Abstract
Portable microfluidic systems provide simple and effective solutions for low-cost point-of-care diagnostics and high-throughput biomedical assays. Robust flow control and precise fluidic volumes are two critical requirements for these applications. We have developed a monolithic polydimethylsiloxane (PDMS) microdevice that allows for storing and mixing subnanoliter volumes of aqueous solutions at various mixing ratios. Filling and mixing is controlled via two integrated PDMS microvalve arrays. The volumes of the microchambers are entirely defined by photolithography, hence volumes from picoliter to nanoliter can be fabricated with high precision. Because the microvalves do not require an energy input to stay closed, fluid can be stored in a highly portable fashion for several days. We have confirmed the mixing precision and predictability using fluorescence microscopy. We also demonstrate the application of the device for calibrating fluorescent calcium indicators. Due to the biocompatibility of PDMS, the device will have broad applications in miniaturized diagnostic assays as well as basic biological studies.
Keywords: Micromixing, Miniaturization, Polydimethylsiloxane, Soft lithography
1 Introduction
Miniaturization of biochemical assays in “lab-on-a-chip” devices is revolutionizing the biomedical benchtop. Microfluidic systems, especially combined with soft lithography techniques (for a review, see [1]), provide simple, low-cost, and high-throughput implementations for these miniaturization platforms. Portable microfluidic systems with precise fluidic dispensing volumes and integrated flow control functions are of great interest in numerous biomedical assays.
Various methods have been reported to construct microvalves for controlling fluid flow. Beebe et al. [2] employed the expansion and contraction of pH-responsive hydrogel components to regulate fluid flow in microchannels. Quake and co-workers demonstrated the integration of pressure-driven polydimethylsiloxane (PDMS) “pinchtype” valves in microfluidic PDMS devices [3] for applications ranging from protein crystallization [4] and cell sorting [5]. The valve is constructed by overlapping two microchannels (a “control” channel atop a fluidic channel) orthogonally, separated by a thin (< 30 µm thick) PDMS membrane. The membrane deflects when the control channel is pressurized, which interrupts flow in the fluidic channel in a manner akin to pinching a flexible tube. Due to the hydrophobic nature of PDMS, the valves are leak-proof because water is naturally excluded from the spacing between the PDMS membrane and the bottom of the channel. A clever multiplexing scheme allows for controlling N channels using only 2 log2 N control channels (e.g., 1000 microvalves using only 18 channels) [6].
However, microvalves in the aforementioned configurations require extra energy source to close the fluidic channel. As a result, the devices are not well suited for applications where the assay is not necessarily performed at the same location as the one where (some or all) reagents were loaded into the device. For valves to completely close, the pinch-type configuration [6] also requires the fluidic microchannels to be fabricated with a rounded cross-section (a square cross-section microchannel cannot be completely flattened at the walls). While satisfactory for many applications, the “photoresist reflow” method for fabricating rounded microchannels is not optimal, because the width of the features influences their final height; hence large heights (> 20 µm) and aspect ratios are not possible for every given feature width, which makes it nonideal for studies where a wide range of designs and heights on one device is critical (e.g., cellular studies [7]).
We have adapted a previously reported PDMS microvalve design that is closed at rest [8–10] to demonstrate the parallel mixing of subnanoliter volumes. The main advantages of this microvalve design are that: (i) No extra energy source is required to close the fluidic path, hence the loaded device is highly portable; and (ii) the device can be built by PDMS replicas from photolithographically patterned SU-8 molds, allowing for microfabricating deep (up to 1 mm) channels with vertical sidewalls (i.e., the height of the features can be specified independently of their width) [11] and resulting in very precise features. The prototype device features two arrays of photolithographically defined nanoliter fluidic chambers and two individually controlled sets of microvalves. We have assessed and verified the precision of the mixing with fluorescence microscopy and applied it to calibrate fluorescent calcium indicators using the micromixer. The device allowed for storage of subnanoliter sized fluid volumes for at least 7 days, which is highly desirable in portable point-of-care diagnostics.
