian migration also corresponds with the distribution of the S4Y-T macrohaplogroup (Karafet et al. 1999). In our study, the S4Y-T haplogroup marker, the RPS4Y-T, was detected in a single Navajo, but it had previously been seen in additional northern Amerind and Na-Dene Native Americans (Bergen et al. 1999; Karafet et al. 1999). Moreover, our data demonstrate that the Native American RPS4Y-T haplogroup originated in the eastern Siberian populations of Kamchatka and the Lower Amur River basin. The extended RPS4Y-T haplotype of our Navajo sample differs from a Lower Amur RPS4Y-T haplotype by just one mutational step but differs from those of southern Middle Siberia by three steps. Thus, the Native American RPS4Y-T Y chromosomes also came from eastern Siberia, along with the M45b chromosomes. These two haplogroups provide compelling evidence that there was a second male migration to North America from the eastern Siberian regions of Kamchatka and the Lower Amur River. This eastern Siberian RPS4Y-T lineage can be traced back to East Asia, where highly diversified RPS4Y-T haplotypes have been found (B. Su and L. Jin, unpublished data).
Comparison of Y-Chromosome and mtDNA Data
Our identification of two major male migrations into the New World—one from southern Middle Siberia, bringing Y-chromosome haplogroups M45a and M3, and a second from eastern Siberia, bringing haplogroups M45b and RPS4Y-T—correlates well with previous conclusions about the maternal migrations that brought mtDNA haplogroups A, B, C, and D to the Americas. Since all Siberian migrations necessarily came through northwestern North America, the more southern distribution of the M45a and M3 lineages versus the M45b and RPS4Y-T lineages indicates that the southern Middle Siberian migration predated the eastern Siberian migration.
The Y-chromosome M45a and M3 lineages, together with the mtDNA haplogroups C and D and the Amerind sublineages of mtDNA haplogroup A, are all found together in southern North America as well as in Central and South America. Furthermore, the M45a Y chromosomes—which are the precursors to the Native American M3 lineage—and the mtDNA haplogroups C and D are at their highest frequencies in southern Middle Siberia, with the M3 lineage and the Amerind mtDNA haplogroup A sublineages both being present in Chukotka. Hence, the first Native American migration must have originated in southern Middle Siberia, traversed Chukotka, and entered the Americas. If we assume that the Amerind Y-chromosome lineages arrived together with mtDNA haplogroups C and D, then this migration occurred ∼20,000–30,000 years before present (YBP) (Schurr et al. 1999).
Similarly, the Y-chromosome haplogroups M45b and RPS4Y-T, along with the sublineage of mtDNA haplogroup A defined by the control-region sequence variant 16192T and the RsaI polymorphism at np16392, are defining features of the Na-Dene of northwestern North America. Furthermore, the M45b and RPS4Y-T Y-chromosome lineages are found at their highest frequencies in the Lower Amur and Sea of Okhotsk regions of eastern Siberia, having originated earlier in Southeast Asia (B. Su and L. Jin, unpublished data). This implies that a major component of the Na-Dene migration arose in southeastern Siberia. Likewise, the precursor of the haplogroup A 16392 RsaI sublineage, defined by the control-region variant 16192T, has been observed in Chukotka and Kamchatka (Schurr et al. 1999), which border on the Sea of Okhotsk. Assuming that the mtDNA haplogroup A 16192T sublineage arrived in the Americas together with the Y-chromosome lineages M45b and RPS4Y-T, then this migration came from southeastern Siberia at ∼7,000–9,500 YBP.
In conclusion, there appears to be a striking correspondence between Siberian and Native American Y-chromosome and mtDNA haplogroup distributions, and, hence, they must have been associated during trans-Beringian migrations. The results strongly suggest that both males and females came to the New World in at least two coherent waves of migration, the first arising in southern Middle Siberia and the second arising later from southeastern Siberia.
