Abstract
Linkage to chromosome 12p for familial Alzheimer disease (AD) has been inconsistent. Using 35 markers near the centromere of chromosome 12, we investigated 79 Caribbean Hispanic families with AD. Two-point linkage analysis using affected sib pairs yielded LOD scores of 3.15 at D12S1623 and 1.43 at D12S1042. The LOD score at D12S1623 decreased to 1.62 in families with late-onset (age >65 years) AD (LOAD), but the LOD score at D12S1042 was unchanged. Among families negative for the apolipoprotein E (APOE-ε4) allele, the LOD score for D12S1623 was lower (1.01), whereas that for D12S1042 increased to 1.73. Among families positive for the APOE-ε4 allele, none of the LOD scores reached 1. Multipoint affected-relative-pair analysis showed peaks at D12S1623 (nonparametric linkage [NPL] score 1.52; P=.028) and near D12S1042, at D12S1057 (NPL score 1.57; P=.027). NPL scores for both D12S1623 and D12S1057 increased in families affected with LOAD, but, in APOE-ε4–negative families, only scores for the region flanking D12S1623 remained elevated (NPL score 1.74; P=.013). This study of Caribbean Hispanics with familial AD extends and provides modest evidence of linkage to loci on chromosome 12p. Linkage varied by age at onset of AD and by the presence or absence of the APOE-ε4 allele.
A putative gene on chromosome 12 (AD5 [MIM 602096]), conferring susceptibility to late-onset Alzheimer disease (LOAD [MIM 104300]), was identified by Pericak-Vance et al. (1997). Subsequent confirmation has been inconsistent and contradictory (Pericak-Vance et al. 1997; Rogaeva et al. 1998; Wu et al. 1998; Scott et al. 1999, 2000). Rogaeva et al. (1998) observed locus heterogeneity attributed to the presence of APOE-ε4. Scott et al. (2000) found stronger linkage in the absence of apolipoprotein E (APOE-ε4 [MIM 107741]) and when Lewy bodies (Lewy-body dementia [MIM 127750]) were detected at autopsy in at least one affected family member.
Compared with other ethnic groups, Caribbean Hispanics have an increased frequency of AD (Gurland et al. 1999; Tang et al. 2001), as do Mexican Americans (Perkins et al. 1997). The current study was designed to extend and confirm the evidence of linkage, on chromosome 12, to AD in Caribbean Hispanics. Patients were identified in the Alzheimer’s Disease Research Center at Columbia University, in an epidemiological study in northern Manhattan, and in clinics in Dominican Republic and Puerto Rico. The Institutional Review Board of Columbia Presbyterian Medical Center and Columbia University Health Sciences and the Bioethics National Committee for Research in the Dominican Republic approved the study. Informed consent was obtained either from the participant or, when the individual was demented, from a family member.
Patients with AD met criteria set by the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s Disease and Related Disorders Association (NINCDS-ADRDA) (McKhann et al. 1984). The Clinical Dementia Rating Scale (CDR) (Hughes et al. 1982) was used to rate disease severity. Brain imaging and other laboratory studies were reviewed and offered when medically required for diagnosis. The neuropsychological tests used included a battery developed and evaluated for use in Hispanic individuals (Pittman et al. 1992; Stern et al. 1992).
Only individuals meeting criteria for probable AD, with a CDR ⩾1.0, were classified as case subjects for analyses. Individuals with other forms of dementia, mild AD (CDR 0.5), and possible AD were classified as having unknown status. Age at onset was based on family report. Blood was obtained from all patients, living siblings, and other family members, when possible. Genotyping of APOE was performed using standard methods (Hixson and Vernier 1990; Maestre et al. 1995).
Multiplex PCR, up to four products per reaction, was performed using 50 ng of DNA in a 10-μl reaction that contained 10 mM Tris-HCl pH 8.3 at 25°C, 50 mM KCl, 1.6–2.0 mM MgCl2, 0.25 mM of each dNTP, variable amounts of each PCR primer to produce even peak heights, and 0.1 units of Platinum Taq DNA Polymerase (Life Technologies), by touchdown PCR (Don et al. 1991) in 384-well plates in MJ Research thermocyclers. The annealing temperature was decreased 2°C per cycle, from 65°C to 55°C (94°C denaturing), and then 32 additional cycles of 90°C denaturing, 55°C annealing, and 72°C elongation were performed. Amplification products were pooled using a TECAN Genesis robot, Tamara-labeled size standard was added (ABI 400HD), and the mixture was resolved using POP4 polymer on a 36-cM capillary array and was detected by an ABI 3100 DNA sequencer. Then the data were analyzed by GENESCAN 3.7 and GENOTYPER 3.0 (Applied BioSystems). Two readers, shielded from clinical diagnoses, independently interpreted genotypes.