2 Materials and methods
2.1 Fabrication of silicon masters
Standard SU-8 photolithography methods were used to create masters for the microfluidic layer and the valve control layer [12]. We designed the masks using Corel-Draw software, and printed transparency masks at 8000 dots per inch (CAT/Art services, Poway, CA, USA). Negative photoresist (SU-8 2050; MicroChem, Newton, MA, USA) was spun on silicon wafers at 500 rpm for 10 s and 2300 rpm for an additional 30 s, prebaked by ramping up at 4°C/min to 95°C (15 min) with a programmable hot plate (Dataplate Hot Plate/Stirrer Series 730; Barnstead International, Dubuque, IA, USA), and exposed for 90 s with collimated UV light (Kaspar–Quintel Model 2001 aligner; the irradiances at 365, 405, and 436 nm are 0.2, 0.4, and 0.3 mW/cm2, respectively). After exposure, the SU-8 substrate was heated by ramping up at 4°C/min to 95°C (5 min), allowed to cool at the same rate, and developed with SU-8 developer (MicroChem) at room temperature for 15 min. The heights of the SU-8 mold features were then measured with a Tencor P15 surface profiler (KLA-Tencor, San Jose, CA, USA). For these parameters, the typical feature height was ~42 µm. Other heights could be obtained by varying the spinning speed according to the SU-8 manufacturer’s instructions.
2.2 Replica molding of PDMS from the master
To facilitate release, prior to PDMS replication the SU-8 master was silanized by exposure to a vapor of a fluorosilane ((tridecafluoro-1,1,2,2-tetrahydrooctyl)-1-trichlorosilane; United Chemical Technologies, Bristol, PA, USA) in house vacuum at room temperature for 30 min. A mixture of PDMS prepolymer and curing agent (10:1 w/w, Sylgard 184, Dow-Corning, Midland, MI, USA) was cast against the SU-8 master and cured at 65°C for 3 h. Pieces of silicone tubing were embedded into PDMS to create access inlets in the molds.
2.3 Volume measurement of each microchamber
The height of each microchamber was obtained by measuring the corresponding posts in the SU-8 master with the profilometer. The area of each microchamber was calculated by calibration from high-magnification images of the PDMS fluidic layer. The measured volumes were then compared with the expected values.
2.4 Thin PDMS membrane
In our microvalve devices, the middle layer consists of a ~12 µm thick PDMS membrane fabricated as in [13]. Briefly, a 10:1 weight ratio of PDMS prepolymer/curing agent mixture was mixed with hexane (3:1 weight ratio) and spun (inside a clean room) on a 3 in. diameter silicon wafer (derivatized with fluorosilane), resulting in a PDMS thin film. To cure the PDMS film, the wafer was heated to 85°C for 4 min on a hot plate. A dust-free environment is critical for ensuring that the PDMS membranes are free of defects; dust particles may result in membranes containing holes and/or defectively bound to the replica mold.
2.5 Device assembly and operation
The PDMS replica of control layer and the PDMS-coated wafer were brought into conformal contact for 10 min. To ensure bonding, the thin film and the replica mold were oxidized in oxygen plasma (Branson/IPC 2000 barrel etcher, 150 W, 1 Torr) for 30 s prior to bringing the two surfaces into contact [14]. The PDMS mold was then peeled from the silicon wafer, with the thin film chemically bound to the replica mold. This two-layer set was then visually aligned and sealed to the fluidic layer under a Nikon SMZ1500 stereoscope; this microscope was equipped with all the accessories necessary for fluorescence imaging. For opening and closing valves, pressures were controlled by a vacuum line (−30 kPa) and an air pressure line (10 kPa) connected through two pressure regulators to an array of miniature three-way solenoid valves (LHDA0511111H; Lee Company, Westbrook, CT, USA). The solenoid valves were connected to National Instruments data acquisition hardware (PCI 6025E, CB-50LP; National Instruments, Austin, TX, USA) controlled via Labview software (National Instruments).
2.6 Image acquisition and analysis
Fluorescein and Fluo-4 potassium salt were purchased from Molecular Probes (Eugene, OR, USA). For fluorescence imaging, light from a Mercury lamp (100 W) was filtered at 480 nm (480DF10; Omega Optical, Brattleboro, VT, USA). Fluorescent emission was collected through a dichroic mirror (510DRLP; Omega Optical) and filtered with a 515EFLP filter (Omega Optical). Images were taken with a SPOT RT CCD camera (Diagnostic Instruments, Sterling Heights, MI, USA) and analyzed with MetaMorph software (Universal Imaging, West Chester, PA, USA). To obtain the intensity profiles, the raw images were first corrected for background light and the unevenness of field illumination using a standard homogeneously doped fluorescent glass slide (Meridian Instruments, Kent, WA, USA). Three 20 × 20 pixel squares were then marked on each chamber pair and the average regional intensity was measured. We noticed that in the areas where the fluidic chambers overlapped with the control layer channels and valves, fluorescence intensity was slightly higher than in other areas, possibly due to the small deflection of thin membrane by pressure built up in the fluidic chambers; therefore the measured squares were picked outside of these overlapping areas.