Acknowledgments
This work was supported by National Institutes of Health grants NS21328, HL45572, and AG13154 (to D.C.W.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Genome Database, The, http://www.gdb.org/ (for DYS388 [accession number G00-365-729)
References
- Bergen AW, Wang CY, Tsai J, Jefferson K, Dey C, Smith KD, Park SC, Tsai SJ, Goldman D (1999) An Asian-Native American paternal lineage identified by RPS4Y resequencing and microsatellite haplotyping. Ann Hum Genet 63:63–80 [DOI] [PubMed] [Google Scholar]
- Bianchi NO, Bailliet G, Bravi CM, Carnese RF, Rothhammer F, Martinez-Marignac VL, Pena SD (1997) Origin of Amerindian Y-chromosomes as inferred by the analysis of six polymorphic markers. Am J Phys Anthropol 102:79–89 [DOI] [PubMed] [Google Scholar]
- Bianchi NO, Catanesi CI, Bailliet G, Martinez-Marignac VL, Bravi CM, Vidal-Rioja LB, Herrera RJ, Lopez-Camelo JS (1998) Characterization of ancestral and derived Y-chromosome haplotypes of New World native populations. Am J Hum Genet 63:1862–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black LT (1988) Peoples of the Amur and maritime regions. In: Fitzhugh WW, Crowell A (eds) Crossroads of continents: cultures of Siberia and Alaska. Smithsonian Institution Press, Washington, DC, pp 24–31 [Google Scholar]
- Bonatto SL, Salzano FM (1997) A single and early migration for the peopling of the Americas supported by mitochondrial DNA sequence data. Proc Natl Acad Sci USA 94:1866–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Silva DR, Santos FR, Hutz MH, Salzano FM, Pena SD (1999) Divergent human Y-chromosome microsatellite evolution rates. J Mol Evol 49:204–214 [DOI] [PubMed] [Google Scholar]
- Cooper G, Amos W, Hoffman D, Rubinsztein DC (1996) Network analysis of human Y microsatellite haplotypes. Hum Mol Genet 5:1759–766 [DOI] [PubMed] [Google Scholar]
- Deka R, Jin L, Shriver MD, Yu LM, Saha N, Barrantes R, Chakraborty R, Ferrell RE (1996) Dispersion of human Y chromosome haplotypes based on five microsatellites in global populations. Genome Res 6:1177–1184 [DOI] [PubMed] [Google Scholar]
- de Knijff P, Kayser M, Caglia A, Corach D, Fretwell N, Gehrig C, Graziosi G, et al (1997) Chromosome Y microsatellites: population genetic and evolutionary aspects. Int J Legal Med 110:134–140 [DOI] [PubMed] [Google Scholar]
- Forster P, Harding R, Torroni A, Bandelt HJ (1996) Origin and evolution of Native American mtDNA variation: a reappraisal. Am J Hum Genet 59:935–945 [PMC free article] [PubMed] [Google Scholar]
- Forster P, Kayser M, Meyer E, Roewer L, Pfeiffer H, Benkmann H, Brinkmann B (1998) Phylogenetic resolution of complex mutational features at Y-STR DYS390 in aboriginal Australians and Papuans. Mol Biol Evol 15:1108–1114 [DOI] [PubMed] [Google Scholar]
- Greenberg J, Turner CG, Zegura SL (1986) The settlement of the Americas: a comparison of linguistic, dental, and genetic evidence. Curr Anthropol 4:477–497 [Google Scholar]
- Hammer MF, Horai S (1995) Y chromosomal DNA variation and the peopling of Japan. Am J Hum Genet 56:951–962 [PMC free article] [PubMed] [Google Scholar]
- Horai S, Kondo R, Nakagawa-Hattori Y, Hayashi S, Sonoda S, Tajima K (1993) Peopling of the Americas, founded by four major lineages of mitochondrial DNA. Mol Biol Evol 10:23–47 [DOI] [PubMed] [Google Scholar]
- Huoponen K, Torroni A, Wickman PR, Sellitto D, Gurley DS, Scozzari R, Wallace DC (1997) Mitochondrial DNA and Y chromosome-specific polymorphisms in the Seminole tribe of Florida. Eur J Hum Genet 5:25–34 [PubMed] [Google Scholar]
- Jobling MA, Samara V, Pandya A, Fretwell N, Bernasconi B, Mitchell RJ, Gerelsaikhan T, Dashnyam B, Sajantila A, Salo PJ, Nakahori Y, Disteche CM, Thangaraj K, Singh L, Crawford MH, Tyler-Smith C (1996) Recurrent duplication and deletion polymorphisms on the long arm of the Y chromosome in normal males. Hum Mol Genet 5:1767–1775 [DOI] [PubMed] [Google Scholar]
