Abstract
Purpose
The expression of an array of signaling molecules, along with the assessment of real-time cell proliferation, has been performed in U87 glioma cell line and in patients’ glioblastoma established cell cultures in order to provide a better understanding of cellular and molecular events involved in glioblastoma pathogenesis. Experimental therapy was performed using a phosphatidylinositol-3′-kinase (PI3K) inhibitor.
Patients and methods
xMAP technology was employed to assess expression levels of several signal transduction molecules and real-time xCELLigence platform for cell behavior.
Results
PI3K inhibition induced the most significant effects on global signaling pathways in patient-derived cell cultures, especially on members of the mitogen-activated protein-kinase family, P70S6 serine-threonine kinase, and cAMP response element-binding protein expression and further prevented tumor cell proliferation.
Conclusion
The PI3K pathway might be a prime target for glioblastoma treatment.
Keywords: personalized medicine, PI3K inhibitor, targeted therapy, xCELLigence, xMAP analysis
Introduction
Intracranial tumors represent a heterogeneous group of neoplasms that appear to underlie distinct patterns of growth, invasion, response to therapy, and prognosis.1,2 Glioblastoma (GB) is one of the most frequently occurring tumors in the central nervous system and the most lethal brain tumor type in adults.3,4 Despite the progress in surgery and radiotherapy, and the addition of temozolomide,5 the prognosis of glioma patients remains poor.6 Among the reasons for therapy failure are therapeutic resistance and frequent recurrence, together with the migratory phenotype and persistence of tumor cancer-initiating stem cells.7,8 Although the initial transforming event(s) remain elusive, the progression of brain tumors has been associated with multiple signaling alterations, which were postulated to confer survival and proliferation advantages.9
Studies regarding glioblastoma pathogenesis point out its molecular complexity; thus, conventional biochemical methods (eg, Western blot, enzyme-linked immunosorbent assay) can overlook critical components and/or signaling profiles of this devastating disease. It has been hypothesized that the combined influence of many genes or biomarkers would make superior identifiers and/or predictors of tumor behavior and patient outcome.10 Mechanistic links of different signaling pathways are still to be uncovered and appear to depend on the cell type. Acknowledging the possibility of having multiple signaling connections, high throughput proteomic profiling is becoming an important approach in cancer research. Moreover, the currently available multiplex analysis, such as xMAP and protein array technologies, offers good premises for human brain tumor research. These technologies may provide simultaneous analysis of a panel of signaling molecules, for improved diagnosis, patient stratification, prognosis, and drug screening. Biomarker discovery for brain tumors is an ongoing pursuit, and the search for the best molecule or combination of molecules is unfolding.11–14
The investigation of molecular mechanisms underlying the behavior of glioblastoma (including their aggressiveness and therapeutic resistance) goes hand in hand with the discovery and development of new inhibitors targeting key actors, such as protein tyrosine kinases.15,16 Reported deregulations of phosphatidylinositol-3-kinase/protein kinase B/mammalian target of rapamycin (PI3K/Akt/mTOR) signaling often represent the results of receptor tyrosine kinase overactivity (involving epidermal growth factor receptor, platelet-derived growth factor receptor, mesenchymal-epithelial transition factor), mutated PI3K subunits, and/or loss of phosphatase and tensin homolog (PTEN) tumor suppressor activity.17 Not only do more than 40% of glioblastoma exhibit a loss of PTEN, which results in excessive PI3K signaling, but a significant number of glioblastoma tumors exhibit functional inactivation of PTEN by post-translational modifications, such as phosphorylation at Y240 and others.17
From the perspective of personalized medicine, assessing signaling deregulation would best benefit from the study of glioblastoma cell cultures established from patient tumors. Hence, patient-derived cell cultures provide the ideal setup to generate results focusing on tumor biology and/or new therapeutic approaches.18
In the present study, we have investigated the expression patterns of several signal transduction molecules in glioblastoma cells using multiplex assay, both in basal conditions and under kinase inhibitor treatment, targeting key signaling molecules. As we have been seeking possible future therapy targets, we have analyzed the midterm effect of inhibitors on signal transduction molecules. We considered it relevant to assess in parallel these patterns in patient-derived cell cultures and standard glioblastoma cell line U87. Furthermore, we have searched for subsequent functional cellular behavior by real-time investigation of cell proliferation, using the xCELLigence approach.
Materials and methods
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval from the Ethics Committee of Victor Babes National Institute of Pathology and the Ethics Committee of Neurology and Neurovascular Diseases National Institute, Bucharest, Romania and have followed the principles outlined in the Declaration of Helsinki for all human experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Samples, cell cultures, and treatments
Samples were collected from 17 patients diagnosed with Stage IV glioma (glioblastoma) undergoing surgery at Neurology and Neurovascular Diseases National Institute, Bucharest, Romania. Patients’ written informed consents were obtained, in accordance with ethical standards included in the current revision of the Declaration of Helsinki.
Paired tumoral and peritumoral tissue samples from all patients were subjected to xMAP analysis for the expression of signal transduction molecules. Tumoral samples from nine cases were processed for cell culture initiation. Each tumor sample was mechanically triturated in cell culture medium and then incubated in enzymatic cocktail solution of collage-nase I 0.25 mg/mL and trypsin 0.6 mg/mL (Sigma-Aldrich, St Louis, MO, USA) for 30 minutes, with gentle stirring, at 37°C. The supernatant was collected and centrifuged for 10 minutes at 250 ×g, then the pellet was resuspended in cell culture medium and passed through a 40 μm mesh. Viability was measured by Trypan blue method (Trypan 0.4%); cells were then seeded at 104 cells/cm2 density and maintained in culture in 5% CO2 atmosphere and 37°C. After 24 hours, the medium was replaced. Cell propagation was then performed in Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin (Sigma-Aldrich) for several passages and monitored for morphological features (by light microscopy) and surface markers: glial fibrillary acidic protein (GFAP); Nestin; and Notch receptors (by immunofluorescence).
Out of the nine cases, seven cell cultures were successfully initiated, two of them maintaining similar morphological features beyond Passage 2; therefore, they were used in further experiments, herein presented as Case A and Case B. For Case B, in some of the real-time assays, cells obtained from the relapse of the original tumor were also studied. No adjuvant therapies (neither radiotherapy nor chemotherapy) were performed on this patient between the two surgical interventions. The mentioned cases were selected to be the best performers in terms of proliferation rates. The reason for our choice was both the need to match U87 proliferation rate and generation from one passage of sufficient cells to obtain a satisfactory cell lysate in terms of total protein concentration to cover all the experimental approaches.
Standard cell line: U87 cells have been purchased from ATCC (Austin, TX, USA) and maintained in culture according to manufacturer’s recommendations: Eagle’s Minimum Essential Medium supplemented with 10% fetal bovine serum (both from ATCC) and 1% antibiotic (Life Technologies, Carlsbad, CA, USA). Cells were cultivated in 5% CO2 humidified atmosphere at 37°C. For maintenance purposes, two to three media changes were required weekly. Culture passages were performed by trypsin-EDTA (Life Technologies) treatment according to the original recommendations of the supplier (ATCC).