3 Results and discussion
3.1 Fabrication and operation of device
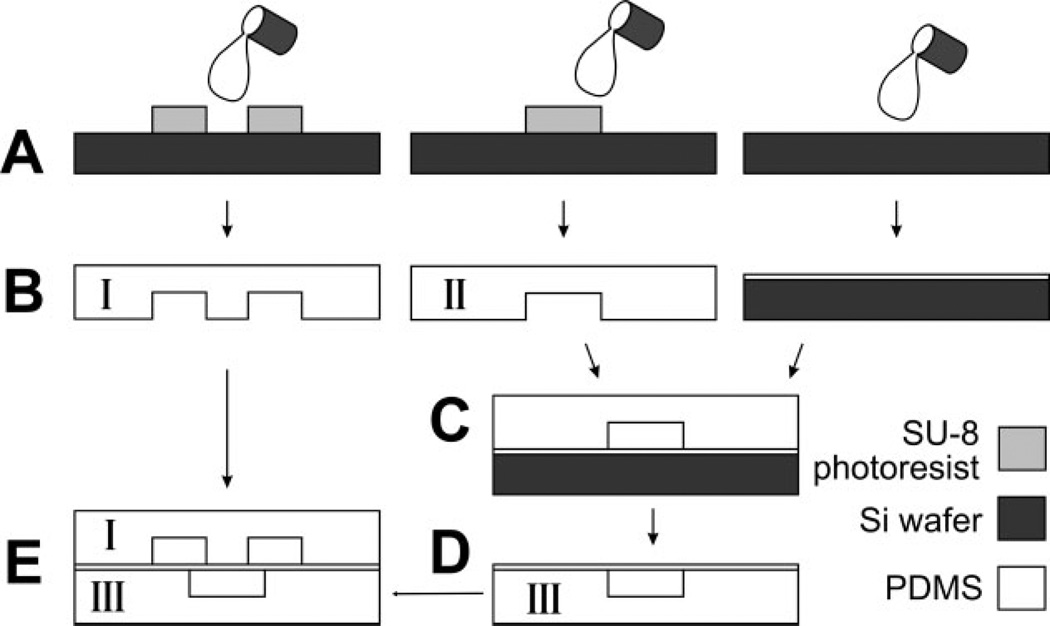
We adapted a previously reported PDMS microvalve design [8–10], with the device consisting of three layers: a fluidic layer containing microchambers of various sizes, a “control layer” containing the microchannels necessary to actuate the microvalves, and a middle thin PDMS membrane that is bound to the control layer. Figure 1 illustrates the modified fabrication procedures to create the three layers and assemble them to a final device (details in Section 2). In the original report from Hosokawa and Maeda [15], the authors had inlets to the fluidic layer on the top side of the device and inlets to the control layer from the bottom side of the device. This inlet placement makes devices difficult to visualize under the microscope. In our device, inlet regions are designed on the masks of both the fluidic and the control layers, but silicone tubing inlets are molded [16] only into one layer (for example, fluidic layer) of the device. To create inlets to the control layer, we manually remove or puncture the few sections of the membrane that cover the inlet regions. Therefore, after alignment and assembly, all microchannels (those that carry flow as well as those that control the valves) are accessible from the top of the device so that the bottom surface is planar, enabling imaging of the device on a conventional microscope stage.
Figure 1.
Device fabrication and assembly. Schematics of soft lithography procedures to fabricate the fluidic layer (“I”, left column), control layer (“II”, middle column), and the thin PDMS membrane (right column). (A) PDMS prepolymer is poured onto a fluorosilanized, photolithographically patterned SU-8 master to mold fluidic and control layers; the thin PDMS membrane is made by spinning PDMS on a fluorosilanized silicon wafer. (B) Cross-section view of the resulting PDMS replicas. (C) Bonding of the control layer to the PDMS membrane after oxygen plasma oxidation of both. (D) All-PDMS control-layer assembly after release from silicon wafer (III). (E) Sealing of the fluidic layer (I) on top of the PDMS control layer (III) so that valve seats overlap two fluidic paths.