- Jobling MA, Tyler-Smith C (1995) Fathers and sons: the Y chromosome and human evolution. Trends Genet 11:449–456 [DOI] [PubMed] [Google Scholar]
- Karafet TM, Zegura SL, Posukh O, Osipova L, Bergen A, Long J, Goldman D, Klitz W, Harihara S, de Knijff P, Wiebe V, Griffiths RC, Templeton AR, Hammer MF (1999) Ancestral Asian source(s) of new world Y-chromosome founder haplotypes. Am J Hum Genet 64:817–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafet T, Zegura SL, Vuturo-Brady J, Posukh O, Osipova L, Wiebe V, Romero F, Long JC, Harihara S, Jin F, Dashnyam B, Gerelsaikhan T, Omoto K, Hammer MF (1997) Y chromosome markers and trans-Bering Strait dispersals. Am J Phys Anthropol 102:301–314 [DOI] [PubMed] [Google Scholar]
- Kayser M, Caglia A, Corach D, Fretwell N, Gehrig C, Graziosi G, Heidorn F, et al (1997) Evaluation of Y-chromosomal STRs: a multicenter study. Int J Legal Med 110:125–133 [DOI] [PubMed] [Google Scholar]
- Krauss ME (1988) Many tongues, ancient tales: peoples of the Amur and maritime regions. In: Fitzhugh WW, Crowell A (eds) Crossroads of continents: cultures of Siberia and Alaska. Smithsonian Institution Press, Washington, DC, pp 145–150 [Google Scholar]
- Lell JT, Brown MD, Schurr TG, Sukernik RI, Starikovskaya YB, Torroni A, Moore LG, Troup GM, Wallace DC (1997) Y chromosome polymorphisms in Native American and Siberian populations: identification of native American Y chromosome haplotypes. Hum Genet 100:536–543 [DOI] [PubMed] [Google Scholar]
- Levin MG, Potapov LP (1964) The peoples of Siberia. University of Chicago Press, Chicago [Google Scholar]
- Merriwether DA, Hall WW, Vahlne A, Ferrell RE (1996) mtDNA variation indicates Mongolia may have been the source for the founding population for the New World. Am J Hum Genet 59:204–212 [PMC free article] [PubMed] [Google Scholar]
- Pena SDJ, Santos FR, Bianchi N, Bravi CM, Carnese FR, Rothhammer F, Gerelsaikhan T, Munkhtuja B, Oyunsuren T (1995) Identification of a major founder Y-chromosome haplotype in Amerindians. Nat Genet 11:15–16 [DOI] [PubMed] [Google Scholar]
- Roewer L, Arneman J, Spurr NK, Grzeschik KH, Epplen JT (1992) Simple repeat sequences on the human Y chromosome are equally polymorphic as their autosomal counterparts. Hum Genet 89:389–394 [DOI] [PubMed] [Google Scholar]
- Ruiz-Linares A, Ortiz-Barrientos D, Figueroa M, Mesa N, Munera JG, Bedoya G, Velez ID, Garcia LF, Perez-Lezaun A, Bertranpetit J, Feldman MW, Goldstein DB (1999) Microsatellites provide evidence for Y chromosome diversity among the founders of the New World. Proc Natl Acad Sci USA 96:6312–6317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos FR, Bianchi NO, Pena SD (1996) Worldwide distribution of human Y-chromosome haplotypes. Genome Res 6:601–611 [DOI] [PubMed] [Google Scholar]
- Santos FR, Pandya A, Tyler-Smith C, Pena SD, Schanfield M, Leonard WR, Osipova L, Crawford MH, Mitchell RJ (1999) The central Siberian origin for native American Y chromosomes. Am J Hum Genet 64:619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurr TG, Ballinger SW, Gan YY, Hodge JA, Merriwether DA, Lawrence DN, Knowler WC, Weiss KM, Wallace DC (1990) Amerindian mitochondrial DNAs have rare Asian mutations at high frequencies, suggesting they derived from four primary maternal lineages. Am J Hum Genet 46:613–623 [PMC free article] [PubMed] [Google Scholar]
- Schurr TG, Sukernik RI, Starikovskaya YB, Wallace DC (1999) Mitochondrial DNA variation in Koryaks and Itel'men: population replacement in the Okhotsk Sea-Bering Sea region during the Neolithic. Am J Phys Anthropol 108:1–39 [DOI] [PubMed] [Google Scholar]
- Scozzari R, Cruciani F, Santolamazza P, Sellitto D, Cole DE, Rubin LA, Labuda D, Marini E, Succa V, Vona G, Torroni A (1997) mtDNA and Y chromosome-specific polymorphisms in modern Ojibwa: implications about the origin of their gene pool. Am J Hum Genet 60:241–244 [PMC free article] [PubMed] [Google Scholar]