Immunofluorescence
Nestin and GFAP expression in cultured cells were investigated by immunofluorescence, according to a modified protocol.19 Anti-GFAP primary antibody was incubated for 2 hours at room temperature, using a rabbit polyclonal IgG (Sigma-Aldrich, G9269; 1:50), followed by detection with Alexa Fluor 488 conjugated goat antirabbit secondary antibody (Life Technologies, A31628; 1:300). For Nestin, we used a mouse monoclonal IgG1 antibody (Santa Cruz Biotechnology, Dallas, TX, USA, SC-23927; 1:50); followed by detection with Alexa Fluor 594 conjugated goat antimouse secondary antibody (Life Technologies, A31624; 1:300). Counterstain for nuclei was performed with 4′,6-diamidino-2-phenylindole (Life Technologies, D1306). Examination was carried out using a Nikon TE300 microscope (Nikon Instruments, Melville, NY, USA) with appropriate fluorescence settings.
Assessment of signal transduction molecules using xMAP analysis
The expression of signal transduction molecules was performed on patient tumoral/peritumoral tissue and on patient-derived and U87 cell cultures.
U87 glioblastoma cells and patient-derived cells were seeded in six-well plates at 90,000 cells/well for the evaluation of protein expression of signal transduction molecules. All experiments were performed in triplicate. After 12 hours, the culture medium was replaced with inhibitor-containing cell media, and further cultivated in standard condition for 24 more hours. The following conditions were examined: untreated cells (control) and treated cells with pharmacological inhibitors, namely PI3K inhibitor LY294002 25 μM, MEK-1 inhibitor PD98059 25 μM, MEK inhibitor U0126 10 μM, and p38 inhibitor SB203580 10 μM. After 24 hours’ incubation with inhibitors, cells were rinsed twice with cold PBS and lysed with lysis buffer provided by the xMAP kit manufacturer. Total protein concentration was assessed by Bradford assay (Sigma-Aldrich).
Multiplex analysis was performed on a Luminex® 200™ platform (Luminexcorp, Austin, TX, USA), using Cell Buffer and Detection Kit and Milliplex MAPmates for total proteins: ERK (extracellular signal-regulated kinases) 1/2, JNK, P70S6K, IκBα, p38, cAMP response element-binding protein (CREB), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; EMD Millipore, Billerica, MA, USA). Briefly, xMAP technology is based on a sandwich-type immunoassay for the detection and quantification of analytes of interest. Capture monoclonal antibodies are covalently immobilized on polystyrene microspheres (beads) of 50 nm or 60 nm diameter, loaded with precise concentrations of two fluorophores; their ratio generates a spectral address, unique for each bead class. The beads in each class are covered with a single type of capture antibody, thus providing the specificity for the analytes. A mixture of beads is incubated with the samples, resulting in the binding of analytes to their corresponding beads, via the capture antibodies on their surface. Biotinylated detection antibodies will bind to a second epitope of the captured analytes. Streptavidin conjugated phycoerythrin is used as a reporter molecule, upon measurement on a flow-cytometric device, which reports and correlates phycoerythrin fluorescence to each bead class.
Cell lysis and sample preparation for multiplex protein detection were performed according to manufacturer instructions (EMD Millipore). GAPDH was used as an internal standard for data normalization. Data measurement was performed using the Luminex 200 software; data acquisition and analysis were performed using STarStation 2.3 (AppliedCytometry, Sheffield, UK).
Assessment of MEK inhibition by MEK inhibitors U0126 and PD98059 effects on phosphorylation of ERK 1/2 was performed on lysates of U87 cell cultures, using Milliplex 8-plex Multi-Pathway Signaling Kit (M8MP kit, EMD Millipore) for phosphoprotein, according to the manufacturer’s instructions. Cell lysis was performed after 1 hour of exposure to inhibitors, in the lysis buffer from the M8MP kit. According to the manufacturer’s protocol, total GAPDH was used for data normalization. Data measurement was performed using the Luminex 200 software; data acquisition and analysis were performed using STarStation 2.3.
Real-time cell monitoring using xCELLigence platform
Briefly, the xCELLigence system measures electrical impedance of culture cells across interdigitated microelectrodes integrated on the bottom of specially designed culture plates. Through this system, cellular events are monitored in real time without any labeling incorporation. The impedance measurement provides quantitative information about the biological status of the cells, including cell number, adhesion, and viability. Electrode impedance changes, automatically displayed as Cell Index, were used to monitor cell viability, number, proliferation, and adhesion.
Both patient-derived cultures and U87 cell line were treated with kinase inhibitors (purchased from Promega, Madison, WI, USA), as follows: LY294002, 25 μM; PD98059, 25 μM; U0126, 10 μM; and SB203580, 10 μM. The inhibitors were solubilized in dimethyl sulfoxide (DMSO), which was also added to the control vehicle (1:400). Starting concentrations for experiments were chosen based on similar approaches in literature.20–24
The tests were performed on E-16 plates (Roche Diagnostics Corporation, Indianapolis, IN, USA; catalog number 05469830001), compatible with RCTA-DP system (Roche Diagnostics Corporation), which are basically 16-well plates covered on the bottom with sensor electrodes capable of reading modification of electric resistivity induced by cell adhesion. Each well was coated with collagen I (Sigma-Aldrich C7661) 5 μg/mL in 0.1M NaHCO3, for 1 hour at 37°C, except for the wells used as negative control. To each well, 5,000–10,000 previously serum-starved cells were added in complete cell media and left to adhere for 2 hours in the RTCA DP device at 37°C and 5% CO2. Kinase inhibitors were added to appropriate concentration, and readings were recorded for 24 hours or more in order to quantify cell proliferation. Controls were incubated with vehicle containing 1/400 DMSO, to match the reconstituted kinase inhibitors solutions. Each well was duplicated or quadruplicated, and each experiment was performed three times.
Western blot
U87 cells were lysed in a buffer containing 2% Triton-X100 and 2% NP-40, 10% protease inhibitors cocktail (Sigma-Aldrich), and 2 mM sodium orthovanadate. Volumes containing the same amount of protein (25 μg) for each experimental condition were loaded on a 10% SDS-PAGE gel. Electrophoresis was run at 20 mA/gel, and separated proteins were subsequently blotted on nitrocellulose membranes at 100 V for 1 hour, on ice. After 1 hour blocking with bovine serum albumin 2.5% in Tris buffered saline, membranes were incubated over night with primary antibodies: CREB1 (C-21, Santa Cruz Biotechnology), ERK 1/2 (sc-135900, Santa Cruz Biotechnology), P70S6K (C-18, Santa Cruz Biotechnology), pAkt1 (S473, 81283, Abcam), and betaactin (Santa Cruz Biotechnology). The following day, after 1 hour incubation in suitable secondary antibody (1:4000) (goat antirabbit and goat antimouse, respectively, Santa Cruz Biotechnology), membranes were incubated for 5 minutes in ECL solution (Thermo Fisher Scientific, Waltham, MA, USA) and exposed to film.
Statistical analysis
For each experiment, data from three independent experiments were analyzed; Student’s t-test was used to calculate statistical significance; in every individual experiment, each experimental condition was reproduced in triplicate. For protein expression analysis using the Luminex system, data acquisition and processing were performed using STarStation 2.3 software (Applied Cytometry, Sheffield, UK), and calibration curves were built using the built-in five-point logistics function of the software.
