Abstract
The genetic background of the mutations that most often cause cystic fibrosis (CF) is different from that of non-CF chromosomes in populations of European origin. It is not known whether these haplotype backgrounds could be found at high frequencies in populations in which CF is, at present, not common; such populations would be candidates for the place of origin of CF mutations. An analysis of haplotypes of CF transmembrane conductance regulator, together with their variation in specific CF chromosomes, in a worldwide survey of normal chromosomes shows (1) a very low frequency or absence of the most common CF haplotypes in all populations analyzed and (2) a strong genetic variability and divergence, among various populations, of the chromosomes that carry disease-causing mutations. The depth of the gene genealogy associated with disease-causing mutations may be greater than that of the evolutionary process that gave rise to present-day human populations. The concept of “population of origin” lacks either spatial or temporal meaning for mutations that are likely to have been present in Europeans before the ethnogenesis of present populations; subsequent population processes may have erased the traces of their geographic origin.
A deep understanding of a genetic disease requires not only a comprehension of its genetic and functional bases but also a description of its natural history. Ideally, such a description would comprise the place and time of origin of the pathogenic genetic variant and an explanation of its presence, frequency, and geographic distribution. Natural history also includes the genetic mechanisms that may explain the appearance of the disease (such as mutation rate and pattern), the population factors that may explain disease frequency and distribution (migration, population size, and genetic drift), and the interaction with the environment (selection in its rich variety).
Cystic fibrosis (CF [MIM 219700]) is the most common severe autosomal recessive disease in populations of European origin, in which it affects 1 in 2,500 individuals. It is caused by mutations in the CF transmembrane conductance regulator (CFTR [MIM 602421]), which was identified and cloned in 1989 (Kerem et al. 1989; Riordan et al. 1989; Rommens et al. 1989). Since then, ∼1,000 mutations have been reported (Cystic Fibrosis Mutation Data Base). Among the various CF mutations, a deletion of 3 bp at codon 508 (ΔF508) is the most frequent, accounting for two-thirds of the global CF chromosomes. Only four other mutations (G542X, N1303K, G551D, and W1282X) have overall frequencies >1% among the CF chromosomes. These five mutations are found throughout Europe, although their distribution shows clear geographic patterns: ΔF508 shows a northwest-to-southeast gradient, with a maximum (87.2% of all CF chromosomes) in Denmark and a minimum (21.3%) in Turkey (European Working Group on CF Genetics 1990). G542X is common in Mediterranean countries and is present in most of Europe, being most frequent (16.7%) in the Balearic Islands (Estivill et al. 1997). N1303K is present around the Mediterranean, and it reaches its highest frequency (17.2%) in Tunisia (Estivill et al. 1997). Mutation G551D is common in northwestern and central Europe, but it is uncommon in other parts of Europe (Estivill et al. 1997). Finally, mutation W1282X is common in most Mediterranean countries, reaching its highest frequency (36.2%) in Israel (Estivill et al. 1997). In addition, 17 other mutations have frequencies of 0.1%–0.9% (Estivill et al. 1997), and most of the remaining mutations are rare or are confined to few populations.
Several short tandem-repeat polymorphisms (STRPs), which are also known as “microsatellites,” and single-nucleotide polymorphisms (SNPs) have been described within the CFTR gene. Both types of markers can be used to trace the origin and evolution of the different CF mutations (Morral et al. 1993; Morral et al. 1994; Bertranpetit and Calafell 1996; Slatkin and Rannala 1997). SNPs can be used to define the stable haplotypic frameworks on which CFTR mutations occurred. Microsatellite markers, which mutate faster and are more diverse among chromosomes, can help to measure genetic variability within the CFTR locus and to estimate the age of CF mutations (Morral et al. 1993; Estivill et al. 1994; Bertranpetit and Calafell 1996).
