Abstract
Objective
High-definition transcranial direct current stimulation (HD-tDCS) and high-density electroencephalography (HD-EEG) require accurate models of current flow for precise targeting and current source reconstruction. At a minimum, such modeling must capture the idiosyncratic anatomy of brain, cerebrospinal fluid (CSF) and skull for each individual subject. Currently, the process to build such high-resolution individualized models from structural magnetic resonance images (MRI) requires labor-intensive manual segmentation, even when leveraging available automated segmentation tools. Also, accurate placement of many high-density electrodes on individual scalp is a tedious procedure. The goal was to develop fully automated techniques to reduce the manual effort in such a modeling process.
Approach
A fully automated segmentation technique based on Statical Parametric Mapping 8 (SPM8), including an improved tissue probability map (TPM) and an automated correction routine for segmentation errors, was developed, along with an automated electrode placement tool for high-density arrays. The performance of these automated routines was evaluated against results from manual segmentation on 4 healthy subjects and 7 stroke patients. The criteria include segmentation accuracy, the difference of current flow distributions in resulting HD-tDCS models and the optimized current flow intensities on cortical targets.
Main results
The segmentation tool can segment out not just the brain but also provide accurate results for CSF, skull and other soft tissues with a field of view (FOV) extending to the neck. Compared to manual results, automated segmentation deviates by only 7% and 18% for normal and stroke subjects, respectively. The predicted electric fields in the brain deviate by 12% and 29% respectively, which is well within the variability observed for various modeling choices. Finally, optimized current flow intensities on cortical targets do not differ significantly.
Significance
Fully automated individualized modeling may now be feasible for large-sample EEG research studies and tDCS clinical trials.
1. Introduction
High-definition transcranial direct current stimulation (HD-tDCS) requires accurate models of current flow for precise targeting (Datta et al. 2009, Mendonca et al. 2011, Datta et al. 2012, Caparelli-Daquer et al. 2012, Minhas et al. 2012). The same could be said about current source reconstruction in high-density electroencephalography (HD-EEG) (Vatta et al. 2010, Gllmar et al. 2010, Akalin Acar amp; Makeig 2013). At the level of accuracy yielded by modern modeling techniques, generic head models are no longer useful: cortical folding is highly idiosyncratic and drastically affects the resulting current flow (Datta et al. 2009, Datta et al. 2012). Secondly, the exact distribution of cerebrospinal fluid (CSF) in the space between brain and skull can significantly affect current flow in EEG (Rice et al. 2013). Finally, while the highly resistive skull is the main attenuating barrier for tDCS and EEG currents, and its thickness is quite variable within and across subjects, the current flow modeling can be greatly affected if the model does not accurately capture the exact skull geometry (Datta et al. 2010, Lanfer et al. 2012). Therefore, one can argue that models that do not include the idiosyncratic anatomy of the brain, CSF and skull for each individual subject cannot provide meaningful localization of tDCS hot-spots or EEG current sources.
The first step to build anatomically accurate head models is the segmentation of structural magnetic resonance images (MR images, MRI). There are a variety of software tools already available for automatic segmentation of the brain and head. Unfortunately, these are either restricted to a limited field of view (FOV) excluding the neck, or they fail to provide sufficient accuracy for CSF, skull and soft tissues (see Section 3.4 for detailed discussion on existing tools). Both, the extended field of view, as well as accurate segmentation of non-brain tissues, are crucially important for correct current flow modeling. Therefore, additional manual segmentation is needed subsequent to any of the currently available automated segmentation procedures. At the desired resolution of 1 mm3 this can be a labor-intensive process typically requiring at least 1 week of manual effort for a 6-tissue segmentation (gray matter, white matter, CSF, skull, scalp and air cavities) as we report here. Additionally, manually placing tens to hundreds of electrodes onto these models can be a tedious process, though some techniques have already been proposed ((Oostenveld et al. 2011), see Section 2.4).
The goal of this work is to fully automate the generation of the required segmentation masks including high-definition electrode placement. We combined an existing fully-automated segmentation tool (Unified Segmentation (Ashburner & Friston 2005), as implemented in the Statical Parametric Mapping software, version 8‡) with a unique tissue probability map (TPM) developed at the Center for Advanced Brain Imaging at Georgia State University. To this we added a routine that corrects obvious morphological errors of the segmentation and developed an automated electrode placement tool. Among the many available MRI segmentation software, we chose New Segment and the mentioned TPM because this combination is unique in that it includes not only gray and white matter, but also CSF, skull, soft tissues, and air cavities, with an FOV extended to the neck. Moreover, it is fully automated and performs better than other available tools (see discussion in Section 3.4).
We compared the deviation between our fully-automated and manual segmentations for 4 healthy individuals and 7 stroke patients. As a reference we also provide results for the SPM8 segmentation routine. We also evaluated the resulting current flow models in the context of HD-tDCS again comparing manual segmentation with the automated technique. We evaluated the electric field throughout the head using standard electrode montages, and the field intensity achieved at cortical targets when employing a recently developed optimized targeting technique (Dmochowski et al. 2011). We make all tools freely available such that additional merits or limitations of the segmentation and electrode placement routines may be evaluated.