Nonpaternity was examined using PEDCHECK (O'Connell and Weeks 1998) and RELATIVE (Goring and Ott 1997), prior to linkage analysis. We excluded individuals from the analysis when the theoretical maximum IBD sharing was estimated to be <50%. Families were considered to be APOE-ε4 positive if 75% of the affected individuals had at least one e4-allele, and all other families were considered to be APOE-ε4 negative (Rogaeva et al. 1998). Families with AD were considered to be affected with LOAD if 75% of the affected individuals reported onset at age >65 years.
We mapped the region extending from 12pter to 12q21.13, using 35 microsatellite markers. Initially, a two-point sib-pair analysis was conducted using ANALYZE (Goring and Terwilliger 2000). The nonparametric model applies the pseudomarker allele-sharing method, under the assumptions that parents are heterozygotes and that the mode of inheritance is autosomal recessive (Knapp et al. 1994). Allele frequencies were based on data derived from all participating family members. Multipoint nonparametric linkage analysis was implemented in GENEHUNTER 2 (Kruglyak et al. 1996). When necessary, noninformative nonfounders (e.g., unaffected children) were excluded, to circumvent the computational limitations of the software. Maps from the Marshfield Medical Research Foundation and the Genome Database were used for locus order and intermarker distance. The sibling transmission/disequilibrium test (Sib-TDT) was used to test allelic association and was expressed as a Z score, with the computed P values based on the normal distribution approximation (Spielman and Ewens 1998).
In the 79 families investigated, there were 320 relatives (table 1). The mean age of living participants was 73.3 years, and the mean age at onset for affected individuals was 74.1 years. The majority of the families (67 [84.8%]) were from the Dominican Republic, 9 (11.4%) were from Puerto Rico, and 3 (3.8%) came from elsewhere in the Caribbean. Twenty-five families have three or more affected members. The majority of families had two affected members.
Table 1.
Description of Caribbean Hispanic Families
| Characteristic | Data |
| Number of families | 79 |
| No. of relatives examined: | 320 |
| Parents | 2 |
| Siblings | 198 |
| Half-siblings | 11 |
| Children | 32 |
| Others | 77 |
| Total | 320 |
| Average no. of relatives examined per family | 4.05 |
| Percent female | 66.1% |
| Mean age at onset (years) | 74.1 (SD 11.1) |
| Affection status: | |
| Probable AD | 51.3% |
| Unaffected | 37.7% |
| Unknown | 11.0% |
| APOE-ε4 allele frequency:a | |
| Overall | 27.6% |
| Probable AD | 31.7% |
| Unaffected | 22.1% |
Individuals with unknown affection status were not included.
There were 15 individuals from these families for whom DNA was not available; thus, only 384 individuals (including probands and relatives) were included in the linkage and association analyses. Fifty-one percent of the members of the families met criteria for probable AD, 38% were unaffected, and 11% were coded as unknown, either because they (a) had milder dementia (CDR 0.5), possible AD, or another cause of dementia or (b) were <40 years of age (table 1). Unaffected individuals were defined as those without dementia at an age comparable to that of the probands (table 1).
The frequency of APOE-ε4 among affected individuals was significantly higher than expected under the null hypothesis of no association (Z=4.79; P=8.3×10-7). On the basis of our classification, 32 families were APOE-ε4 positive and 47 families were APOE-ε4 negative; 8 families had predominantly early-onset disease, and 71 families had LOAD.
The two-point analysis showed the strongest evidence for linkage for markers D12S1623 (LOD 3.15) and D12S1042 (LOD 1.43), ∼30 cM apart (fig. 1). D12S1623 is located ∼4 cM telomeric of alpha-2-macroglobulin (A2M [MIM 103950]), but analysis of A2M showed no evidence for linkage (LOD 0.75). When the analysis was restricted to families affected with LOAD, the D12S1623 LOD score decreased to 1.63, whereas that for D12S1042 did not change. Among APOE-ε4–negative families, the LOD score for D12S1623 decreased further, to 1.01, but the LOD for D12S1042 increased to 1.73. The LOD scores for D12S1057 and D12S398, near D12S1042, also were >1. None of the LOD scores reached 1.0 in the APOE-ε4–positive families.