When nondisease chromosomes (usually nondisease chromosomes of carriers) were analyzed in the populations in which CF is present, it became evident that, among Europeans, the general genetic background was very different from that of CF chromosomes (Estivill et al. 1994; Morral et al. 1996). Because CF is mostly confined to populations of European ancestry, it seemed likely that the origin of the most frequent CF mutations should be non-European (Morral et al. 1994) and that subsequent gene flow had brought CF chromosomes into Europe.
A hypothesis based on natural selection has been suggested to explain the high prevalence of CF: heterozygotes for CF mutations would be protected against the dehydrating action of Vibrio cholerae (Romeo et al. 1989; Gabriel et al. 1994) or Salmonella typhi (Pier et al. 1998; Pier 1999, 2000) enterotoxins, although this has been disputed (Högenauer et al. 2000). Protection against cholera or typhoid fever would make it more likely that selective pressures raised the CF frequency outside Europe, where these diseases are more prevalent; this is an argument for a non-European origin of CF.
Because the haplotypic backgrounds on which major CF mutations occurred have been clearly identified (Morral et al. 1996), we undertook a worldwide survey, to find populations in which specific haplotypes linked to CF mutations would be present at high frequencies. This could give hints about past specific selective pressures that could explain the spread of the mutations. Given that recombination rates in 7q31.2 (the chromosome region where CFTR maps) are one-fifth of the genome average (Payseur and Nachman 2000), it is unlikely that recombination disrupted the allelic associations in this region. Moreover, recombinant haplotypes, because of their high polymorphism, are easily detected with these markers (Morral et al. 1994).
We analyzed four STRPs and two SNPs located within CFTR (fig. 1). These are, in 5′→3′ chromosomal order, IVS1CA (Moulin et al. 1997; Mateu et al. 1999), IVS6aGATT (Chehab et al. 1991), IVS8CA (Morral et al. 1991), T854 (Zielenski et al. 1991b), IVS17bTA (Zielenski et al. 1991a), and TUB20 (Quere et al 1991). We typed samples from 949 unrelated autochthonous healthy individuals (1,898 chromosomes) from 17 populations that represented all major geographic areas (fig. 2). Appropriate informed consent was obtained from human subjects. DNA from five populations (Basque, Catalan, Tanzanian, Kazakh, and Saharawi) was extracted from fresh blood of donors. DNA samples for the other populations were obtained from lymphoblastoid cell lines maintained in K. K. Kidd’s laboratory at Yale University. Because no disease chromosomes had yet been typed for the more recently described IVS1CA marker, a total of 126 patients (252 chromosomes), who carried the ΔF508, G542X, and/or N1303K mutations, were typed for this locus. Allele frequencies for this locus are reported here, for the first time, in chromosomes carrying CF mutations. The samples were obtained at the Institut de Recerca Oncològica, Barcelona.
Figure 1.
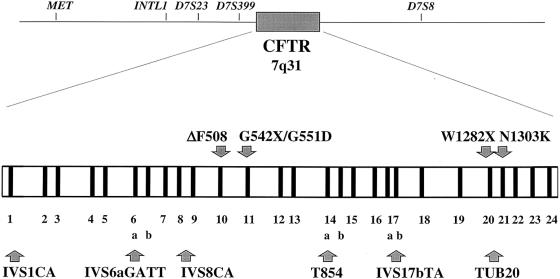
Polymorphisms in the CFTR region (IVS1CA, IVS6aGATT, IVS8CA, T854, IVS17bTA, and TUB20), and location of the five most common CF mutations (ΔF508, G542X, N1303K, G551D, and W1282X). CFTR exons are numbered 1–24.
Figure 2.