2. Methods
2.1. MRI acquisition and pre-processing
MRI scans of the head were performed on four healthy subjects (three males, average age 38 years, range 33–45 years). Head 1 was scanned on a 3T Siemens Trio scanner (Erlangen, Germany). The T1-weighted images were collected using a gradient echo (GRE) sequence with TE = 2.3 ms, TR = 1900 ms, 280 × 320 matrix scan with 208 sagittal slices. Head 2 and Head 3 were scanned on a 3T General Electric Signa Excite HD scanner (Fairfield, CT). The T1-weighted images were acquired using a GRE sequence with TE = 2.2 ms, TR = 7.3 ms, 256 × 256 matrix scan with 212 axial slices for Head 2, and with 252 axial slices for Head 3. Head 4 was also scanned on a 3T Siemens Trio scanner. The T1-weighted images were obtained using a GRE sequence with TE = 4.2 ms, TR = 2250 ms, 256 × 256 matrix scan with 176 sagittal slices. All images had an isotropic resolution of 1 mm3.
To test the proposed automated routine on pathological heads, we also obtained MRI scans on seven stroke patients at the McCausland Center for Brain Imaging at University of South Carolina. The seven patients (three males, average age 60 years, range 48–75 years) all had lesions in the left hemisphere. MRI scanning relied on a 3T Siemens Trio system. Both T1- and T2-weighted images were collected for improved segmentation of the lesions. T1 images utilized a turbo field echo sequence (MP-RAGE) with TE = 5.7 ms, TR = 9.5 ms, 256 × 256 matrix scan with 160 sagittal slices. T2 images utilized a SPACE (Sampling Perfection with Application optimized Contrasts by using different flip angle Evolutions) protocol with TE = 352 ms, TR = 3200 ms, 256 × 256 matrix scan with 160 sagittal slices. All images had an isotropic resolution of 1 mm3.
The raw MR images were saved in DICOM (Digital Imaging and Communications in Medicine) format and converted to NIfTI (Neuroimaging Informatics Technology Initiative) format (.nii, .hdr and .img) using MRIcro (version 1.40, Center for Advanced Brain Imaging (CABI), Georgia State University and Georgia Institute of Technology, Atlanta, GA). The images were then bias corrected in Statistical Parametric Mapping 8 (SPM8, Wellcome Trust Centre for Neuroimaging, London, UK).
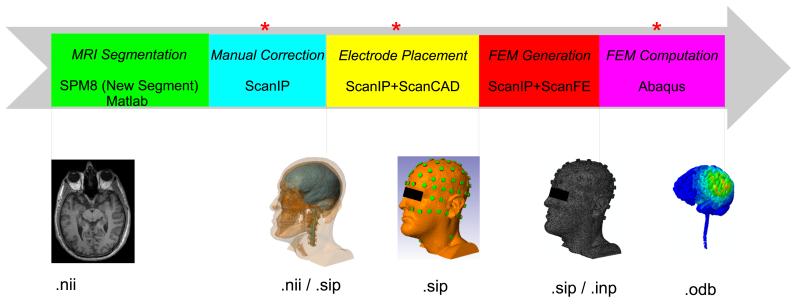
The traditional workflow for individualized HD-tDCS modeling is shown in Figure 1. We will elaborate on each step in the following subsections where we describe our automated procedure.
Figure 1.
Traditional workflow for individualized HD-tDCS modeling. The manual steps we attempted to automate are indicated by red asterisks. Data formats of our current workflow are indicated below the images. (FEM: finite element model)
2.2. Intensity-based automatic MRI segmentation
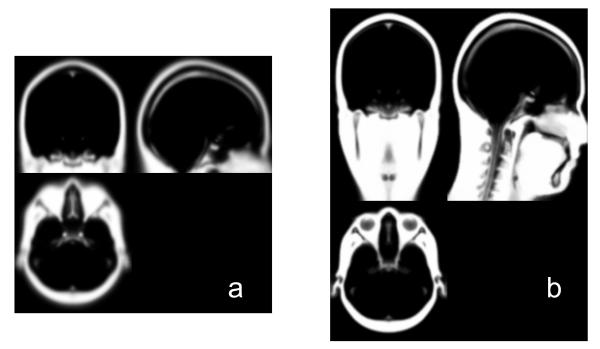
The pre-processed MR images (T1 only for healthy subjects, T1 and T2 for stroke heads) were segmented by a probabilistic segmentation routine (New Segment, an extension of Unified Segmentation (Ashburner & Friston 2005), and a function in SPM8). The New Segment routine estimates the posterior probability of each image voxel belonging to a specific tissue type based on the intensity of each voxel and a prior probability distribution of different tissue types (i.e., the tissue probability map, TPM). The default TPM provided by SPM8 for New Segment has six tissue types: gray matter (GM), white matter (WM), CSF, skull, scalp and air (the air cavities§). However, its field of view (FOV) only contains the brain area (Figure 2a). Here we used an improved TPM developed at CABI. It has an extended FOV including the neck (Figure 2b). Using New Segment with the improved TPM, the head was also segmented into six tissues: GM, WM (including the brain stem), CSF (including surface CSF, CSF inside ventricles, and the eyeballs), skull, scalp (soft tissue) and air. Note that for stroke heads, the lesions are automatically classified as CSF by the TPM due to their similar intensity distribution. It takes approximately 20 minutes∥ to segment an MR image with a typical size of 256 mm × 256 mm × 160 mm and 1 mm3 resolution.