Figure 1.
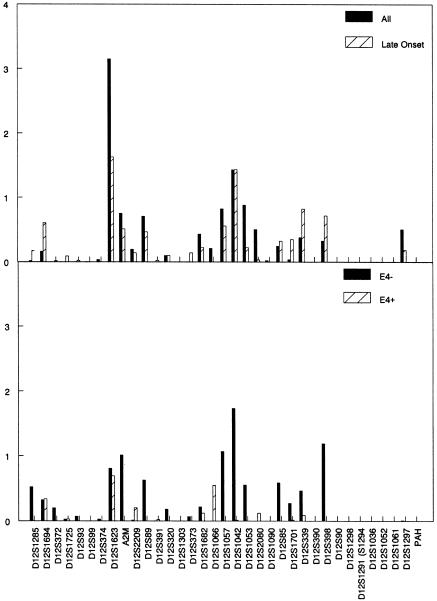
Results of the two-point analysis showing linkage for markers D12S1623 (LOD 3.15) and D12S1042 (LOD 1.43) on chromosome 12. The two markers are ∼30 cM apart. The upper graph shows the results overall and when restricted by age at onset >65 years. The lower graph shows the results stratified by the presence or absence of an APOE-ε4 allele.
Multipoint affected-relative-pair analysis showed peaks at D12S1623 (NPL score 1.52; P=.028) and near D12S1042 and D12S1057 (NPL score 1.57; P=.027) (fig. 2). In families affected with LOAD, the NPL score for D12S1623 was 2.01 (P=.006), whereas that for D12S1057 was 1.63 (P=.021). Among APOE-ε4–negative families, scores for the region flanking D12S1623 remained elevated (NPL score 1.74; P=.013), but, among APOE-ε4–positive families, there was no evidence of linkage for the region flanking D12S1623 (NPL score 0.25; P=.379). The NPL score for the D12S1042 region remained unchanged (NPL score 1.34; P=.053) regardless of APOE-ε4 status.
Figure 2.
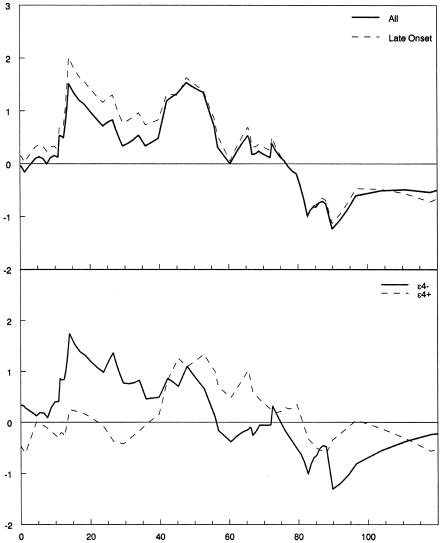
Results of multipoint affected-relative-pair analyses, showing peaks near the same two markers, D12S1623 (NPL score 1.52; P=.028) and D12S1057 (NPL score 1.57; P=.027). Among late-onset families, the NPL score for D12S1623 was 2.01 (P=.006), whereas that for D12S1057 was 1.63 (P=.021). The upper graph shows the results overall and when restricted by age at onset >65 years. The lower graph shows the results stratified by the presence or absence of an APOE-ε4 allele.
The Sib-TDT was used to test for linkage in the presence of association flanking the markers D12S1623 and D12S1042. Allele 5 of D12S374, near the first peak (1.46 cM from D12S1623), was associated with LOAD (Z=2.27; P=.0116). Allele 10 of D12S1090, 7.68 cM from D12S1042, showed the strongest evidence for linkage (Z=2.93; P=.0017). The strength of association varied with age at onset and with the presence or absence of the APOE-ε4 allele, for several markers (table 2).
Table 2.
Sib-TDT Analysis of AD among Caribbean Hispanics[Note]
|
Results in Families |
|||||||||
| All |
Affected with LOAD |
APOE-ε4 Negative |
APOE-ε4 Positive |
||||||
| Marker | Allele | Z | P | Z | P | Z | P | Z | P |
| D12S1694 | 10 | 1.68 | .0465 | ||||||
| D12S374 | 5 | 1.80 | .0359 | 2.27 | .0116 | 1.80 | .0359 | ||
| D12S2209 | 12 | 1.72 | .0427 | ||||||
| D12S89 | 12 | 1.65 | .0495 | ||||||
| D12S1682 | 6 | 2.07 | .0192 | ||||||
| D12S1042 | 4 | 2.02 | .0217 | 2.27 | .0116 | ||||
| D12S1090 | 10 | 2.93 | .0017 | 2.93 | .0017 | 1.84 | .0329 | 1.86 | .0314 |
| D12S1701 | 4 | 1.77 | .0384 | ||||||
| D12S339 | 8 | 1.65 | .0495 | ||||||
| D12S390 | 6 | 2.32 | .0101 | 2.01 | .0222 | 2.12 | .0170 | ||
Note.— Only the alleles with one-sided P value (unadjusted) of ⩽.05 are listed.