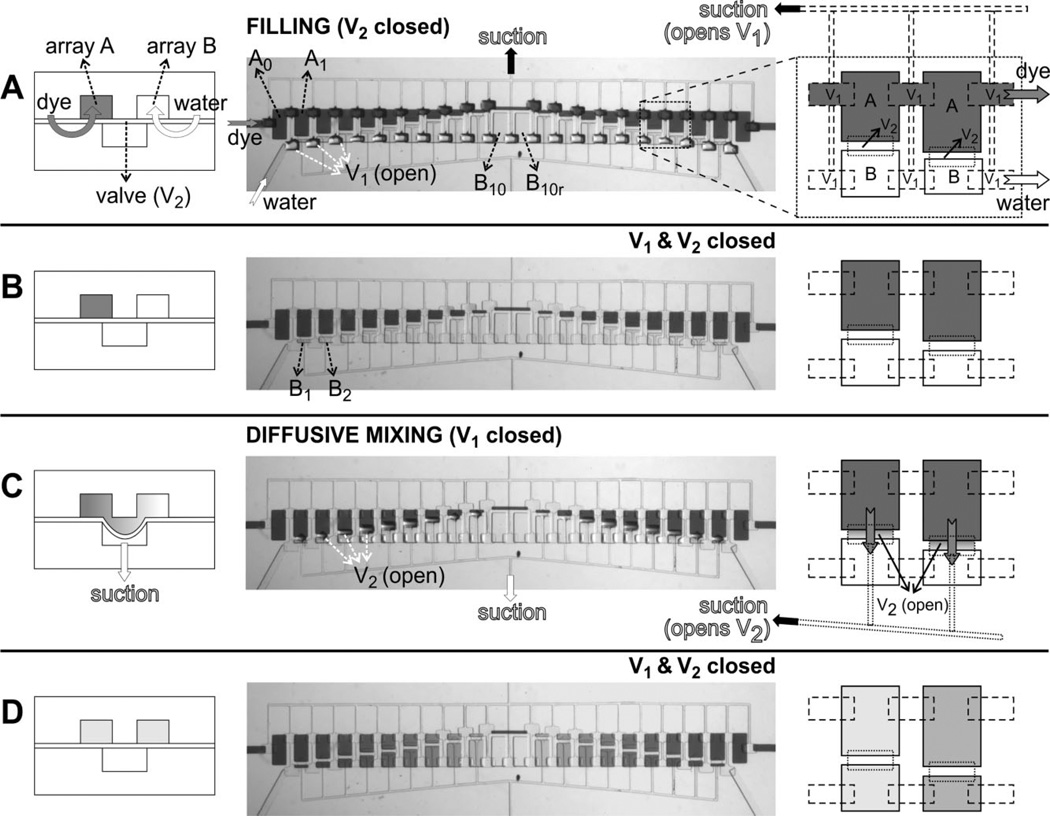
In our prototype micromixer, the fluidic layer contains two arrays of microchambers (termed “array A” and “array B,” see Fig. 2A). Along array A, the size of the microchambers (labeled A0, A1, …, A9) decreases, starting from the left, from 200 µm × 400 µm (A0) to 200 µm × 40 µm (A9); A10 is a 500 µm × 40 µm chamber and is used just for fluidic connection in Array A; to the right of chamber A10 is a set of chambers (A9r, A8r, …, A0r) symmetrically increasing in sizes from A9’s size to A0’s size. The chambers in array B are designed such that the added volume of any two adjacent chambers in different rows always equals to the volume of A0, i.e., volume (A0) = volume (B10) = volume (A1) + volume (B1) = volume (A2) + volume (B2) = …, etc. A0, A0r and B10, B10r are designed as respective controls for solutions A and B without mixing. The heights of microchambers measured from the SU-8 master were ~40–42 µm, resulting in a microchamber volume progression from 0.32 to 3.36 nL. The control layer has two independently controlled sets of valves (Figs. 2A and C). A set of valves {V1} is used to connect arrays A and B with their respective inlets, whereas a second set of valves {V2} is used to connect each pair of chambers in the two arrays (i.e., between A1 and B1, A2 and B2, …, A1r and B1r, etc.).
Figure 2.