Figure 2.
Scalp TPM employed by the New Segment routine. (a) the default TPM in SPM8; (b) the improved TPM developed at CABI.
2.3. Segmentation errors and the automated correction routine
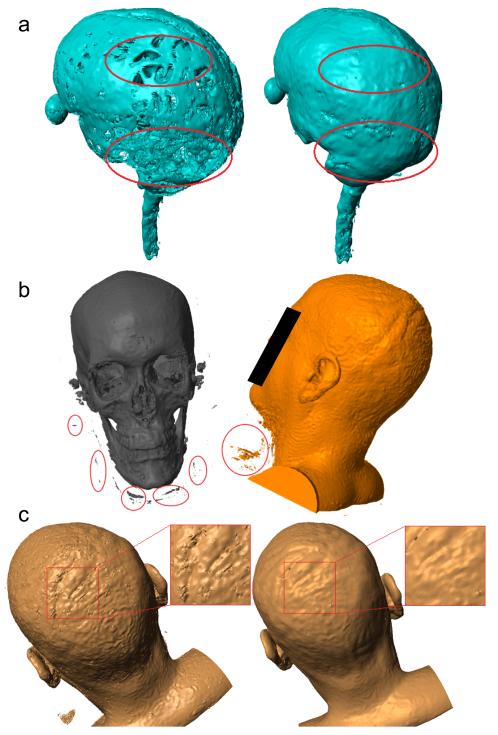
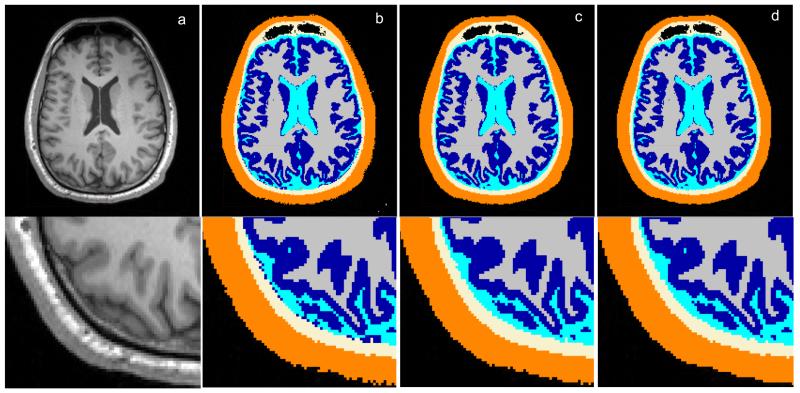
The output from New Segment is a probability distribution indicating how likely each image voxel belongs to a specific tissue type. For subsequent finite element modeling (FEM) of the current flow inside the head, binary tissue masks are needed. Therefore, these continuous-valued probabilities were converted into discrete assignments by selecting for each voxel the tissue with the highest probability. This resulted in 6 non-overlapping masks, one for each tissue. The masks are of high quality (Figure 3) but have a number of minor errors that result primarily from image noise and low contrast in some areas. These errors can be categorized into the following four types:
Discontinuities in the CSF: the surface CSF should be a continuous layer of fluid surrounding the brain and spinal cord. However, the CSF layer is thin in some areas, and may be mislabeled due to limited MRI resolution (1 mm3), leading to mask discontinuities (Figure 3a).
“Disconnected” voxels: disjoint voxels of one tissue type, erroneously within another tissue typically resulting from background noise (Figure 3b).
Unassigned voxels: after segmentation with SPM8 some voxels are assigned a zero probability of belonging to all tissue types¶.
Rough tissue masks: noise in the original MR images can lead to rough surfaces, e.g. scalp mask shown in Figure 3c, which are less smooth than expected based on normal anatomy.
Figure 3.
Examples showing errors in the binarized segmentations from SPM8 and the improvements after automated corrections, as indicated by red circles/squares. (a) discontinuities in CSF; (b) “disconnected” voxels; (c) rough tissue surface.
CSF discontinuities are of particular importance for current flow modeling as the high conductivity of CSF is erroneously displaced in those locations, leading to significant changes in predicted current flow inside the brain. The remaining errors could cause convergence problems when generating the FEM in the meshing software. Fortunately, these errors can be readily identified during visual inspection based on the original MRI and thus they are traditionally corrected by hand. This manual correction of segmentation errors can produce masks with very high anatomical accuracy. However, the process is very labor intensive (typically 1–3 weeks for one head depending on the image quality) and requires a familiarity with anatomical MRI. Therefore, it would be beneficial to develop an automated correction technique.