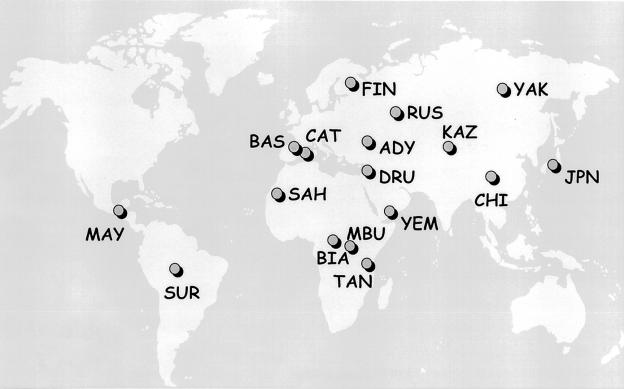
Distribution of 17 populations analyzed. Population names (sample sizes [no. of chromosomes]) are as follows: ADY = Adygei (106); BAS = Basques (222); BIA = Biaka Pygmies (138); CAT = Catalans (176); CHI = Han Chinese (124); DRU = Druze (126); FIN = Finns (70); JPN = Japanese (96); KAZ = Kazakhs (80); MAY = Maya (106); MBU = Mbuti Pygmies (78); RUS = Russians (96); SAH = Saharawi (118); SUR = Surui (94); TAN = Tanzanians (80); YAK = Yakut (102); YEM = Yemenites (86). Further information on these population samples can be found in the Allele Frequency Database and in a report by Mateu et al. (2001).
Typing methods for six loci are as described elsewhere (Mateu et al. 2001), in an article in which we have reported allele and haplotype frequencies for the population set. Mean maximum-likelihood estimates of haplotype frequencies and the jackknife standard errors (SEs) were calculated from the multisite marker–typing data, by the HAPLO program (Hawley and Kidd 1995). We used PHYLIP (Felsenstein 1989) to produce maximum-likelihood population trees of allele frequencies of five polymorphisms (no data were available for IVS1CA, for chromosomes carrying G551D or W1282X) of normal and CF chromosomes. IVS6aGATT, IVS8CA, T854, IVS17bTA, and TUB20 allele frequencies for CF chromosomes (with ΔF508, G542X, N1303K, G551D, and W1282X mutations) were obtained from the literature (Morral et al. 1994, 1996). IVS8CA and IVS17bTA allele frequencies for ΔF508 chromosomes in geographically defined samples for European populations were from Morral et al. (1994).
Allele frequencies for the intron 1 CA repeat in CF chromosomes are given in table 1. The three most common mutations are clearly associated with the 21-repeat allele, which is likely to have been present in the haplotype backgrounds where these mutations originated.
Table 1.
Allele Frequencies for the Intron 1 CA Repeat in CF Chromosomes[Note]
|
Frequency of |
|||
| No. ofRepeats | ΔF508(n = 148) | G542X(n = 56) | N1303K(n = 17) |
| 21 | .993 | 1 | .941 |
| 22 | 0 | 0 | .059 |
| 23 | .007 | 0 | 0 |
Note.— Numbers of chromosomes studied appear in parentheses.
The most frequent CF-causing mutations are found in two groups of haplotype backgrounds (table 2). When all six markers are considered in their chromosomal order (i.e., IVS1CA, IVS6aGATT, IVS8CA, T854, IVS17bTA, and TUB20), these haplotype background groups are: (1) 21-6-(17/22/23/24)-1-(31/32/33)-2, of which the most frequent are 21-6-23-1-31-2 (for ΔF508 and N1303K mutations) and 21-6-23-1-33-2 (for the G542X mutation) it is evident that these three different CF mutations (which have independent origins) are found in very closely related haplotypes, since they differ only by a few repeat units at the fast-evolving STRP sites; and (2) 7-(16/17)-2-7-1, in which G551D and W1282X are found. For these mutations, no data are available for IVS1CA, the first marker.
Table 2.
Most Frequent CFTR Haplotype(s) for the Five Most Common CF Mutations
|
Haplotype(s) at Markera |
||||||
| CF Mutation | IVS1CA | IVS6aGATT | IVS8CA | T854 | IVS17bTA | TUB20 |
| ΔF508 | 21 | 6 | 23/17 | 1 | 31/32 | 2 |
| G542X | 21 | 6 | 23 | 1 | 33/32 | 2 |
| N1303K | 21 | 6 | 23/22/24 | 1 | 31 | 2 |
| G551D | NA | 7 | 16 | 2 | 7 | 1 |
| W1282X | NA | 7 | 17 | 2 | 7 | 1 |
IVS1CA was typed in the present study. The remaining markers were typed by Morral et al. (1996). NA = not available.