Device operation. Operation of the device consists of four steps (A–D) shown in cross-sectional schematics (left column), top-view optical micrographs (middle column), and top-view schematics (right column). Left schematics correspond to a line orthogonal to both arrays and the right-column schematics correspond to an area comprising 2 × 2 microchambers, as indicated by the dashed-line square in the top-view optical micrograph in (A). For clarity, the pneumatic lines are shown only when suction is applied. (A) Initial filling of microchambers. Suction is applied to the top pneumatic lines (black arrow) in order to open valve set {V1} while valve set {V2} is kept closed; then array A is filled with colored dye (dark gray fluid, dark gray arrows) and array B is filled with de-ionized water (white arrows). Note the curvature of the {V1} membranes in the optical micrograph. (B) Isolation of each fluidic chamber after {V1} is closed. (C) Diffusive mixing between each A–B pairs after suction is applied to the bottom pneumatic lines (which open valve set {V2}). Note the gradient forming in the fluidic connection after mixing starts. (D) State of the device at the end of a mixing experiment (after closing valve set {V2}) where a sequence of 11 dilutions, one on each pair of isolated microchambers, is created due to differences in microchamber volumes between arrays A and B; the right-column schematics depict two of these 11 dilutions (dark and light shades of gray).
The operation principle and sequences are illustrated in Fig. 2. The microvalve seat is the portion of the PDMS membrane that overlaps the bottom surface of the walls between two fluidic compartments (such as chamber A0 and A1 in the case of microvalve set {V1}). At rest, due to the compliance and hydrophobicity of PDMS, the membrane seals (reversibly) against its seat, therefore the chambers remain isolated from each other without energy input. Valves can be opened by applying negative pressure (e.g., house vacuum), so the PDMS membrane deflects down and separates from the surface that supports the wall between A0 and A1, thus connecting A0 and A1. Valve closure is then achieved by switching the pressure setting from vacuum to atmospheric pressure. (Additional positive pressure, e.g., from pressurized air, accelerates valve closure.) Since the positive and negative pressures are adjustable, and opening and closing of the valves can be automated using pneumatic valves [4], the device is straightforwardly computer-operated with conventional scientific control software. To fill the microchambers, valve set {V1} is opened to allow the flow of dye and de-ionized water to arrays A and B, respectively (Fig. 2A). The valve set {V1} is then closed, thereby isolating each chamber in both arrays (Fig. 2B). Next, valve set {V2} is opened to allow fluid mixing between adjacent chambers in different arrays (e.g., mixing between A1 and B1, etc.; see Fig. 2C). Due to the thinness of PDMS membrane (10–12 µm), upon vacuum/suction application the valve seats deflect down and touch the floor of the control layer (42 µm deep); the area of overlap between a microvalve and the chambers it connects ranges from 100 µm × 40 µm to 200 µm × 40 µm; therefore we estimate that the fluidic connection between two chambers is approximately 100 µm × 40 µm × 42 µm = 0.168 nL to 2 × 0.168 nL = 0.336 nL when the valve is open, 1/20 to 1/10 of the added volume of the pairing fluidic chambers (i.e., the volume of A0, 3.36 nL). We believe the fluidic chambers must have deformed to produce this new volume of fluidic connection. Mixing only takes ~1 min to complete for these volumes. Closing {V2} then pushes the fluid above back to each fluidic chamber and chambers deform back to their original shape. Since the two fluidic arrays are designed with chambers of 11 different sizes, 11 different dilution ratios are produced (Fig. 2D). Because the time required for the membrane to reseal is very small compared to the mixing times, we have not tested the maximum switching frequency during operation; we anticipate that our valves are much slower than those based on the pinch-off design [4] because the area of the valve seat is much larger.