The proposed automated correction routine is based on morphological and Boolean operations, and was implemented in Matlab+ (R2010b, MathWorks, Natick, MA). The details of the procedure are as follows (there are three free parameters that will be listed subsequently):
Smoothing of tissue masks: to avoid complications in the FEM generation, the resulting tissue probabilities from New Segment were smoothed before conversion into binary masks using a Gaussian low-pass filter with s mm standard deviation.
Discontinuities in the CSF: to fill potential CSF discontinuities, we reclassified all GM voxels that are adjacent to the skull, interpreting them instead as CSF with the following operation: CSF ← dilate(skull, x mm) ∩ GM (← and ∩ are assignment and set intersection, respectively). We opted to reassign GM (as opposed to the skull), as intuition suggests that this will perturb current flow less severely, because the skull is a thinner layer compared to GM, lending itself less well to displacement (Datta et al. 2010). Moreover, WM and GM have similar electrical conductivity (Section 2.5). Therefore, ensuring continuities in skull is more important than the integrity of GM.
“Disconnected” voxels: the connected component * for each mask was determined and all components with less than N voxels were categorized as unassigned voxels.
Unassigned voxels: each of the resulting unassigned voxels was assigned the tissue type which is nearest to its location. This was implemented by processing all tissue masks using a Gaussian low-pass filter with 1 mm standard deviation. For a given unassigned voxel, the smoothed tissue mask with the highest value at that voxel serves to identify the category of that voxel.
The three free parameters involved in this automated processing are:
Smoothing length constant s: a higher value will make the masks smoother, and thus guarantee the convergence as well as reduce the size in the following FEM modeling. Aggressive smoothing, however, compromises anatomical details, and thus, segmentation accuracy. Typical values for s are: GM–0.2, WM–0.1, CSF–0.1, skull–0.4, scalp–1, air–1.
Size (x) of the structural element to fill in CSF discontinuities: the trade-off in selecting x is between filling all the discontinuities (higher value) and preserving the integrity of GM (lower value). We set x = 3 in all instances.
Size threshold (N) for identification of disconnected voxels: The trade-off here is comparable to choosing the smoothing length constant. Typical values for N are: GM–30, WM–20, CSF-the size of the fourth largest component (thus all components smaller than the eye balls are categorized as unassigned voxels), skull– 300, scalp–the size of the second largest component (thus all components smaller than the scalp itself are classified as unassigned voxels), air–20.
This routine takes approximately 2–4 minutes to correct the segmentation errors, representing a significant reduction of manual labor. We will discuss its performance in Section 2.6.
2.4. Electrode placement
In order to simulate the current flow in the brain, gel-based electrodes need to be virtually fitted onto the scalp. Previously, this was done manually using ScanIP (version 4.2, Simpleware Ltd, Exeter, UK) with the ScanCAD Module, which typically requires hours to place only few electrodes (4 × 1 configuration) (Datta et al. 2009), motivating the availability of an automated electrode placement tool. Although free electrode placement tools are available (e.g., FieldTrip (Oostenveld et al. 2011), which allows for alignment to template coordinates), they cannot automatically fit the electrodes exactly on the local scalp surface, barring manual adjustment of the interactive interface. Moreover, no function module is provided to model the fitted electrodes as small cylindrical disks. Therefore, we developed an automated algorithm for electrode placement which can place and model multiple electrodes following a standard montage in several minutes (some ideas adapted from (Koessler et al. 2008)).
The montage of electrodes follows the convention of the standard 10–10 international system (Klem et al. 1999). Pre-defined coordinates of 74 electrodes from EasyCap # were used. Any other standard configuration available in a similar format can also be used for electrode placement. An additional row of electrodes was placed to potentially allow stimulation of deeper or lower-lying cortical targets. As Head 1 has a large FOV that covers the entire neck, we also placed four additional electrodes around the neck. To avoid complications in automatically placing electrodes near or behind the ear-lobes, we omitted positions TP9 and TP10 in all heads. Thus, we placed 93 electrodes on the scalp mask for Head 1 (72 in the original 10–10 system + 17 in additional row + 4 on the neck), and 89 electrodes for each of other heads.