The haplotypes associated specifically with ΔF508, even if several alleles for the STRs are pooled, are not found in frequencies that would suggest a place of likely origin for ΔF508 in any of the populations sampled (table 3). These haplotypes have been found—at very low frequencies—only in one Middle Eastern population (Druze, 2.4%) and in one European population (Finns, 1.6%). The most common haplotypes in European and Middle Eastern populations are 21-7-16-2-7-1 and 22-7-16-1-30-2, which have allele frequencies of 9.4% and 19.7%, respectively (Mateu et al. 2001). Only minor traces of chromosomes that bear phylogenetically related haplotypes are found in several European and Asian populations (table 3).
Table 3.
Frequencies, in Normal Chromosomes, of Haplotypes Associated with CF Mutations ΔF508, G542X, and N1303K[Note]
|
Mean Frequency (SE) in Haplotype(%)a |
||||
| Population | A | B | C | D |
| Druze | 2.4 ± 1.4 | 0 | 4 ± 1.7 | 0 |
| Basque | 0 | 0 | 4.2 ± 1.4 | 0 |
| Catalan | 0 | 0 | 1.4 ± .9 | .6 ± .6 |
| Finnish | 0 | 1.6 ± 1.6 | 3.2 ± 2.2 | 1.6 ± 1.6 |
| Russian | 0 | 0 | 1.7 ± 1.7 | 1.7 ± 1.7 |
| Adygei | 0 | 0 | 2.0 ± 1.4 | 0 |
| Japanese | 0 | 0 | 0 | 1.2 ± 1.2 |
Note.— The frequency in the populations not listed is zero.
Haplotype A = 21-6-23-1-31-2; haplotype B = 21-6-17-1-31-2; haplotype C = (20/21/22)-6-(22/23/24)-1-(30/31/32/33/34)-2; and haplotype D = (20/21/22)-6-(16/17/18)-1-(30/31/32)-2.
Haplotypes associated with CF mutations G542X and N1303K are closely related to those of ΔF508, and the situation is therefore similar. This suggests that all three mutations arose in the same population. When the three haplotype backgrounds are considered together (a strategy that permits examination of a deeper branch in the gene tree of haplotypes), frequencies in normal chromosomes remain at very low levels (table 3). Given the sample size, some of these haplotypes could be present in the populations but not included in the sample. For sample sizes of 70–222 chromosomes, a nonobserved haplotype could have, with a 95% probability, an upper frequency of 4.19%–1.34% (Rohlf and Sokal 1995). Thus, haplotypes found at frequencies of the same order of magnitude as the CF carrier frequency in the population would be detected. It should be noted that a signal of the origin of any of the three major mutations in a population would be a frequency clearly above that, since all three mutations seem to have occurred on the same background, an event that would be quite improbable if the haplotype frequency were so low. Alternatively, that particular haplotype may be mutagenic, which, in absence of other kinds of biological evidence, seems implausible; it has been clearly demonstrated that they have a single origin (Morral et al. 1993; Bertranpetit and Calafell 1996).
The situation is very different for the two other frequent mutations (G551D and W1282X). Both are associated with closely related haplotypes, now found at high frequency in populations of different continents (table 4): Africa (Mbuti, 12%; Saharawi, 20.8%), western Asia (Adygei, 17.3%), Europe (Catalans, 16.7%), and Siberia (Yakut, 2.6%). They have not been found in other populations of eastern Asian or in American Indians.
Table 4.