The main findings of this study are consistent with the linkage of LOAD to chromosome 12p in Caribbean Hispanics. Linkage was stronger in APOE-ε4–negative families. Despite our previous findings of a weak association (Romas et al. 2000), there was no evidence of linkage to A2M in a larger set of families. The strongest support for linkage in these Caribbean Hispanic families was on 12p, telomeric to the sites with highest LOD scores reported by others (Rogaeva et al. 1998; Wu et al. 1998). However, we also obtained modest support for linkage at a second site—corresponding to D12S1057 and D12S1042, closer to the centromere—first reported by Scott et al. (1999, 2000).
Prior studies of chromosome 12 have been primarily in non-Hispanic whites of American or European origin (Pericak-Vance et al. 1997; Blacker et al. 1998; Rogaeva et al. 1998; Scott et al. 1999, 2000). Attempts to fine-map this candidate region of chromosome 12 have met with mixed success (Clatworthy et al. 1997; Blacker et al. 1998; Wu et al. 1998; Scott et al. 1999, 2000; Zubenko et al. 1999; Dodel et al. 2000). Wu et al. (1998) could not confirm linkage, and Rogaeva et al. (1998) found evidence of linkage to an adjacent region. Scott et al. (1999, 2000) showed that linkage was conditional on the presence or absence of APOE-ε4 and on the finding of Lewy bodies in post mortem material from family members. Our results among Caribbean Hispanics also provide support for two sites potentially linked to AD. These may be (a) two independent peaks, ∼30 cM away from each other, possibly representing two separate genes, (b) a single gene with chance variation, or (c) two false-positive peaks. A number of independent investigations using different populations and different analytic approaches show modest peaks on 12p (albeit not at exactly the same location). Although the likelihood of a false-positive peak on 12p is reduced, it is not eliminated.
In the two-point analysis, support for linkage at D12S1623 was substantially higher than that for the multipoint affected-relative-pair analysis. The polymorphism information content for D12S1623 was low (0.42). When the information content improved in the multipoint analysis, the support for linkage may have declined because of truly weaker linkage. However, in this type of analysis, errors in tightly linked regions can be exaggerated, reducing the evidence favoring linkage (Risch and Giuffra 1992).
These regions on chromosome 12 contains two interesting genes: that for A2M and that for the low-density lipoprotein receptor–related protein (LRP1 [MIM 107770]) (Blacker et al. 1998; Liao et al. 1998). With some exceptions, neither association has been confirmed in subsequent studies (Clatworthy et al. 1997; Baum et al. 1998; Hollenbach et al. 1998; Beffert et al. 1999; Dodel et al. 2000; Gibson et al. 2000; Romas et al. 2000).
The strengths of our study are the unique population and the criterion-based, conservative diagnoses. Although we have had only three autopsies to date, all have confirmed clinical diagnoses of probable AD. The difficulty encountered in refining the candidate region on chromosome 12 has been attributed to clinical and locus heterogeneity. The exact location has not been replicated in any study, but the identification of families from different ethnic backgrounds who exhibit linkage to approximately the same region of 12p strengthens the possibility that a susceptibility gene may exist on this chromosome.