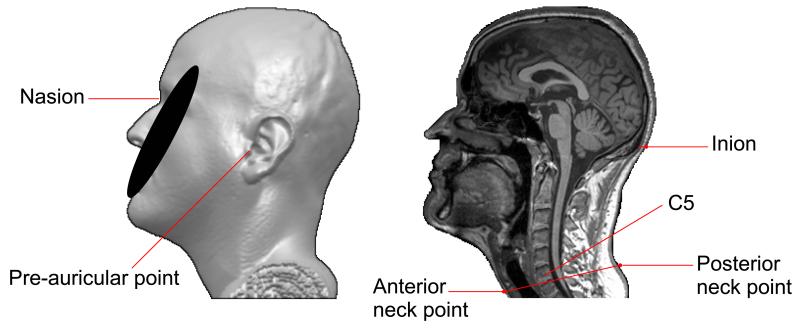
Electrode placement was implemented in Matlab††, based on the commonly used anatomical landmarks: nasion (Nz), inion (Iz), pre-auricular right (PAR) and pre-auricular left (PAL). For the neck electrodes, two additional points are needed: one in the middle neck anterior, with the other located in the center of the neck posterior. The two neck points should have similar z–coordinates as the intervertebral disc between cervical vertebrae C5 and C6 (Figure 4). These fiducial points are provided by the user by visual inspection of the MRI (i.e., using MRIcro 3D viewer).
Figure 4.
The anatomical landmarks provided by the user to initialize electrode placement.
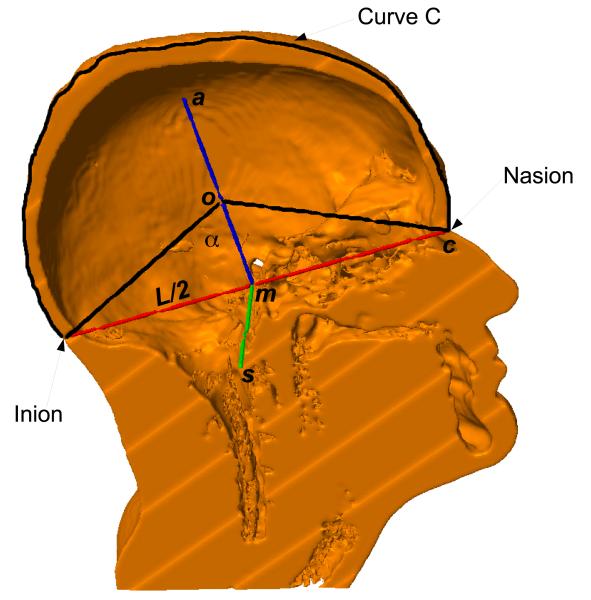
The line connecting from PAL to PAR defines a unit vector s. Similarly, the line from Iz to Nz gives a unit vector c. The cross product of s and c generates a third unit vector a = s × c. Assuming a spherical head, the origin o of the coordinate system can be obtained by
| (1) |
where m and L are the midpoint and distance between Nz and Iz, respectively, and α is an angle computed from the pre-defined spherical electrode coordinates, as shown in Figure 5. These three vectors, together with the origin, define an affine transformation which is used to transform the pre-defined 10–10 electrode coordinate system to the RAS (right–anterior–superior) coordinate system of the MRI volume. The transformed electrode coordinates are then projected from the origin o onto the scalp surface.
Figure 5.
The three vectors s, c, a, and the origin o of the coordinate system of electrode placement. m is the midpoint of Nz and Iz, and L is the distance between Nz and Iz. The sagittal plane normal to s and passing through o intersects the scalp, generating the curve C (the black tracing).
The projected locations for the electrodes on the scalp cannot be equidistant from one another unless the head is a sphere. To make them as equidistant as possible for any individual head, we slightly adjusted the origin o as follows: A sagittal plane perpendicular to s and passing through the origin o intersects with the scalp surface (Figure 5). This intersection defines a curve C that, according to the 10–10 system, is to be divided in 10 equal intervals between Nz and Iz. These are the intervals, di, i = 1, 2, … 10, between adjacent electrodes on the middle line: Iz, Oz, POz, Pz, CPz, Cz, FCz, Fz, AFz, Fpz, Nz. These intervals are equidistant and span the length of curve C (denoted by DC). The origin o was adjusted along the direction of vector a to minimize the mismatch from this idealized definition:
| (2) |
With the optimized origin o, all electrode coordinates defined on a unit sphere are affine transformed by T and projected from the origin o onto the scalp surface to obtain the final 3D coordinates for the electrodes.
For the neck, we calculated the midpoint between the two neck points as provided by the user. From this midpoint, projections onto the scalp surface in the anterior, posterior, left and right directions give the four additional electrode locations on the neck.
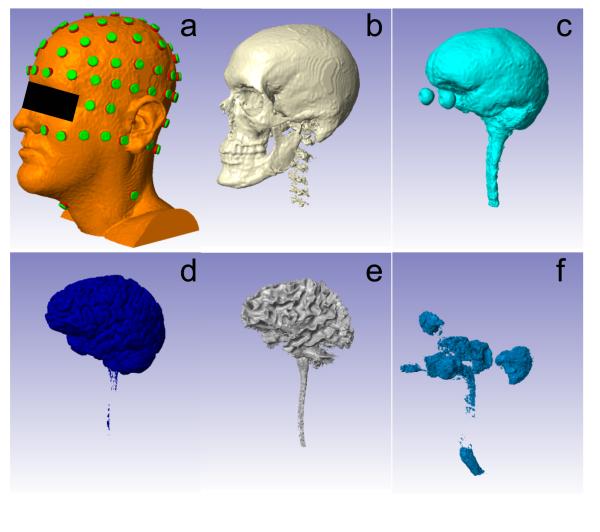
All electrodes were modeled as cylindrical disks of 2 mm height and 6 mm radius parallel to the local scalp surface (Figure 6a). A layer of conducting gel with the same radius as the electrode was inserted into the 1–2 mm gap between scalp and electrode. After electrode placement, electrodes and gel were exported as binary masks.