Frequencies, Normal Chromosomes, of Haplotypes 7-16/17-2-7-1, Associated with CF Mutations G551D and W1282X[Note]
| Population | Mean Frequency ± SE(%) |
| Tanzanians | 3.2 ± 2.2 |
| Biaka | 9 ± 2.6 |
| Mbuti | 12 ± 4 |
| Saharawi | 20.8 ± 3.9 |
| Druze | 15.1 ± 3.2 |
| Yemenites | 7.5 ± 2.9 |
| Basques | 11.2 ± 2.1 |
| Catalans | 16.7 ± 2.9 |
| Finns | 16.1 ± 4.7 |
| Russians | 15 ± 4.6 |
| Adygei | 17.3 ± 3.8 |
| Kazakhs | 6.7 ± 3.2 |
| Yakut | 2.6 ± 1.8 |
Note.— The frequency in the populations not listed is zero.
To further explore the population-genetics relationships, a tree was built using maximum likelihood, including allele frequencies for non-CF chromosomes and for chromosomes carrying specific CF mutations; in the case of ΔF508, it has been possible to consider different European populations separately (fig. 3). The structure of the variation is mainly based on haplotype background (not on population). Thus, it appears that ΔF508, G542X, and N1303K are closely related to each other, as are G551D and W1282X, independently of the population from which chromosomes were sampled. A similar pattern has been observed for other genome regions (Bosch et al. 1999). The main stratifying factor is genetic background (i.e., presence or absence of CF mutations), rather than population, even when very distant populations are considered.
Figure 3.
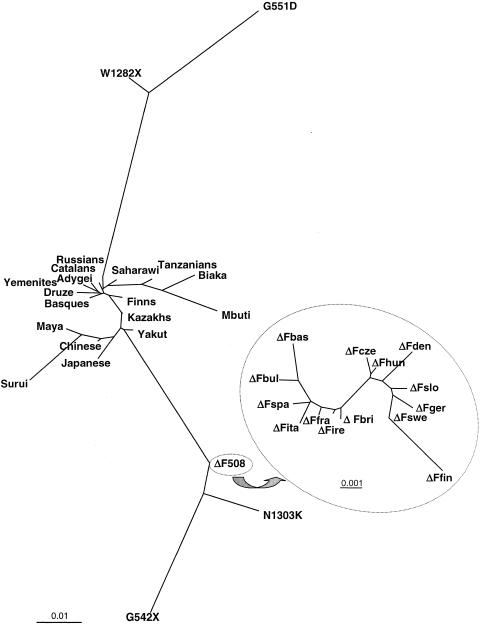
Maximum-likelihood tree of allele frequencies of five loci (IVS6aGATT, IVS8CA, T854, IVS17bTA and TUB20) among normal chromosomes, from worldwide populations, and among CF chromosomes (ΔF508, G542X, N1303K, G551D and W1282X chromosomes). The inset shows an enlarged maximum-likelihood tree of allele frequencies of two loci (IVS8CA and IVS17bTA) among ΔF508 chromosomes in different European populations (Fbas = Basque; Fbri = British; Fbul = Bulgarian; Fcze = Czech; Fden = Danish; Ffin = Finnish; Ffra = French; Fger = German; Fhun = Hungarian; Fire = Irish; Fita = Italian; Fslo = Slovakian; Fspa = Spanish; Fswe = Swedish). Bars show the scale in genetic-distance units. Several trees have been built, using either other methods (e.g., Nei-Kimura distance and the neighbor-joining algorithm) or different sets of chromosomes. In all cases, results are similar.
The present results of a worldwide genetic survey show that the main CF-mutation haplotype backgrounds either are found at very low frequencies or, more usually, are altogether absent. Although no population has them in frequencies high enough to suggest a likely place of origin, it should be kept in mind that mutations ΔF508, G542X, and N1303K are independent unique events and that their occurrence in a similar background gives support to the hypothesis that the three mutations arose in a single population in which these haplotypes were frequent. In that hypothetical population, demographic events (such as expansion) or selective agents could have raised those deleterious mutations to relatively high frequencies and could have spread them to other areas.