Results
Having personalized therapy as a future goal and searching for the best experimental conditions to assess effects of inhibitors, we chose patient-derived primary cell cultures.
Cell morphological features of initial cultures were maintained during several passages, as revealed by cell shape, size, and density. Immunofluorescence examination for GFAP, Nestin, and Notch receptors 1–4 detected a relatively constant presence of these biomarkers from one passage to another. GFAP was expressed in over 90% of the cells in all situations, attesting the tumor origin of the cultivated cells. Examples of GFAP and Nestin distribution in Passages 1 and 4 are provided in Figure 1.
Figure 1.
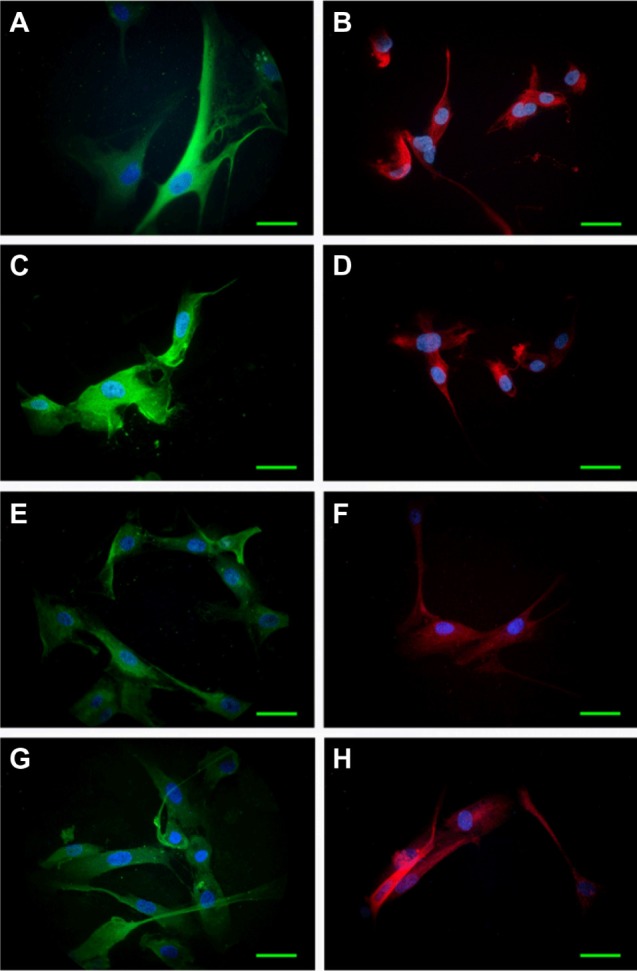
Expression of GFAP (green) and Nestin (red) in cultivated cells derived from tumor specimens.
Notes: Immunofluorescence, examined on Nikon TE300, 60× obj. Case A: Passage 1, GFAP (A), Nestin (B); Passage 4, GFAP (C), Nestin (D). Case B: Passage 1, GFAP (E), Nestin (F); Passage 4, GFAP (G), Nestin (H). Counterstain for nuclei: DAPI (blue). Scale bar: 20 μm.
Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; GFAP, glial fibrillary acidic protein; obj, objective.
Upon prolonged cultivation, Case B culture established neurospheres similar to those generated by the U87 cell line.
Qiu et al reported a similar approach using GFAP as a marker for glioma cells in culture.25 Witusik-Perkowska et al reported adherent cells and spheroid cultures derived from glioblastoma, displaying similar morphologies with our results.18
Assessment of signal transduction molecules modulated by protein kinase inhibitors: xMAP analysis
Our study examined the expression pattern of several signal transduction molecules that act as key players in various cancers, including glioblastoma. The expression levels of key molecules involved in signaling pathways relevant to brain carcinogenesis: ERK 1/2, JNK, P70S6K, IκBα, p38, and CREB were determined in tissue samples from glioblastoma patients. The expression profile is described in Figure 2A. Significant differences were obtained in tumoral compared with peritumoral samples: the kinases ERK 1/2, P70S6K, JNK, and p38 were 1.7-fold to 4.0-fold increased, and the transcription factor CREB had a twofold increment.
Figure 2.
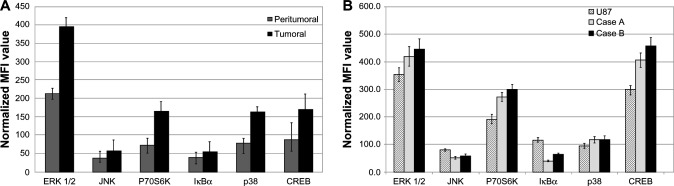
Expression profiles of signal transduction proteins quantified by xMAP analysis.
Notes: (A) Expression level of signaling molecules on tumoral compared to peritumoral tissue. (B) Basal expression of signaling molecules on patient-derived and U87 cell cultures. MFI estimated by xMAP-array. Normalization was based on GAPDH. The data represent the mean and SD of three independent experiments, each performed in triplicate.
Abbreviations: CREB, cAMP response element-binding protein; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; MFI, mean fluorescence intensities; SD, standard deviation; ERK, extracellular signal-regulated kinase; JNK, jun amino-terminal kinase.
The basal (unstimulated) expression pattern of the same molecules was examined in cell cultures established from patients and in the U87 glioblastoma cell line and are presented in Figure 2B. The displayed profile of expression appears rather similar for most of the examined molecules, between case-derived and U87 cells.
The expression profile of signal transduction molecules in patient-derived cell cultures closely follows the trend found in the originating tumor tissue, but at a higher concentration. This may be explained by a higher ratio of the physiologically active cells and to a lower contamination with stromal, endothelial, and inflammatory cells compared to the originating tissue (Figure 2A and B).
Similar results were obtained using xMAP and Western blot technologies for determining protein levels in a linear fashion (Figure S1).
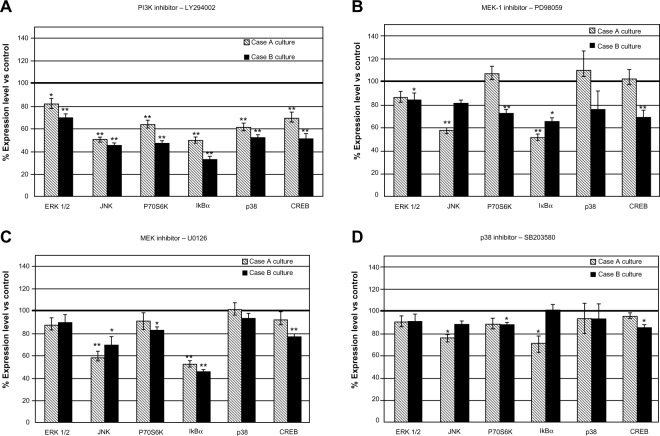
Treating the cells with protein kinase inhibitors resulted in an array of responses of signaling molecule expression, a pattern that appears to be specific for each cell culture (Figure 3).
Figure 3.
Modulation of protein expression of signal transduction molecules in case-derived cell cultures treated with pharmacological inhibitors: LY294002 (A), PD98059 (B), U0126 (C) and SB203580 (D).