Figure 6.
Head model established from six tissue types and electrodes. From (a) to (f): scalp (with electrodes placed), skull, CSF, GM, WM, air. Note that the air outside the head was removed in the air mask since it is not needed to model transcranial current flow.
2.5. Finite element model generation and computation
The six tissue masks, along with the placed electrodes, constitute a volume conduction model for the whole head (Figure 6). To simulate the current flow distribution inside the head, the Laplace’s Equation (−▽.(σ▽V) = 0, V: electric potential distribution in the volume; σ: tissue conductivity) needs to be solved (Griffiths 1999, Datta et al. 2009). The conductivity values used for each tissue type are as follows (in S/m): GM: 0.276; WM: 0.126; CSF: 1.65; skull: 0.01; scalp: 0.465; air: 2.5 × 10−14; gel: 0.3; electrode: 5.9 × 107 (Wagner et al. 2007, Datta et al. 2009). For simplicity, we did not model anisotropic conductivity, see Section 3.4.
For arbitrary shaped media, i.e., arbitrary distributions of σ, no closed form solution exists and one typically resorts to numerical techniques. Therefore, the volume is discretized into a set of finite elements (FE), each with uniform conductivity, and Laplace’s Equation is solved at all discrete elements (Logan 2007). We used ScanIP (+ScanFE Module) to generate the FEM with tetrahedral elements. Given the size and resolution of the model (1 mm3), it is necessary to use adaptive meshing (ScanFE-Free algorithm), which chooses the size of each element adaptively based on the required detail in the structure. This provides a more compact mesh size without compromising computational accuracy. In this study, the FEM we generated consists of approximately 7 million tetrahedral elements, with a generation time of 1–2 hours.
Abaqus (version 6.9, SIMULIA, Providence, RI) was used to solve Laplace’s Equation on the FEM. It takes approximately 1–1.5 hours to solve the FEM for a specific bipolar electrode configuration (e.g., Fp1–Iz, see Section 2.6). The solution is the distribution of electric field E induced by stimulation with a current density of 1 A/m2. This solution is the output of the workflow for individualized HD-tDCS modeling.
To perform optimized steering of the applied current (Dmochowski et al. 2011), one needs to generate the electric field distribution for all bipolar electrode configurations (with one fixed reference electrode). We generated a Matlab script to run FEM simulations in Abaqus automatically for all the bipolar configurations (i.e., 92 configurations for Head 1 and 88 configurations for each of other heads).
2.6. Evaluation of the automated routine
After automatically correcting segmentation errors, placing electrodes and solving the FEM, the time needed to construct a high-resolution (1 mm3) model of HD-tDCS for a specific electrode configuration is reduced from weeks to several hours, with the majority of this time consumed by the FEM generation (1–2 hours) and solving (1–1.5 hours). The majority of the gain stems from the automation of segmentation correction. However, significant human expertise in establishing tissue boundaries is lost in this automation. This is important in particular for chronic stroke subjects whose anatomies deviate significantly from normal anatomy. To assess how much the automated correction affects the results, we also constructed manually corrected models for all heads (performed using ScanIP). We evaluated the differences by comparing both the segmentation results and the predicted electric field distributions from these two methods. For the segmentation, we calculated the deviation of automated results from manual results as the rate of incongruent voxel assignments (number of missing or extraneous voxels compared to manual results, Nerr) relative to the number of total voxels (Ntot) in a given manual mask:
| (3) |
dseg was computed for all tissue types.
To compare the electric fields, we implemented the bipolar configuration and also simulated a conventional large pad electrode on the generated FEM. In the bipolar configuration, we chose Iz as the cathode, and selected the anode from three different regions on the scalp: forehead (Fp1), motor area (C3) and occipital area (O1). We also simulated a distant reference in Head 1: Cz as the anode and the frontal neck electrode (Nk1) as the cathode. In all other heads, since no electrode was placed on the neck, electrode pair FT9–FT10 was solved as the simulation of distant reference. In the pad simulation, the contralateral M1-SO configuration was solved by simulating an anode pad placed on right primary motor cortex (M1, modeled here by a cluster of 9 electrodes: FC2, FC4, FC6, C2, C4, C6, CP2, CP4, CP6), and a corresponding cathode in the left supra-orbital (SO) area (modeled by a cluster of 8 electrodes: Fp1, AF3, AF7, F1, F3, F5, F7, FC3). The electric field computed from Abaqus is defined at all mesh nodes. For the ease of comparing the fields from the two methods, the solutions on this irregular mesh grid were imported into Matlab, interpolated onto a regular grid with the same dimension and resolution (i.e., 1 mm3) as the original MRI data, and recalibrated to correspond to a 1 mA current injection. We then calculated the deviation of automated results from the results obtained with the manual segmentation. Denoting the automatically and manually obtained electric fields by Ea and Em, respectively, the deviation follows as:
| (4) |
dfield was calculated for each voxel (see Figure 9 (the third row) for example), and also for each tissue type (the difference ∥Ea–Em∥ averaged across voxels in that tissue first, then divided by the average ∥Em∥.