Acknowledgments
Support was provided by grants AG15473, AG08702, and AG07232, from the National Institute of Aging; by the Charles S. Robertson Memorial Gift for Alzheimer's Disease Research, from the Banbury Fund; and by a grant from the Blanchette Hooker Rockefeller Foundation. We also thank the members of Estudio Familiar de Influencia Genetica en Alzheimer, The Sociedad Dominicana de Geriatria y Gerontologia, The Sociedad Dominicana de Neurologia y Neurocirugia, The Sociedad Dominicana de Psiquiatria, and the Associacion Dominicana Alzheimer y Similares, Inc.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Genome Database, The, http://www.gdb.org/
- Marshfield Medical Research Foundation, Center for Medical Genetics, http://research.marshfieldclinic.org/genetics/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for AD [MIM 104300], AD5 [MIM 602096], APOE-ε4 [MIM 107741], Lewy body dementia [MIM 127750], A2M [MIM 103950], and LRP1 [MIM 107770])
References
- Baum L, Dong ZY, Choy KW, Pang CP, Ng HK (1998) Low density lipoprotein receptor related protein gene amplification and 766T polymorphism in astrocytomas. Neurosci Lett 256:5–8 [DOI] [PubMed] [Google Scholar]
- Beffert U, Arguin C, Poirier J (1999) The polymorphism in exon 3 of the low density lipoprotein receptor- related protein gene is weakly associated with Alzheimer's disease. Neurosci Lett 259:29–32 [DOI] [PubMed] [Google Scholar]
- Blacker D, Wilcox MA, Laird NM, Rodes L, Horvath SM, Go RC, Perry R, Watson B, Jr., Bassett SS, McInnis MG, Albert MS, Hyman BT, Tanzi RE (1998) Alpha-2 macroglobulin is genetically associated with Alzheimer disease. Nat Genet 19:357–360 [DOI] [PubMed] [Google Scholar]
- Clatworthy AE, Gomez-Isla T, Rebeck GW, Wallace RB, Hyman BT (1997) Lack of association of a polymorphism in the low-density lipoprotein receptor-related protein gene with Alzheimer disease. Arch Neurol 54:1289–1292 [DOI] [PubMed] [Google Scholar]
- Dodel RC, Du Y, Bales KR, Gao F, Eastwood B, Glazier B, Zimmer R, Cordell B, Hake A, Evans R, Gallagher-Thompson D, Thompson LW, Tinklenberg JR, Pfefferbaum A, Sullivan EV, Yesavage J, Alstiel L, Gasser T, Farlow MR, Murphy GM Jr, Paul SM (2000) Alpha2 macroglobulin and the risk of Alzheimer's disease. Neurology 54:438–442 [DOI] [PubMed] [Google Scholar]
- Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS (1991) “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res 19:4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson AM, Singleton AB, Smith G, Woodward R, McKeith IG, Perry RH, Ince PG, Ballard CG, Edwardson JA, Morris CM (2000) Lack of association of the alpha2-macroglobulin locus on chromosome 12 in AD. Neurology 54:433–438 [DOI] [PubMed] [Google Scholar]
- Goring HH, Ott J (1997) Relationship estimation in affected sib pair analysis of late-onset diseases. Eur J Hum Genet 5:69–77 [PubMed] [Google Scholar]
- Goring HH, Terwilliger JD (2000) Linkage analysis in the presence of errors III: marker loci and their map as nuisance parameters. Am J Hum Genet 66:1298–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurland BJ, Wilder DE, Lantigua R, Stern Y, Chen J, Killeffer EH, Mayeux R (1999) Rates of dementia in three ethnoracial groups. Int J Geriatr Psychiatry 14:481–493 [PubMed] [Google Scholar]
- Hixson JE, Vernier DT (1990) Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J Lipid Res 31:545–548 [PubMed] [Google Scholar]
- Hollenbach E, Ackermann S, Hyman BT, Rebeck GW (1998) Confirmation of an association between a polymorphism in exon 3 of the low-density lipoprotein receptor-related protein gene and Alzheimer's disease. Neurology 50:1905–1907 [DOI] [PubMed] [Google Scholar]
- Hughes CP, Berg L, Danziger WL, Coben LA, Martin RL (1982) A new clinical scale for the staging of dementia. Br J Psychiatry 140:566–572 [DOI] [PubMed] [Google Scholar]