3.2 Accuracy of mixing using fluorescein
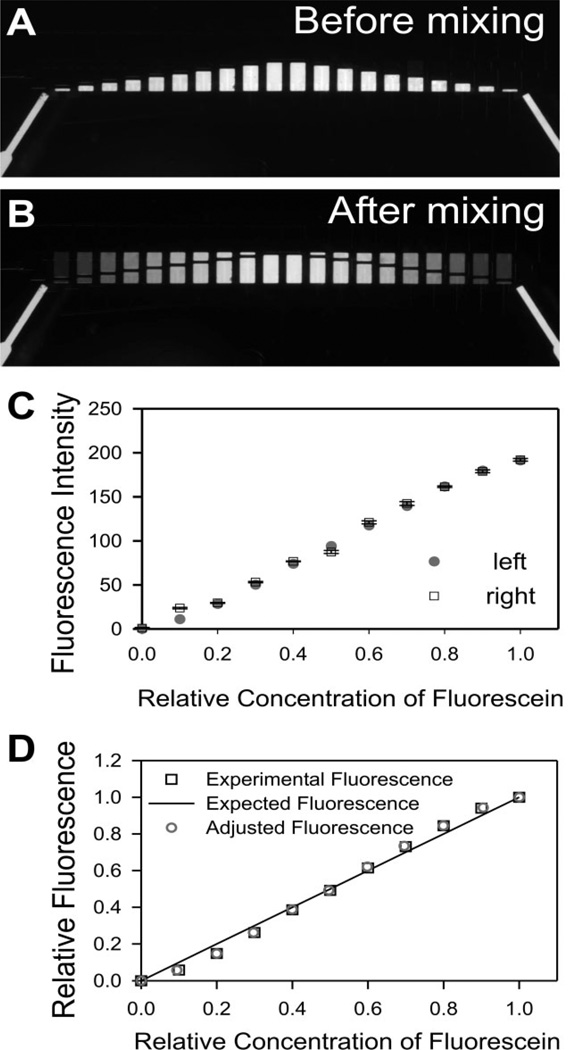
A key advantage of using photolithographically defined chambers is that the final concentrations after mixing can be predicted accurately from the designed chamber dimensions. To quantitate the accuracy of such predictions, we analyzed the fluorescence profiles of mixing fluorescein with water. (For this experiment the device was placed on the stage with the fluidic layer on top so as to avoid optical artifacts caused by emitted fluorescence light crossing features of the control layer; excitation light comes from above in this particular microscope.) Experimental procedures were the same as in the previous experiment with coloring dyes, except that arrays A and B were initially filled with de-ionized water and fluorescein (1 mg/mL), respectively (Fig. 3A). After mixing, the 11 dilution ratios from pure water to 1 mg/mL fluorescein are evident (Fig. 3B). We measured the after mixing fluorescence of each microchamber as described in Section 2. Since we have designed chamber A0, A0r and B10, B10r as internal calibration references (they have no connecting pairs in the opposite array, hence cannot be diluted with the other solution), the average intensity values in chambers A0 and A0r were subtracted from all the intensity values in the other chambers. These calibrated values were plotted against putative dilution factors (0, 0.1, …, 1) in Fig. 3C.We designed the left half and the right half to serve as internal mirror experiments in one chip, and found they yielded similar results (Fig. 3C). Since the R2 values for left and right halves are both 0.992, the experimental values were very linear as we expected. To evaluate the precision of the experimental results, we compared the relative fluorescence intensity (pure water set as 0, undiluted 1 mg/mL fluorescein set as 1) from the left half to the expected values which should be a linear curve from 0 to 1 (Fig. 3D, “experimental fluorescence” and “expected fluorescence”, respectively).
Figure 3.
Quantitative analysis of mixing precision with fluorescein. (A) Before mixing. Fluorescence image of isolated fluidic chambers, with array A containing deionized water and array B containing 1 mg/mL fluorescein. (B) After mixing. Fluorescence image showing results of 11 different dilutions. (C) Measured fluorescence intensities (after background correction) in the left (circles) and the right (squares) half of the device versus relative dye dilution. Error bars are SEM for three different regions from each chamber. (D) Comparison of the experimental results with predictions. Experimental fluorescence graph shows the relative intensity values (intensity from chambers of undiluted 1 mg/mL fluorescein solution is assigned the arbitrary value of 1) of the left half of the device as a function of the dilution factors (0, 1:10, 2:10, to 1:1); the expected fluorescence was calculated from the original design parameters; the adjusted fluorescence curve was obtained from the experimental fluorescence after factoring in the actual height and area of each microchamber (see text for details).
Due to the microfabrication process, each chamber in an SU-8 master may have a slightly different height and the actual areas of features may also deviate slightly from design, so the volume ratios of actual chamber pairs and thus the actual dilution factors might be slightly different from design (0, 0.1, …, 1). A systematic error could derive from this difference and reduce the precision of the mixing. This error was very small in our microdevice, as we found that the actual volume of each microchamber was very close to the designed volume, with only 0.1–1.0% deviation from expected values. The exceptions were the smallest (200 µm × 40 µm) chambers (5.8% deviation), due to their slightly (~2–3 µm) smaller heights than that of all the other chambers, possibly because they are located at the edge of the device. By using the actual volume ratios and compensating the fluorescence intensity values with the measured heights (rather than those predicted from design), we plotted the “adjusted fluorescence” data in Fig. 3D. Note the data points in “adjusted fluorescence” were very close to the points in “experimental fluorescence” and both very close to the “expected fluorescence” curve, which confirmed that we can increase the precision in our prediction of mixing by factoring in the fabrication-resulted deviations, but due to the high precision of photolithography procedures, the deviations were very small.