The current widespread distribution of haplotypes related to G551D and W1282X is compatible with an origin in Europe, although a geographic distribution that would allow us to identify the birthplace of these mutations is not evident. Moreover, given the low frequency of these mutations, no localized selection pressures need to be invoked to explain their origin and current distribution.
The maximum-likelihood tree for CF and non-CF chromosomes (fig. 3) stresses the importance of the genetic background as well as the lesser importance of the population in which the individual (or, more precisely, a chromosome) belongs. The age of ΔF508 is controversial, with estimates ranging from 3,000 years ago (i.e., post-Neolithic era) (Serre et al. 1990) to >40,000 years ago (in the Upper Paleolithic era, and clearly pre-Neolithic) (Morral et al. 1994). A recent study, using coalescence theory (Wiuf 2001), has confirmed an ancient age for the mutation, with estimates ranging from 11,000 to 34,000 years ago. Using standard population-genetics models, Reich and Lander (2001) have also estimated—on the basis of the overall frequency of CF alleles—that ΔF508 may predate the expansion of anatomically modern humans. Even though the estimates are strongly dependent on genetic (e.g., mutation rate and selection) and demographic (e.g., expansion dynamics and population size) parameters, the mutation is clearly pre-Neolithic and is an ancient mutation in human history. Other CF mutations are slightly younger (Morral et al. 1993). The estimates for older mutations are in agreement with the stronger genetic-variation differentiation shown by CF mutations.
The problem of the place or population of origin of the most common CF mutations may lack sense, because several of them may be older (Morral et al. 1994; Wiuf 2001) than the ethnogenesis process that originated the present European populations (Cavalli-Sforza et al. 1994). Thus, population processes such as genetic drift and gene flow, may have had sufficient time to reshape the genetic structure of the population in which the major CF mutations arose. Indeed, they may have erased the traces that could lead to the identification of a population as the cradle of CF mutations. Given that the mutations are ancient and that genetic variation in CF chromosomes is structured by lineage rather than by population origin, a population-genetic approach may not produce results adequate for a detailed comprehension of the evolutionary processes that gave rise to CF mutations.
Acknowledgments
This research was supported by the Fundació La Marató de TV3-1998 (project “La Història Natural de la Fibrosi Quística: Interpretació Geogràfica de la Variació Genètica”), by Dirección General de Investigación Científica y Técnica (Spain) grant PB98-1064, and by Direcció General de Recerca, Generalitat de Catalunya, grant 1998SGR00009. This work was also possible thanks to fellowships to E.M., from Universitat de Barcelona and Universitat Pompeu Fabra. Druze and Yemenite samples were kindly supplied by Dr. Batsheva Bonné-Tamir from Sackler Faculty of Medicine (Tel Aviv University, Tel Aviv); samples from Tanzania were kindly supplied by Dr. Clara Menéndez from Unitat d’Epidemiologia i Bioestadística (Hospital Clínic, Barcelona); and samples from Kazakhstan were kindly supplied by Dr. Davide Pettener from Unità di Antropologia (Università di Bologna, Bologna). We also especially thank Judith R. Kidd and Kenneth K. Kidd (Department of Genetics, Yale University School of Medicine, New Haven) for supplying Pygmy, Finn, Russian, Adygei, Chinese, Japanese, Yakut, Mayan, and Surui DNA samples. Finally, we would like to thank Montgomery Slatkin for useful comments on an earlier version of the manuscript.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Allele Frequency Database (ALFRED), http://info.med.yale.edu/genetics/kkidd (for further information on population samples)