- Knapp M, Seuchter SA, Baur MP (1994) Linkage analysis in nuclear families. 2. Relationship between affected sib-pair tests and lod score analysis. Hum Hered 44:44–51 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Liao A, Nitsch RM, Greenberg SM, Finckh U, Blacker D, Albert M, Rebeck GW, Gomez-Isla T, Clatworthy A, Binetti G, Hock C, Mueller-Thomsen T, Mann U, Zuchowski K, Beisiegel U, Staehelin H, Growdon JH, Tanzi RE, Hyman BT (1998) Genetic association of an alpha2-macroglobulin (Val1000lle) polymorphism and Alzheimer's disease. Hum Mol Genet 7:1953–1956 [DOI] [PubMed] [Google Scholar]
- Maestre G, Ottman R, Stern Y, Gurland B, Chun M, Tang MX, Shelanski M, Tycko B, Mayeux R (1995) Apolipoprotein E and Alzheimer's disease: ethnic variation in genotypic risks. Ann Neurol 37:254–259 [DOI] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM (1984) Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology 34:939–944 [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pericak-Vance MA, Bass MP, Yamaoka LH, Gaskell PC, Scott WK, Terwedow HA, Menold MM, Conneally PM, Small GW, Vance JM, Saunders AM, Roses AD, Haines JL (1997) Complete genomic screen in late-onset familial Alzheimer disease: evidence for a new locus on chromosome 12. JAMA 278:1237–1241 [PubMed] [Google Scholar]
- Perkins P, Annegers JF, Doody RS, Cooke N, Aday L, Vernon SW (1997) Incidence and prevalence of dementia in a multiethnic cohort of municipal retirees. Neurology 49:44–50 [DOI] [PubMed] [Google Scholar]
- Pittman J, Andrews H, Tatemichi T, Link B, Struening E, Stern Y, Mayeux R (1992) Diagnosis of dementia in a heterogeneous population: a comparison of paradigm-based diagnosis and physician's diagnosis. Arch Neurol 49:461–467 [DOI] [PubMed] [Google Scholar]
- Risch N, Giuffra L (1992) Model misspecification and multipoint linkage analysis. Hum Hered 42:77–92 [DOI] [PubMed] [Google Scholar]
- Rogaeva E, Premkumar S, Song Y, Sorbi S, Brindle N, Paterson A, Duara R, Levesque G, Yu G, Nishimura M, Ikeda M, O'Toole C, Kawarai T, Jorge R, Vilarino D, Bruni AC, Farrer LA, St George-Hyslop PH (1998) Evidence for an Alzheimer disease susceptibility locus on chromosome 12 and for further locus heterogeneity. JAMA 280:614–618 [DOI] [PubMed] [Google Scholar]
- Romas SN, Mayeux R, Rabinowitz D, Tang MX, Zadroga HR, Lantigua R, Medrano M, Tycko B, Knowles JA (2000) The deletion polymorphism and Val1000Ile in alpha-2-macroglobulin and Alzheimer disease in Caribbean Hispanics. Neurosci Lett 279:133–136 [DOI] [PubMed] [Google Scholar]
- Scott WK, Grubber JM, Abou-Donia SM, Church TD, Saunders AM, Roses AD, Pericak-Vance MA, Conneally PM, Small GW, Haines JL (1999) Further evidence linking late-onset Alzheimer disease with chromosome 12. JAMA 281:513–514 [DOI] [PubMed] [Google Scholar]
- Scott WK, Grubber JM, Conneally PM, Small GW, Hulette CM, Rosenberg CK, Saunders AM, Roses AD, Haines JL, Pericak-Vance MA (2000) Fine mapping of the chromosome 12 late-onset Alzheimer disease locus: potential genetic and phenotypic heterogeneity. Am J Hum Genet 66:922–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman RS, Ewens WJ (1998) A sibship test for linkage in the presence of association: the sib transmission/disequilibrium test. Am J Hum Genet 62:450–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern Y, Andrews H, Pittman J, Sano M, Tatemichi T, Lantigua R, Mayeux R (1992) Diagnosis of dementia in a heterogeneous population: development of a neuropsychological paradigm-based diagnosis of dementia and quantified correction for the effects of education. Arch Neurol 49:453–460 [DOI] [PubMed] [Google Scholar]
- Tang MX, Cross P, Andrews H, Jacobs DM, Small S, Bell K, Merchant C, Lantigua R, Costa R, Stern Y, Mayeux R (2001) Incidence of AD in African-Americans, Caribbean Hispanics, and Caucasians in northern Manhattan. Neurology 56:49–56 [DOI] [PubMed] [Google Scholar]
- Wu WS, Holmans P, Wavrant-DeVrieze F, Shears S, Kehoe P, Crook R, Booth J, Williams N, Perez-Tur J, Roehl K, Fenton I, Chartier-Harlin MC, Lovestone S, Williams J, Hutton M, Hardy J, Owen MJ, Goate A (1998) Genetic studies on chromosome 12 in late-onset Alzheimer disease. JAMA 280:619–622 [DOI] [PubMed] [Google Scholar]
- Zubenko GS, Hughes HB 3d, Stiffler JS (1999) Neurobiological correlates of a putative risk allele for Alzheimer's disease on chromosome 12q. Neurology 52:725–732 [DOI] [PubMed] [Google Scholar]