Notes: The data represent the mean and SD of three independent experiments, each performed in triplicate. Expression of all tested signaling molecules was consistently inhibited after exposure to PI3K inhibitor LY294002 (P<0.05). MEK-1 inhibitor PD98059 determined also statistically significant inhibition on Case B, at moderate levels, while on Case A it was effective only for fewer molecules. JNK MAP-kinase and IκBα expressions were affected similarly by U0126 and PD98059. *P<0.05; **P<0.01.
Abbreviations: PI3K, phosphatidylinositol-3′-kinase; SD, standard deviation; vs, versus; CREB, cAMP response element-binding protein; ERK, extracellular signal-regulated kinase; JNK, jun amino-terminal kinase.
PI3K inhibitor treatment induced the strongest response in patient-derived cell cultures, demonstrated by a decreased expression level of all tested signaling molecules (Figure 3A). Case B-derived cells showed an overall more prominent response, displaying decreased expression levels in the range of 30.47%–66.61% in comparison with untreated control cells; for Case A-derived cells, the range was 18.66%–50.44%. The other inhibitors proved to be less effective when compared to PI3K.
Similar results were obtained using xMAP and Western blot technologies for determining protein levels and phosphorylation status in a linear fashion (Figures S2 and S3). The inhibitor did, as expected, completely inhibit the level of pAkt (Figure S3).
The effects of kinase inhibition were demonstrated on the components of the targeted signaling cascades (PI3K → P70S6K, MEK-ERK 1/2 → P70S6K) and also on molecules involved in other signal transduction cascades (CREB). P70S6K was significantly down-regulated after the inhibition of PI3K by LY294002 (Figure 3A); since P70S6K is a major player in the control of protein synthesis, a general decline in protein synthesis is expected as a consequence of the inhibition of one of its main activators.
MEK-1 inhibition of Case B cells induced after PD98059 treatment was associated with a moderate decrease in expression level of all investigated signaling molecules. This finding did not apply to Case A, where PD98059 inhibitor displayed more selective effects. MEK-1 and MEK-1/2 inhibitors exerted more significant effects than the p38 inhibitor (Figure 3B–D), suggesting ERK 1/2 as another therapeutic target for glioblastoma cases. Inhibition of ERK 1/2 phosphorylation by MEK inhibitors U0126 and PD98059 in U87 cell cultures was detected as expected (Figure S3), showing that inhibitors acted on their primary target pathways.
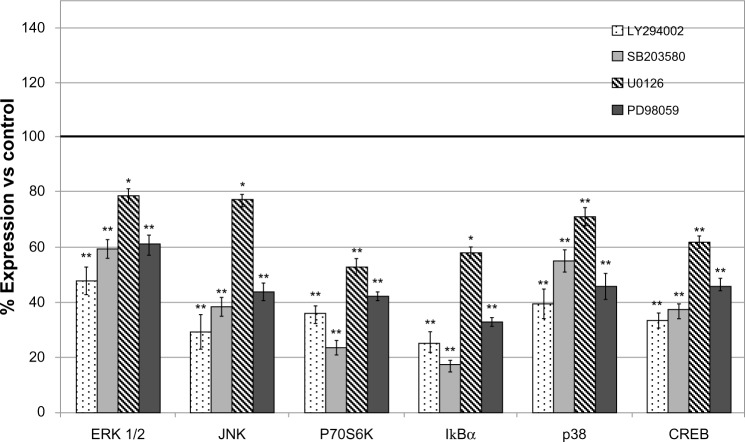
Compared to the patient-derived cell cultures, the U87 cell line was sensitive in variable degrees to all used inhibitors, pointing to PI3K inhibitor as the most active (Figure 4).
Figure 4.
Modulation of protein expression of signal transduction molecules in U87 glioblastoma cell line treated with pharmacological inhibitors LY294002, PD98059, U0126, and SB203580.
Notes: The data represent the mean and SD of three independent experiments. LY294002 produced, overall, the strongest inhibition on protein expression. *P<0.05; **P<0.01.
Abbreviations: CREB, cAMP response element-binding protein; vs, versus; ERK, extracellular signal-regulated kinase; JNK, jun amino-terminal kinase; SD, standard deviation.
The results of xMAP expression analysis were confirmed by Western blot for total ERK, P70S6K, and CREB (Figure S2). Confirmation of effects of MEK inhibitor and PI3K inhibitor on phosphorylation levels of their targets were obtained by xMAP and Western blot (Figure S3).
Real-time monitoring using xCELLigence platform
Following xMAP assay results, we assessed whether these molecular modifications impact tumor cell behavior. In basal conditions, impedance registered for patient-derived culture cells tends to plateau within the first 24 hours of cultivation, similar to the U87 glioblastoma cell line. The choice to use only cells in basal conditions (steady state growth in normal tissue culture medium, without starvation and stimulation) for the assessment of effects displayed by pharmacological inhibitors is supported by Bartscht et al.26 This group showed that both stimulated cells and nonstimulated cells have a similar scattering behavior related to inhibition.26
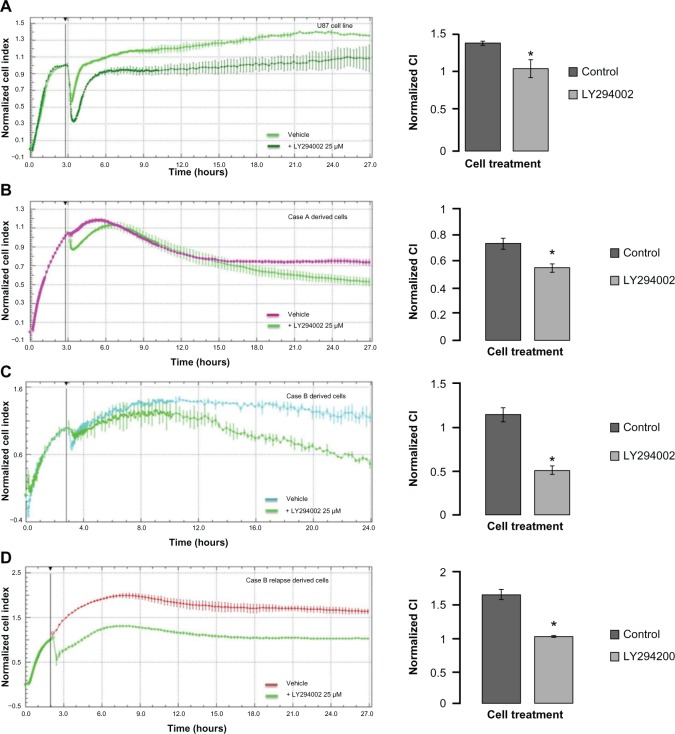
Under all inhibitors, adhesion and proliferation of U87 glioblastoma cells decreased, with the PI3K inhibitor having the most prominent effect (data not shown), in compliance with the inhibition obtained on xMAP analysis. In these circumstances, we have further investigated if the molecular modifications yielded by PI3K inhibition induced significant alteration of patient-derived cell culture behavior. Decreased scattering and proliferation was observed with 25 μM of PI3K inhibitor for both the U87 cell line and the patient-derived cultures (Figure 5). Case A derived cells showed a delayed response to kinase inhibition (Figure 5B); however, at 24 hours, this response also reached statistical significance, as indicated in the right panel.
Figure 5.
Real-time impedance analysis of cell proliferation under PI3K inhibition.