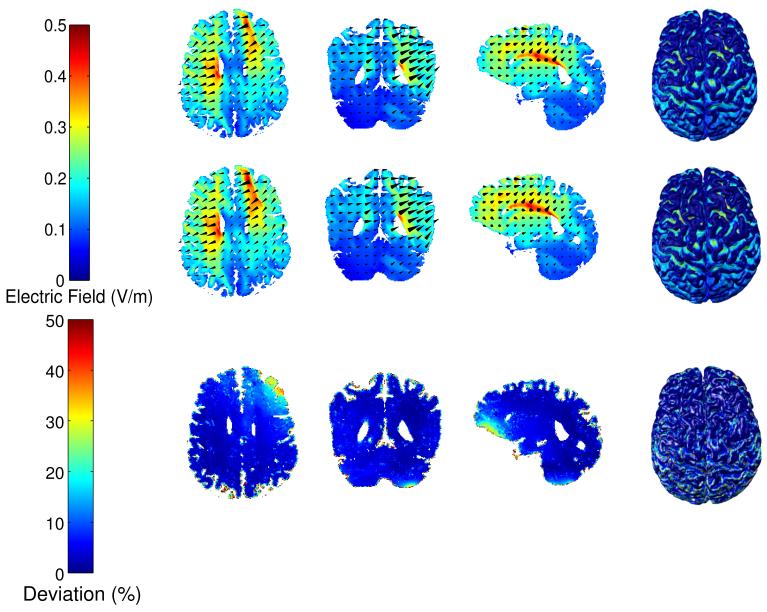
Figure 9.
The distributions of electric field within GM and WM in Head 1 for the manual (first row) and automated method (second row) under M1–SO configuration, and the deviation of automated results from manual results (third row). The four columns are the axial, coronal, sagittal slices, and the surface plot, respectively.
Since clinicians are particularly interested in the areas of peak activation (presumably corresponding to desired neurophysiological effects), we also compared the spatial patterns of these peaks between manual and automated results (in GM and WM only). The peak area was defined as those voxels in the brain with electric field magnitude in the upper quartile, and the Jaccard index (Jaccard 1901) was used to compare the similarity of the peak areas from manual and automated results. It is given by
| (5) |
where Pm, Pa are the peak masks from manual and automated results, respectively. A Jaccard index close to 1 indicates higher similarity while an index close to 0 denotes low similarity. It was evaluated on each subject for each electrode configuration.
The final purpose of current flow modeling is to select a specific electrode configuration for treatment. From a practical point of view, two modeling techniques can be considered equivalent if they lead to similar electrode configurations and current flow intensities at the target. Here we are interested in the optimal configuration of multiple high-definition electrodes for a realistic treatment scenario. We therefore use the optimization technique recently developed for a clinical trial to treat aphasia in chronic stroke (Dmochowski et al. 2013). Optimal electrodes were computed for the automated and manual segmentation results for all 7 stroke patients. The targets are at eloquent peri-lesional cortical areas (Figure 11), and were determined from functional MRI (Baker et al. 2010). An optimal electrode montage for each subject was calculated by maximizing the electric field intensity at the target while limiting total current to 2 mA with no more than 1 mA in each electrode (to ensure safety and comfort). This approach leads in all instances to a pair of positive and a pair of negative polarity electrodes (Dmochowski et al. 2013).
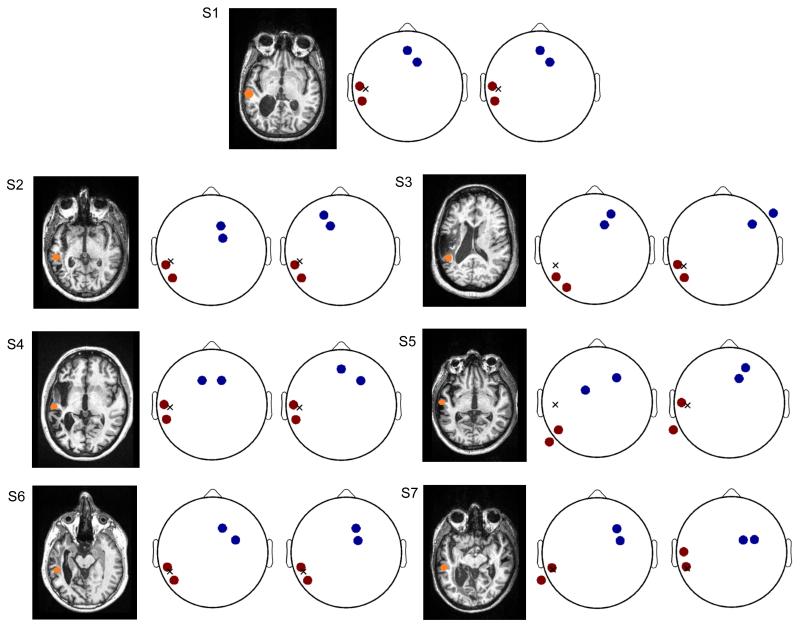
Figure 11.