- Cystic Fibrosis Mutation Data Base, http://www.genet.sickkids.on.ca/cftr
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CF [MIM 219700] and CFTR [MIM 602421])
References
- Bertranpetit J, Calafell F (1996) Genetic and geographical variability in cystic fibrosis: evolutionary considerations. In: Cardew G (ed) Variation in the human genome. Ciba Foundation Symposium 197. Wiley & Sons, Chichester, England, pp 97–118 [DOI] [PubMed] [Google Scholar]
- Bosch E, Calafell F, Santos FR, Pérez-Lezaun A, Comas D, Benchemsi N, Tyler- Smith C, Bertranpetit J (1999) Variation in short tandem repeats is deeply structured by genetic background on the human Y chromosome. Am J Hum Genet 65:1623–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton, NJ [Google Scholar]
- Chehab EF, Johnson J, Louie E, Goossens M, Kawasaki E, Erlich H (1991) A dimorphic 4-bp repeat in the cystic fibrosis gene is in absolute linkage disequilibrium with the ΔF508 mutation: implications for prenatal diagnosis and mutation origin. Am J Hum Genet 48:223–226 [PMC free article] [PubMed] [Google Scholar]
- Estivill X, Bancells C, Ramos C, Biomed CF Mutation Analysis Consortium (1997) Geographic distribution and regional origin of 272 cystic fibrosis mutations in European populations. Hum Mutat 10:135–154 [DOI] [PubMed] [Google Scholar]
- Estivill X, Morral N, Bertranpetit J (1994) Age of the ΔF508 cystic fibrosis mutation. Nat Genet 8:216–218 [DOI] [PubMed] [Google Scholar]
- European Working Group on CF Genetics (1990) Gradient of distribution in Europe of the major CF mutation and of its associated haplotypes. Hum Genet 85:436–441 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (1989) PHYLIP: phylogeny inference package. Cladistics 5:164–166 [Google Scholar]
- Gabriel SE, Brigman KN, Koller BH, Boucher RC, Stutts MJ (1994) Cystic fibrosis heterozygote resistance to cholera toxin in the cystic fibrosis mouse model. Science 266:107–109 [DOI] [PubMed] [Google Scholar]
- Hawley ME, Kidd KK (1995) HAPLO: a program using the EM algorithm to estimate frequencies of multi-site haplotypes. J Hered 86:409–411 [DOI] [PubMed] [Google Scholar]
- Högenauer C, Santa Ana CA, Porter JL, Millard M, Gelfand A, Rosenblatt RL, Prestidge CB, Fordtran JS (2000) Active intestinal chloride secretion in human carriers of cystic fibrosis mutations: an evaluation of the hypothesis that heterozygotes have subnormal active intestinal chloride secretion. Am J Hum Genet 67:1422–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerem BS, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, Buchwald M, Tsui L-C (1989) Identification of the cystic fibrosis gene: genetic analysis. Science 245:1073–1080 [DOI] [PubMed] [Google Scholar]
- Mateu E, Calafell F, Bonné-Tamir B, Kidd JR, Casals T, Kidd KK, Bertranpetit J (1999) Allele frequencies in a worldwide survey of a CA repeat in the first intron of the CFTR gene. Hum Hered 49:15–20 [DOI] [PubMed] [Google Scholar]
- Mateu E, Calafell F, Lao O, Bonné-Tamir B, Kidd JR, Pakstis A, Kidd KK, Bertranpetit J (2001) Worldwide genetic analysis of the CFTR region. Am J Hum Genet 68:103–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morral N, Bertranpetit J, Estivill X, Nunes V, Casals T, Giménez J, Reis A, et al (1994) The origin of the major cystic fibrosis mutation (ΔF508) in European populations. Nat Genet 7:169–175 [DOI] [PubMed] [Google Scholar]