Notes: Five thousand U87 cells (A), Case A derived culture cells (B), Case B derived culture cells (C), and Case B relapse derived culture cells (D) were plated on collagen-coated cells and left to adhere for 2 hours. After the plating period, vehicle comprising 1/400 DMSO or 25μM LY294002, PI3K inhibitor, were added to the wells; cells were further incubated in standard cell culture conditions for 48 hours. LY294002 inhibited proliferation in all experimental setups, yet at different amplitudes and times. Proliferation CI were normalized after an initial 2-hour plating period. Readings were collected every minute for the first 2 hours, then every 15 minutes for the remainder of the experiment. Statistical analysis was carried out using Student’s t-test (two-tailed) for normalized cell indexes at 24 hours *(P<0.01). The data represent the mean and SD of three independent experiments.
Abbreviations: CI, cell indexes; DMSO, dimethyl sulfoxide; SD, standard deviation.
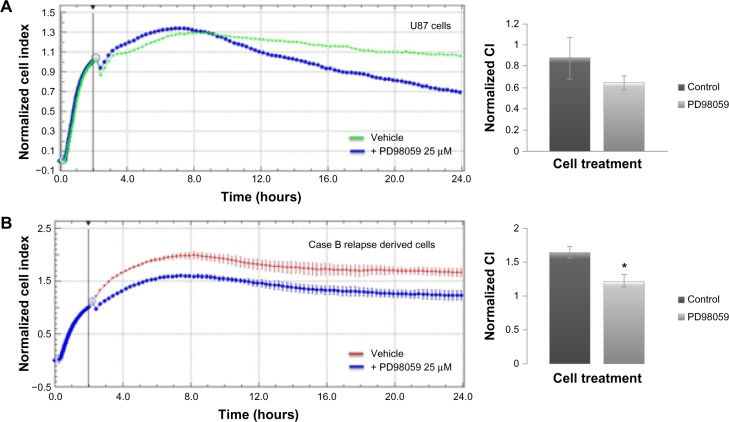
We have further assessed the adherence and proliferation patterns for U87 and patient B relapse derived cells in response to MEK-1 inhibition by PD98059 25 μM (Figure 6). Twenty-four hours of incubation in cell culture standard condition, with MEK-1 inhibitor, displayed a decreased proliferation under kinase inhibition for both the standardized cell line (Figure 6A) and patient-derived cells (Figure 6B).
Figure 6.
Real-time impedance analysis of cell proliferation under MEK-1 inhibition.
Notes: Five thousand U87 cells (A), and Case B relapse derived culture cells (B) were plated on collagen-coated cells and left to adhere for 2 hours. After the plating period, vehicles containing 1/400 DMSO or MEK-1 inhibitor to a final concentration of 25 μM were added to wells and cells were further incubated in standard cell culture conditions for 48 hours. Both cell types displayed inhibited proliferation upon treatment. Proliferation CI were normalized after an initial 2-hour plating period. Readings were collected at every minute for the first 2 hours, then every 15 minutes for the remainder of the experiment. Statistical analysis was carried out using Student’s t-test (two-tailed) for normalized cell indexes at 24 hours (P<0.01)*. The data represent the mean and SD of three independent experiments.
Abbreviations: CI, cell indexes; DMSO, dimethyl sulfoxide; SD, standard deviation.
Discussion
Our main results reveal that inhibiting the PI3K pathway in patient-derived glioblastoma cells impacts the expression of signaling proteins significantly, along with the cell proliferation pattern. A combination of real-time monitoring of cell behavior and screening for expression levels of signaling proteins can highlight important differences between primary tumors of similar histological type, and can possibly allow comparisons of primary and relapse tumor samples from the same patient.
Improving overall understanding of the fundamental mechanisms that govern cell proliferation and invasiveness is vital for the future development of therapeutic approaches for treating devastating cancers, such as glioblastoma. As in any other type of cancer, glioblastoma shows increased cell survival due to mutations, gene amplifications, and deletions, often (at least 40%) targeting the PI3K signaling pathway.27,28 Although the PI3K signaling pathway is being investigated as a target for treatment of glioblastoma,28–31 it is still essential to study the effect of inhibitors on patient-derived cell cultures and carefully characterize sensitivity to inhibitors or combinations of inhibitors. The global effects we notice in signaling studies might support the dependence of these tumors to the PI3K signaling pathway; therefore, PI3K inhibitors as therapeutic agents might change the clinical treatment landscape of glioblastoma patients.32–34
Our study revealed that treatment of glioblastoma cell cultures from patient tumors with PI3K inhibitor induced significant decrease in the expression level of several key signaling molecules involved in cell survival (p38), proliferation (ERK 1/2, IκBα, p38 MAP[mitogen-activated protein]-kinase, CREB), differentiation (ERK 1/2, CREB), migration (ERK 1/2, CREB), and apoptosis (ERK 1/2, P70S6K, IκBα, JNK, CREB). The simultaneous detection of several molecules involved in various signaling pathways has not been previously reported in glioblastoma. Further studies are needed in order to assess whether this regulation is transcriptional or post-transcriptional. The effects of PI3K inhibitors on the expression of key signal transduction molecules were also reflected in cell proliferation and are in accordance with similar findings regarding the role of PI3K as a therapy target, disclosed by Boller et al.34 This group demonstrated that specific pharmacological inhibitors or RNA interferences impaired cell proliferation in several tumor types, including glioblastoma.34 The maximal efficacy of PI3K inhibitor recorded by us on the cellular responses is consistent with other studies that point out the major relevance of PI3K signaling in glioblastoma and that PI3K inhibitors can complement, enhance, or override the limited success of epidermal growth factor receptor inhibitors in clinical trials.35
One of the molecules affected by inhibitors, P70S6K, can be activated by loops emerging from PI3K and ERK pathways. This offers an “escape” route that may diminish the efficacy when targeting a single pathway, and supports some suggestions regarding the simultaneous use of PI3K and MAP-kinase inhibitors.36 In our study, down-regulation of P70S6K was observed for Case B with both PI3K and MEK-1 inhibitors, in accordance with decreased proliferation profile revealed by real-time monitoring assay. A similar interference between signaling pathways was evidenced for PI3K and MEK/ERK pathways by Sunayama et al.37
The distinct patterns of reactivity to inhibitors in our experiments point to the differences between patients for similar histological grades. Distinct sensitivity to growth inhibition by pharmacological MEK inhibitors was demonstrated by See et al on several glioblastoma cell lines.38
The modification of protein expression for several signal transduction molecules under inhibition of one pathway (in our case, PI3K) is consistent with the previous demonstration by Han et al, that PI3K inhibition affects β-catenin mediated transcription.39 Furthermore, precedents also exist in the cytokine receptor signaling field, where, for instance, it was shown that MAP-kinase ERK activation by IL(interleukin)-2 via IL-2R β and γ chain is decreased by PI3K inhibitors. Reif et al demonstrated that there is a crucial link between the IL-2 receptor, the proto-oncogene PKB, and P70S6K.40
Down-regulation of key signal transducers generates a potential desensitization of cells towards extracellular signals, as well as towards several endogenous oncogenic molecules. The down-regulated molecules affect downstream signaling for cytokines, growth, and angiogenic factors, and belong (in most cases, simultaneously) to several gene ontologies (antiapoptosis, cell cycle, transcription, regulation of transcription, angiogenesis, cell adhesion, cell growth, cell differentiation).41 Moreover, Cox et al have investigated gene expression for proteins distributed in various transduction pathways. The inhibition of a single molecule (SOX-2) revealed significant modification of expression (over twofold up- or down-regulation) of a large number of genes, reporting a total of 620 transcripts with altered expression.42
Using different inhibitors to disrupt tyrosine kinase signaling, our study resulted in changes in proliferation rates of glioblastoma cells in culture, as assessed by xCELLigence studies. In vitro analyses indicate that patient B would benefit mostly from PI3K inhibition therapy. The differences in responses between cases (as outlined by both proteomic analysis and functional assays) make an argument for personalized medicine in a sense extending genetic testing to functional and signaling assessments. The difference in sensitivity between different patient-derived cultures is not unexpected, since differences in expression profiles of several genes have been demonstrated for several glioblastoma derived cell lines.43
Importantly, a considerable consistency could be found between the effects of various protein-tyrosine-kinase inhibitors on protein expression of signal transduction molecules and cell behavior (adhesion and proliferation), as measured in real-time experiments. Thus, the best-ranked inhibitors (as established by protein expression experiments) also proved to be the most effective tools in decreasing cell growths.