3.3 Ca2+ indicator calibration using the microdevice
As a proof of the concept, we used the parallel mixing microdevice for calibrating a fluorescent calcium indicator, a key procedure in calcium imaging measurements. A standard method to calibrate the dissociation constant (Kd) of an ion indicator has been described by Tsien and Pozzan [17]. Briefly, the protocol requires preparing two dilute samples of the indicator, one sample in 10 mm K2EGTA (zero Ca2+ sample) and the other in 10 mm CaEGTA (high Ca2+ sample). These two samples are then cross-diluted to produce a series of 11 solutions with the amount of total Ca2+ increasing by 1 mm CaEGTA with each dilution. The [Ca2+]free in each dilution can be calculated from the Kd of CaEGTA. If indicator concentration, temperature, ionic strength, and pH are kept constant, the Kd of Ca2+ indicator can then be calculated from the fluorescence intensity curve against these known Ca2+ concentrations. Since a total of 11 different dilutions have to be carried out and 11 different images (or fluorometer measurements) have to be acquired, this method is time consuming (~1 h). In addition, for fluorescence imaging, care has to be taken to ensure that the thickness of each sample is constant. Our microdevice is ideal for this procedure, due to its parallel (automated) mixing capability and to the precise mixing profiles created from photolithographically fabricated chambers. The heights of the chambers are very close and can be measured in advance if more precise fluorescent intensity values are required.
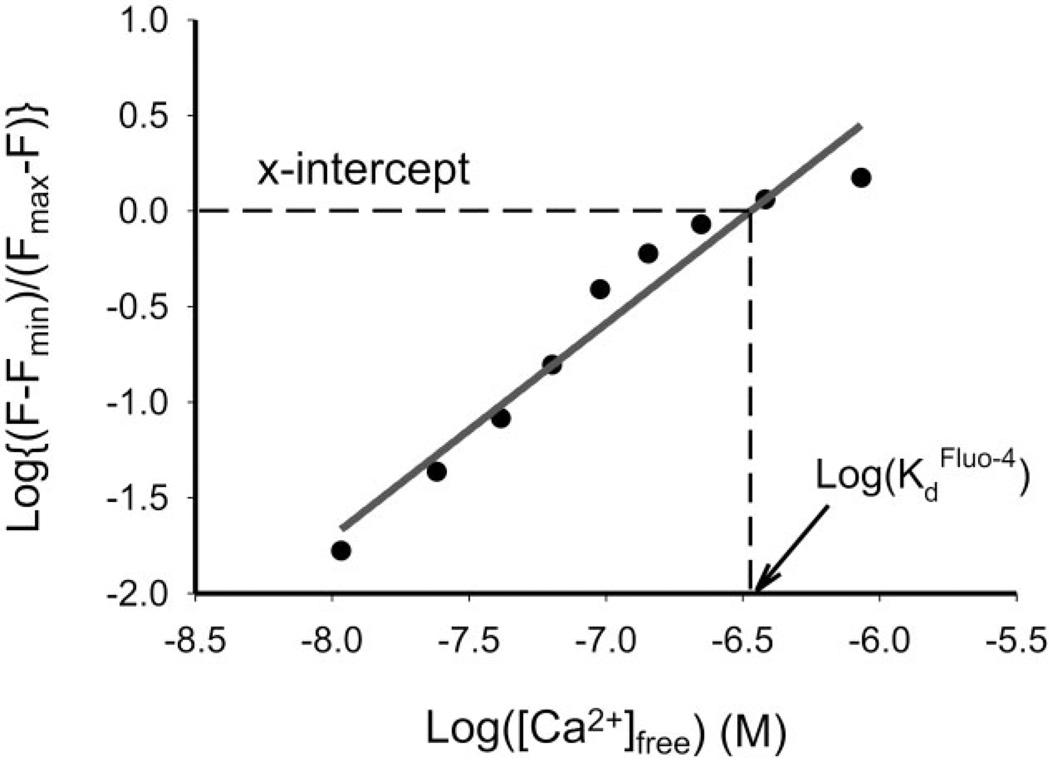
Figure 4 shows the fluorescence plot that serves to calibrate Fluo-4 (pentapotassium salt) in just one step using the microdevice. The operation procedures were similar as previous experiments with fluorescein. In this case, the chamber array A was filled with 10 mm K2EGTA, and chamber array B was filled with 10 mm CaEGTA; both also contain 10 µm Fluo-4, 100 mm KCl, and 10 mm MOPS buffer (pH 7.3). After mixing the two solutions in different ratios, the free Ca2+ concentrations in these microchambers were estimated by the WinMaxC program (version 2.50, Chris Patton, Hopkins Marine Station, Stanford University, CA, USA) to correct estimates of free Ca2+ for temperature and ionic strength. We calculate the Kd of Fluo-4 to be 338 nm, within 2% of the value reported by Molecular Probes (345 nm at pH 7.2).