- Morral N, Dörk T, Llevadot R, Dziadeek V, Mercier B, Férec C, Costes B, Girodon E, Zielenski J, Tsui L-C, Tümmler B, Estivill X (1996) Haplotype analysis of 94 cystic fibrosis mutations with seven polymorphic CFTR DNA markers. Hum Mutat 8:149–159 [DOI] [PubMed] [Google Scholar]
- Morral N, Nunes V, Casals T, Chillón M, Giménez J, Bertranpetit J, Estivill X (1993) Microsatellite haplotypes for cystic fibrosis: mutation frameworks and evolutionary tracers. Hum Mol Genet 2:1015–1022 [DOI] [PubMed] [Google Scholar]
- Morral N, Nunes V, Casals T, Estivill X (1991) CA/GT microsatellite alleles within the cystic fibrosis transmembrane conductance regulator (CFTR) gene are not generated by unequal crossingover. Genomics 10:692–698 [DOI] [PubMed] [Google Scholar]
- Moulin DS, Smith AN, Harris A (1997) A CA repeat in the first intron of the CFTR gene. Hum Hered 47:295–297 [DOI] [PubMed] [Google Scholar]
- Payseur BA, Nachman MW (2000) Microsatellite variation and recombination rate in the human genome. Genetics 156:1285–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pier GB (1999) Evolution of the ΔF508 CFTR mutation: response. Trends Microbiol 7:56–58 [DOI] [PubMed] [Google Scholar]
- ——— (2000) Role of the cystic fibrosis transmembrane conductance regulator in innate immunity to Pseudomonas aeruginosa infections. Proc Natl Acad Sci USA 97:8822–8828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pier GB, Grout M, Zaidi T, Meluleni G, Mueschenborn SS, Banting G, Ratcliff R, Evans MJ, Colledge WH (1998) Salmonella typhi uses CFTR to enter intestinal epithelial cells. Nature 393:79–82 [DOI] [PubMed] [Google Scholar]
- Quere I, Guillermit H, Mercier B, Audrezet MP, Ferec C (1991) A polymorphism in intron 20 of the CFTR gene. Nucleic Acids Res 19:5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DE, Lander ES (2001) On the allelic spectrum of human disease. Trends Genet 17:502–510 [DOI] [PubMed] [Google Scholar]
- Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL, Drumm ML, Iannuzzi MC, Collins FS, Tsui L-C (1989) Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 245:1066–1073 [DOI] [PubMed] [Google Scholar]
- Rohlf FJ, Sokal RR (1995) Statistical tables. WH Freeman, New York [Google Scholar]
- Romeo G, Devoto M, Galietta LJV (1989) Why is the cystic fibrosis gene so frequent? Hum Genet 84:1–5 [DOI] [PubMed] [Google Scholar]
- Rommens JM, Iannuzzi MC, Kerem BS, Drumm ML, Melmer G, Dean M, Rozmahel R, Cole JL, Kennedy D, Hidaka N, Zsiga M, Buchwald M, Riordan JR, Tsui L-C, Collins FS (1989) Identification of the cystic fibrosis gene: chromosome walking and jumping. Science 245:1059–1065 [DOI] [PubMed] [Google Scholar]
- Serre JL, Simon-Bouy B, Mornet E, Jaume-Roig B, Balassopoulou A, Schwartz M, Taillandier A, Boué J, Boué A (1990) Studies of RFLP closely linked to the cystic fibrosis locus throughout Europe lead to new considerations in population genetics. Hum Genet 84:449–454 [DOI] [PubMed] [Google Scholar]
- Slatkin M, Rannala B (1997) Estimating the age of alleles by use of intraallelic variability. Am J Hum Genet 60:447–458 [PMC free article] [PubMed] [Google Scholar]
- Wiuf C (2001) Do ΔF508 heterozygotes have a selective advantage? Genet Res 78:41–47 [DOI] [PubMed] [Google Scholar]
- Zielenski J, Markiewicz D, Rininsland F, Rommens JM, Tsui L-C (1991a) A cluster of highly polymorphic dinucleotide repeats in intron 17b of the CFTR gene. Am J Hum Genet 49:1256–1262 [PMC free article] [PubMed] [Google Scholar]
- Zielenski J, Rozmahel R, Bozon D, Kerem BS, Grzelczak Z, Riordan JR, Rommens JM, Tsui L-C (1991b) Genomic DNA sequence of the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Genomics 10:214–228 [DOI] [PubMed] [Google Scholar]