The cellular pattern of signaling molecules in basal (unstimulated) cultivation conditions is in good correlation with the profile obtained in originating tissue samples (data not shown). In our view, this can represent a valuable tool for the assessment of in vitro-to-in vivo translatability. A further step in reducing this gap would be a better reproduction of some in vivo microenvironmental determinants, such as partial oxygen pressure, since hypoxia and members of hypoxia-inducible factors family are described among the major actors in glioblastoma, as well as in other tumors.44–46
Assessing key signaling molecules through the rapid and efficient screening method represented by xMAP technology can be useful for the diagnosis of brain tumors and for the discovery of new potential therapeutic targets or therapy monitoring. Furthermore, using molecular profiling, via high-throughput technologies such as proteomics, genomics, and metabolomics, it is currently possible to identify unique differences within an individual tumor, or between tumors of same histological grade.47
Conclusion
Our results show that in comparison to standard cell lines, evaluating tumor-derived cells by a combination of real-time monitoring of cellular behavior and multiplex analysis of several signaling pathways can be used for individualized therapy in patients with both primary and relapsed tumors.
PI3K inhibition proved to have the greatest capacity to influence the signaling proteins pattern, which would suggest that this pathway might be a prime target for glioblastoma treatment, alone or in combination, depending on the molecular association appending to each individual tumor.
Supplementary materials
Validation of xMAP assays for signal transduction molecule.
Notes: Serial dilutions of U87 cell lysates 1, 1/2, 1/4, and 1/8 for xMAP and 1, 1/2, 1/4, 1/8, and 1/16 for Western blot, respectively, were used for the estimation of the expression levels of P70S6K, ERK 1/2, and CREB. For xMAP assay, the MFI were normalized using GAPDH as a “housekeeping molecule.” (A) Dependence of normalized MFI with concentration of sample. (B) Western blot evaluation of protein levels of P70S6K, ERK 1/2, and CREB at different dilutions. Both methods demonstrated similar trends between sample concentration and the specific signal.
Abbreviations: CREB, cAMP response element-binding protein; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; MFI, mean fluorescence intensities; ND, non diluted; ERK, extracellular signal-regulated kinase.
Validation of modulation of signal transduction molecules expression by WB.
Notes: U87 cell lysates, from untreated cells and cells treated with PI3K inhibitor LY294002 were analyzed by WB. Expression levels of P70S6K, ERK 1/2, and CREB were specifically detected. Depletion of protein expression for each molecule was demonstrated in treated cells. Actin was used as a loading control. U87 cells were grown with vehicle (1/400 DMSO – untreated cells) or 25 μM PI3K inhibitor (LY294002) for 24 hours, lysed, and the same amount of protein was loaded on a 10% SDS-PAGE gel, transferred onto a nitrocellulose membrane, and examined for total protein expression. Betaactin reblotting was used as loading control. PI3K inhibitor treatment lowered total protein levels for all three investigated proteins.
Abbreviations: CREB, cAMP response element-binding protein; DMSO, dimethyl sulfoxide; PI3K, phosphatidylinositol-3′-kinase; WB, Western blot; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; ERK, extracellular signal-regulated kinase.
Verification of inhibitory effects of some inhibitors on the activity of targeted signal transduction molecules.
Notes: (A) xMAP determination of pERK 1/2, in U87 cells treated with the MEK inhibitors U0126 and PD98059. U87 controls were cultivated under similar conditions and administered an equal amount of vehicle as that used for the administration of inhibitor. Both inhibitors affected the levels of phosphorylation of ERK 1/2, as demonstrated by Luminex assay using the pERK MAP-mates. (B) WB determination of inhibition of Akt phosphorylation by inhibition of PI3K by treatment of U87 cells with PI3K inhibitor LY294002. U87 cells were treated with vehicle (1/400 DMSO) or 25 μM PI3K inhibitor (LY294002) for 24 hours, lysed, and the same amount of protein was loaded on a 10% SDS-PAGE gel, transferred onto a nitrocellulose membrane, and examined for Akt activation. Betaactin reblotting was used as loading control. Cell treatment with PI3K inhibitor significantly lowered pAkt levels.
Abbreviations: Akt, protein kinase B; Ctrl, control; DMSO, dimethyl sulfoxide; MFI, mean fluorescence intensities; PI3K, phosphatidylinositol-3′-kinase; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; WB, Western blot; pAkt, phospho Akt.
Acknowledgments
The research presented in this paper was supported by Grant POS-CCE 685-152/2010, Romania. Stefan N Constantinescu was supported by the Ludwig Institute for Cancer Research, ARC, and Fondation contre le cancer, Belgium. The authors present special thanks to Dr Daniel Petrescu from Neurology and Neurovascular Diseases National Institute, Bucharest, Romania, for providing the samples of patients’ tumors.