Figure 4.
Fluo-4 calibration. The graph shows the result of calibrating Fluo-4 (pentapotassium salt) from one experiment using the microdevice. x-Intercept of the log plot, i.e., the Kd of Fluo-4, is extrapolated from a linear regression of the data, yielding a value of 338 nm.
4 Concluding remarks
In conclusion, we have presented a prototype of a parallel subnanoliter micromixer. The device contains arrays of isolated microchambers of different sizes (ranging from 200 µm × 40 µm × 40 µm = 0.32 to 3.36 nL) and was controlled by automated individual elastomeric microvalves. The device is easy to fabricate and simple to control. The volumes are photolithographically defined and, thus, very precise, as demonstrated in a parallel dilution experiment. Fluid and reagents can be stored in the microdevice for several days, allowing for highly portable assays. Notably, PDMS is biocompatible, so the device has broad applicability in miniaturized diagnostic assays as well as in cell-based assays such as drug screening and enzyme-based biomolecule detection.
Acknowledgments
This work was supported by the National Science Foundation (CAREER Award to Albert Folch) and by NIH Grant No. 61-0295. Clean-room processing was done at the Washington Technology Center.
Abbreviation
- PDMS
polydimethylsiloxane
References
- 1.Xia YN, Whitesides GM. Angew. Chem. Int. Ed. Engl. 1998;5:551–575. [Google Scholar]
- 2.Beebe DJ, Moore JS, Bauer JM, Yu Q. Nature. 2000;6778:588–590. doi: 10.1038/35007047. [DOI] [PubMed] [Google Scholar]
- 3.Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Science. 2000;5463:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 4.Hansen CL, Skordalakes E, Berger JM, Quake SR. Proc. Natl. Acad. Sci. USA. 2002;26:16531–16536. doi: 10.1073/pnas.262485199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu AY, Chou HP, Spence C, Arnold FH, Quake SR. Anal. Chem. 2002;11:2451–2457. doi: 10.1021/ac0255330. [DOI] [PubMed] [Google Scholar]
- 6.Thorsen T, Maerkl SJ, Quake SR. Science. 2002;5593:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 7.Tourovskaia A, Figueroa-Masot X, Folch A. Lab Chip. 2005;1:14–19. doi: 10.1039/b405719h. [DOI] [PubMed] [Google Scholar]
- 8.Hosokawa K, Hanada K, Maeda R. J. Micromech. Microeng. 2002;1:1–6. [Google Scholar]
- 9.Jo BH, Moorthy J, Beebe DJ. Micro Total Analysis Systems 2000. The Netherlands: Kluver Academic Publishers, Dordrecht; 2000. pp. 335–338. [Google Scholar]
- 10.Jo BH, Van Lerberghe LM, Motsegood KM, Beebe DJ. J. Microelectromech. Syst. 2000;1:76–81. [Google Scholar]
- 11.Lee KY, LaBianca N, Rishton SA, Zolgharnain S. J. Vac. Sci. Technol. B. 1995;6:3012–3016. [Google Scholar]
- 12.Chen C, Hirdes D, Folch A. Proc. Natl. Acad. Sci. USA. 2003;4:1499–1504. doi: 10.1073/pnas.0435755100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffman J, Shao J, Hsu C-H, Folch A. Adv. Mater. 2004:23–24. 2201–2206. [Google Scholar]
- 14.McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu H, Schueller OJA, Whitesides GM. Electrophoresis. 2000;21:27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 15.Hosokawa K, Maeda R. J. Micromech. Microeng. 2000;3:415–420. [Google Scholar]
- 16.Folch A, Toner M. Biotechnol. Prog. 1998;3:388–392. doi: 10.1021/bp980037b. [DOI] [PubMed] [Google Scholar]
- 17.Tsien R, Pozzan T. Methods Enzymol. 1989;172:230–262. doi: 10.1016/s0076-6879(89)72017-6. [DOI] [PubMed] [Google Scholar]