- Semino O, Passarino G, Oefner, PJ, Lin AA, Arbuzova, S, Beckman LE, De Benedictis G, Francalacci P, Kouvatsi A, Limborska S, Marcikiae M, Mika A, Mika B, Primorac D, Santachiara-Benerecetti AS, Cavalli-Sforza LL, Underhill PA (2000) The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science 290:1155–1159 [DOI] [PubMed] [Google Scholar]
- Shields GF, Schmiechen AM, Frazier BL, Redd A, Voevoda MI, Reed JK, Ward RH (1993) mtDNA sequences suggest a recent evolutionary divergence for Beringian and northern North American populations. Am J Hum Genet 53:549–562 [PMC free article] [PubMed] [Google Scholar]
- Starikovskaya YB, Sukernik RI, Schurr TG, Kogelnik AM, Wallace DC (1998) mtDNA diversity in Chukchi and Siberian Eskimos: implications for the genetic history of Ancient Beringia and the peopling of the New World. Am J Hum Genet 63:1473–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su B, Xiao J, Underhill P, Deka R, Zhang W, Akey J, Huang W, Shen D, Lu D, Luo J, Chu J, Tan J, Shen P, Davis R, Cavalli-Sforza L, Chakraborty R, Xiong M, Du R, Oefner P, Chen Z, Jin L (1999) Y-chromosome evidence for a northward migration of modern humans into Eastern Asia during the last Ice Age. Am J Hum Genet 65:1718–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukernik RI, Schurr TG, Starikovskaya EB, Wallace DC (1996) Mitochondrial DNA variation in native Siberians with special reference to the evolutionary history of American Indians. I. Studies on restriction polymorphism. Genetika 32:432–439 [PubMed] [Google Scholar]
- Torroni A, Chen YS, Semino O, Santachiara-Beneceretti AS, Scott CR, Lott MT, Winter M, Wallace DC (1994) mtDNA and Y-chromosome polymorphisms in four Native American populations from southern Mexico. Am J Hum Genet 54:303–318 [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Schurr TG, Cabell MF, Brown MD, Neel JV, Larsen M, Smith DG, Vullo CM, Wallace DC (1993a) Asian affinities and continental radiation of the four founding Native American mtDNAs. Am J Hum Genet 53:563–590 [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Schurr TG, Yang C-C, Szathmary EJ, Williams RC, Schanfield MS, Troup GA, Knowler WC, Lawrence DN, Weiss KM (1992) Native American mitochondrial DNA analysis indicates that the Amerind and the Nadene populations were founded by two independent migrations. Genetics 130:153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Sukernik RI, Schurr TG, Starikorskaya YB, Cabell MF, Crawford MH, Comuzzie AG, Wallace DC (1993b) mtDNA variation of aboriginal Siberians reveals distinct genetic affinities with Native Americans. Am J Hum Genet 53:591–608 [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Jin L, Lin AA, Mehdi SQ, Jenkins T, Vollrath D, Davis RW, Cavalli-Sforza LL, Oefner PJ (1997) Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res 7:996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Jin L, Zemans R, Oefner PJ, Cavalli-Sforza LL (1996) A pre-Columbian Y chromosome-specific transition and its implications for human evolutionary history. Proc Natl Acad Sci USA 93:196–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonne-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ (2000) Y chromosome sequence variation and the history of human populations. Nat Genet 26:358–361 [DOI] [PubMed] [Google Scholar]
- Wallace DC, Garrison K, Knowler WC (1985) Dramatic founder effects in Amerindian mitochondrial DNAs. Am J Phys Anthropol 68:149–155 [DOI] [PubMed] [Google Scholar]
- Zerjal T, Dashnyam B, Pandya A, Kayser M, Roewer L, Santos FR, Schiefenhovel W, Fretwell N, Jobling MA, Harihara S, Shimizu K, Semjidmaa D, Sajantila A, Salo P, Crawford MH, Ginter EK, Evgrafov OV, Tyler-Smith C (1997) Genetic relationships of Asians and Northern Europeans, revealed by Y-chromosomal DNA analysis. Am J Hum Genet 60:1174–1183 [PMC free article] [PubMed] [Google Scholar]