Author contributions
Maria Linda Cruceru, Ana-Maria Enciu, and Adrian Claudiu Popa performed the experiments; Radu Albulescu, Maria Linda Cruceru, and Ana-Maria Enciu analyzed the data; and Cristiana Pistol Tanase, Radu Albulescu, Monica Neagu, and Stefan N Constantinescu designed the research study and analyzed data. All authors took part in either drafting the article or revising it critically for important intellectual content.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Schwartz SA, Weil RJ, Johnson MD, Toms SA, Caprioli RM. Protein profiling in brain tumors using mass spectrometry: feasibility of a new technique for the analysis of protein expression. Clin Cancer Res. 2004;10(3):981–987. doi: 10.1158/1078-0432.ccr-0927-3. [DOI] [PubMed] [Google Scholar]
- 2.Iwami K, Natsume A, Wakabayashi T. Cytokine networks in glioma. Neurosurg Rev. 2011;34(3):253–263. doi: 10.1007/s10143-011-0320-y. discussion 263–264. [DOI] [PubMed] [Google Scholar]
- 3.Ohka F, Natsume A, Wakabayashi T. Current trends in targeted therapies for glioblastoma multiforme. Neurol Res Int. 2012;2012:878425. doi: 10.1155/2012/878425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dresemann G. Temozolomide in malignant glioma. Onco Targets Ther. 2010;3:139–146. doi: 10.2147/ott.s5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoon CH, Kim MJ, Kim RK, et al. c-Jun N-terminal kinase has a pivotal role in the maintenance of self-renewal and tumorigenicity in glioma stem-like cells. Oncogene. 2012;31(44):4655–4666. doi: 10.1038/onc.2011.634. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Li Y, Yu TS, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sai K, Wang S, Balasubramaniyan V, et al. Induction of cell-cycle arrest and apoptosis in glioblastoma stem-like cells by WP1193, a novel small molecule inhibitor of the JAK2/STAT3 pathway. J Neurooncol. 2012;107(3):487–501. doi: 10.1007/s11060-011-0786-z. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, McKay RM, Parada LF. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012;149(1):36–47. doi: 10.1016/j.cell.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheck AC, Stafford P. Design and use of biomarkers for the current and future clinical management of brain tumors. Biomark Med. 2012;6(3):293–295. doi: 10.2217/bmm.12.18. [DOI] [PubMed] [Google Scholar]
- 11.Iwadate Y, Sakaida T, Hiwasa T, et al. Molecular classification and survival prediction in human gliomas based on proteome analysis. Cancer Res. 2004;64(7):2496–2501. doi: 10.1158/0008-5472.can-03-1254. [DOI] [PubMed] [Google Scholar]
- 12.Kumar HR, Zhong X, Sandoval JA, Hickey RJ, Malkas LH. Applications of emerging molecular technologies in glioblastoma multiforme. Expert Rev Neurother. 2008;8(10):1497–1506. doi: 10.1586/14737175.8.10.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olar A, Aldape KD. Biomarkers classification and therapeutic decision-making for malignant gliomas. Curr Treat Options Oncol. 2012;13(4):417–436. doi: 10.1007/s11864-012-0210-8. [DOI] [PubMed] [Google Scholar]
- 14.Albulescu R, Codrici E, Popescu ID, et al. Cytokine patterns in brain tumour progression. Mediators Inflamm. 2013;2013:979748. doi: 10.1155/2013/979748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60(3):166–193. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukas RV, Boire A, Nicholas MK. Targeted therapy in the treatment of malignant gliomas. Onco Targets Ther. 2009;2:115–133. doi: 10.2147/ott.s3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fenton TR, Nathanson D, Ponte de Albuquerque C, et al. Resistance to EGF receptor inhibitors in glioblastoma mediated by phosphorylation of the PTEN tumor suppressor at tyrosine 240. Proc Natl Acad Sci U S A. 2012;109(35):14164–14169. doi: 10.1073/pnas.1211962109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Witusik-Perkowska M, Rieske P, Hułas-Bigoszewska K, et al. Glioblastoma-derived spheroid cultures as an experimental model for analysis of EGFR anomalies. J Neurooncol. 2011;102(3):395–407. doi: 10.1007/s11060-010-0352-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popescu LM, Manole E, Serboiu CS, et al. Identification of telocytes in skeletal muscle interstitium: implication for muscle regeneration. J Cell Mol Med. 2011;15(6):1379–1392. doi: 10.1111/j.1582-4934.2011.01330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ping YF, Yao XH, Jiang JY, et al. The chemokine CXCL12 and its receptor CXCR4 promote glioma stem cell-mediated VEGF production and tumour angiogenesis via PI3K/AKT signalling. J Pathol. 2011;224(3):344–354. doi: 10.1002/path.2908. [DOI] [PubMed] [Google Scholar]
- 21.Kambe A, Yoshioka H, Kamitani H, Watanabe T, Baek SJ, Eling TE. The cyclooxygenase inhibitor sulindac sulfide inhibits EP4 expression and suppresses the growth of glioblastoma cells. Cancer Prev Res (Phila) 2009;2(12):1088–1099. doi: 10.1158/1940-6207.CAPR-09-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin HY, Sun M, Tang HY, et al. L-Thyroxine vs 3,5,3′-triiodo-L-thyronine and cell proliferation: activation of mitogen-activated protein kinase and phosphatidylinositol 3-kinase. Am J Physiol Cell Physiol. 2009;296(5):C980–C991. doi: 10.1152/ajpcell.00305.2008. [DOI] [PubMed] [Google Scholar]
- 23.Zohrabian VM, Forzani B, Chau Z, Murali R, Jhanwar-Uniyal M. Rho/ROCK and MAPK signaling pathways are involved in glioblastoma cell migration and proliferation. Anticancer Res. 2009;29(1):119–123. [PubMed] [Google Scholar]
- 24.Pore N, Jiang Z, Shu HK, Bernhard E, Kao GD, Maity A. Akt1 activation can augment hypoxia-inducible factor-1alpha expression by increasing protein translation through a mammalian target of rapamycin-independent pathway. Mol Cancer Res. 2006;4(7):471–479. doi: 10.1158/1541-7786.MCR-05-0234. [DOI] [PubMed] [Google Scholar]
- 25.Qiu B, Zhang D, Tao J, Wu A, Wang Y. A simplified and modified procedure to culture brain glioma stem cells from clinical specimens. Oncol Lett. 2012;3(1):50–54. doi: 10.3892/ol.2011.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartscht T, Lehnert H, Gieseler F, Ungefroren H. The Src family kinase inhibitors PP2 and PP1 effectively block TGF-beta1-induced cell migration and invasion in both established and primary carcinoma cells. Cancer Chemother Pharmacol. 2012;70(2):221–230. doi: 10.1007/s00280-012-1904-0. [DOI] [PubMed] [Google Scholar]
- 27.Kita D, Yonekawa Y, Weller M, Ohgaki H. PIK3CA alterations in primary (de novo) and secondary glioblastomas. Acta Neuropathol. 2007;113(3):295–302. doi: 10.1007/s00401-006-0186-1. [DOI] [PubMed] [Google Scholar]
- 28.Park CM, Park MJ, Kwak HJ, et al. Ionizing radiation enhances matrix metalloproteinase-2 secretion and invasion of glioma cells through Src/epidermal growth factor receptor-mediated p38/Akt and phosphatidylinositol 3-kinase/Akt signaling pathways. Cancer Res. 2006;66(17):8511–8519. doi: 10.1158/0008-5472.CAN-05-4340. [DOI] [PubMed] [Google Scholar]
- 29.Höland K, Salm F, Arcaro A. The phosphoinositide 3-kinase signaling pathway as a therapeutic target in grade IV brain tumors. Curr Cancer Drug Targets. 2011;11(8):894–918. doi: 10.2174/156800911797264743. [DOI] [PubMed] [Google Scholar]
- 30.Wick W, Weller M, Weiler M, Batchelor T, Yung AW, Platten M. Pathway inhibition: emerging molecular targets for treating glioblastoma. Neuro Oncol. 2011;13(6):566–579. doi: 10.1093/neuonc/nor039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pu P, Kang C, Zhang Z, Liu X, Jiang H. Downregulation of PIK3CB by siRNA suppresses malignant glioma cell growth in vitro and in vivo. Technol Cancer Res Treat. 2006;5(3):271–280. doi: 10.1177/153303460600500308. [DOI] [PubMed] [Google Scholar]
- 32.Wen PY, Lee EQ, Reardon DA, Ligon KL, Alfred Yung WK. Current clinical development of PI3K pathway inhibitors in glioblastoma. Neuro Oncol. 2012;14(7):819–829. doi: 10.1093/neuonc/nos117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koul D, Shen R, Kim YW, et al. Cellular and in vivo activity of a novel PI3K inhibitor, PX-866, against human glioblastoma. Neuro Oncol. 2010;12(6):559–569. doi: 10.1093/neuonc/nop058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boller D, Doepfner KT, De Laurentiis A, et al. Targeting PI3KC2β impairs proliferation and survival in acute leukemia, brain tumours and neuroendocrine tumours. Anticancer Res. 2012;32(8):3015–3027. [PubMed] [Google Scholar]
- 35.Opel D, Westhoff MA, Bender A, Braun V, Debatin KM, Fulda S. Phosphatidylinositol 3-kinase inhibition broadly sensitizes glioblastoma cells to death receptor- and drug-induced apoptosis. Cancer Res. 2008;68(15):6271–6280. doi: 10.1158/0008-5472.CAN-07-6769. [DOI] [PubMed] [Google Scholar]
- 36.Kim YW, Liu TJ, Koul D, et al. Identification of novel synergistic targets for rational drug combinations with PI3 kinase inhibitors using siRNA synthetic lethality screening against GBM. Neuro Oncol. 2011;13(4):367–375. doi: 10.1093/neuonc/nor012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sunayama J, Matsuda K, Sato A, et al. Crosstalk between the PI3K/mTOR and MEK/ERK pathways involved in the maintenance of self-renewal and tumorigenicity of glioblastoma stem-like cells. Stem Cells. 2010;28(11):1930–1939. doi: 10.1002/stem.521. [DOI] [PubMed] [Google Scholar]
- 38.See WL, Tan IL, Mukherjee J, Nicolaides T, Pieper RO. Sensitivity of glioblastomas to clinically available MEK inhibitors is defined by neurofibromin 1 deficiency. Cancer Res. 2012;72(13):3350–3359. doi: 10.1158/0008-5472.CAN-12-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han L, Yang Y, Yue X, et al. Inactivation of PI3K/AKT signaling inhibits glioma cell growth through modulation of β-catenin-mediated transcription. Brain Res. 2010;1366:9–17. doi: 10.1016/j.brainres.2010.09.097. [DOI] [PubMed] [Google Scholar]
- 40.Reif K, Burgering BM, Cantrell DA. Phosphatidylinositol 3-kinase links the interleukin-2 receptor to protein kinase B and p70 S6 kinase. J Biol Chem. 1997;272(22):14426–14433. doi: 10.1074/jbc.272.22.14426. [DOI] [PubMed] [Google Scholar]
- 41.Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S, AmiGO Hub. Web Presence Working Group AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25(2):288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cox JL, Wilder PJ, Desler M, Rizzino A. Elevating SOX2 levels deleteriously affects the growth of medulloblastoma and glioblastoma cells. PLoS One. 2012;7(8):e44087. doi: 10.1371/journal.pone.0044087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clark MJ, Homer N, O’Connor BD, et al. U87MG decoded: the genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet. 2010;6(1):e1000832. doi: 10.1371/journal.pgen.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heddleston JM, Wu Q, Rivera M, et al. Hypoxia-induced mixed-lineage leukemia 1 regulates glioma stem cell tumorigenic potential. Cell Death Differ. 2012;19(3):428–439. doi: 10.1038/cdd.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bar EE, Lin A, Mahairaki V, Matsui W, Eberhart CG. Hypoxia increases the expression of stem-cell markers and promotes clonogenicity in glioblastoma neurospheres. Am J Pathol. 2010;177(3):1491–1502. doi: 10.2353/ajpath.2010.091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Méndez O, Zavadil J, Esencay M, et al. Knock down of HIF-1alpha in glioma cells reduces migration in vitro and invasion in vivo and impairs their ability to form tumor spheres. Mol Cancer. 2010;9:133. doi: 10.1186/1476-4598-9-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosser CJ, Goodison S. Today’s discoveries to tomorrow’s care: cancer biomarkers revisited. Foreword. Biomark Med. 2010;4(4):491–493. doi: 10.2217/bmm.10.78. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Validation of xMAP assays for signal transduction molecule.
Notes: Serial dilutions of U87 cell lysates 1, 1/2, 1/4, and 1/8 for xMAP and 1, 1/2, 1/4, 1/8, and 1/16 for Western blot, respectively, were used for the estimation of the expression levels of P70S6K, ERK 1/2, and CREB. For xMAP assay, the MFI were normalized using GAPDH as a “housekeeping molecule.” (A) Dependence of normalized MFI with concentration of sample. (B) Western blot evaluation of protein levels of P70S6K, ERK 1/2, and CREB at different dilutions. Both methods demonstrated similar trends between sample concentration and the specific signal.
Abbreviations: CREB, cAMP response element-binding protein; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; MFI, mean fluorescence intensities; ND, non diluted; ERK, extracellular signal-regulated kinase.
Validation of modulation of signal transduction molecules expression by WB.
Notes: U87 cell lysates, from untreated cells and cells treated with PI3K inhibitor LY294002 were analyzed by WB. Expression levels of P70S6K, ERK 1/2, and CREB were specifically detected. Depletion of protein expression for each molecule was demonstrated in treated cells. Actin was used as a loading control. U87 cells were grown with vehicle (1/400 DMSO – untreated cells) or 25 μM PI3K inhibitor (LY294002) for 24 hours, lysed, and the same amount of protein was loaded on a 10% SDS-PAGE gel, transferred onto a nitrocellulose membrane, and examined for total protein expression. Betaactin reblotting was used as loading control. PI3K inhibitor treatment lowered total protein levels for all three investigated proteins.
Abbreviations: CREB, cAMP response element-binding protein; DMSO, dimethyl sulfoxide; PI3K, phosphatidylinositol-3′-kinase; WB, Western blot; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; ERK, extracellular signal-regulated kinase.
Verification of inhibitory effects of some inhibitors on the activity of targeted signal transduction molecules.
Notes: (A) xMAP determination of pERK 1/2, in U87 cells treated with the MEK inhibitors U0126 and PD98059. U87 controls were cultivated under similar conditions and administered an equal amount of vehicle as that used for the administration of inhibitor. Both inhibitors affected the levels of phosphorylation of ERK 1/2, as demonstrated by Luminex assay using the pERK MAP-mates. (B) WB determination of inhibition of Akt phosphorylation by inhibition of PI3K by treatment of U87 cells with PI3K inhibitor LY294002. U87 cells were treated with vehicle (1/400 DMSO) or 25 μM PI3K inhibitor (LY294002) for 24 hours, lysed, and the same amount of protein was loaded on a 10% SDS-PAGE gel, transferred onto a nitrocellulose membrane, and examined for Akt activation. Betaactin reblotting was used as loading control. Cell treatment with PI3K inhibitor significantly lowered pAkt levels.
Abbreviations: Akt, protein kinase B; Ctrl, control; DMSO, dimethyl sulfoxide; MFI, mean fluorescence intensities; PI3K, phosphatidylinositol-3′-kinase; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; WB, Western blot; pAkt, phospho Akt.