The optimized electrode montages maximizing the electric field intensity at the target for all stroke subjects (S1–S7). Left: Axial view of the anatomical MRI showing the lesion anatomy and the target at peri-lesional area (orange spot). Middle: the optimal electrode montage obtained from manual modeling process. Right: the optimal montage from automated modeling method. The anodes and cathodes are marked in red and blue, respectively, and the target projected on the scalp is represented by an “X”.
3. Results and Discussion
3.1. MRI segmentation performance
Automatic segmentation was performed on MR images from 11 subjects (4 healthy and 7 stroke), as described in Section 2.2. Briefly, the SPM8 New Segment routine was used in combination with an improved TPM with an extended FOV. Subsequently, the proposed automated correction routine was applied to correct the errors in segmentation results (see Section 2.3). To establish ground-truth segmentation, we also manually corrected these errors using ScanIP. Figure 7 shows one slice of the segmentation results in Head 1 after different methods. We then calculated the deviation of the automated segmentation from these manually corrected segmentations (see Eq. 3). The deviation dseg is shown in Figure 8 for various tissue types, comparing results for different procedures and across subjects. Figure 8 also evaluates the effect of smoothing, as we wanted to know whether smoothing – which is required for convergence when generating the FEM in ScanIP – would significantly deteriorate the automated segmentation results.
Figure 7.
Tissue segmentations for Head 1. (a) Raw MRI slice; (b) Binarized results from SPM8; (c) Results after automated correction; (d) Results after manual correction.
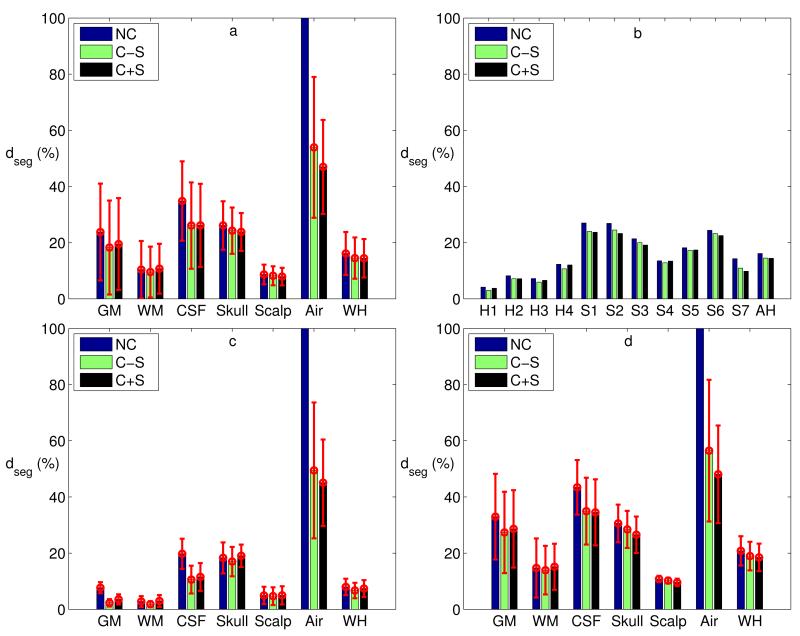
Figure 8.
(a) Segmentation deviations averaged across all subjects for each tissue type and the whole head (WH) (red error bars indicate standard deviations across subjects). (b) Segmentation deviations computed across the whole head for each subject’s head (healthy heads: H1–H4, stroke heads: S1–S7) and averaged across all heads (AH). (c & d) Segmentation deviations averaged across healthy (c) and stroke (d) subjects for each tissue type and the whole head. (NC: no correction, C−S: correction without smoothing, C+S: correction with smoothing.)
As shown in Figure 8a, after automated correction (without smoothing), the segmentation deviation of the whole head averaged across all subjects is 15%. While the deviations of CSF, skull and air are somewhat higher (26%, 24% and 54% respectively), the overall deviations are dominated by the large volume of GM, WM and scalp, which have lower deviations (18%, 10% and 8% respectively). For each tissue type and the whole head (WH), we conducted a repeated measures Analysis of Variance (ANOVA) with the three conditions (no correction [NC], correction without smoothing [C−S], correction with smoothing [C+S]). The results suggest that the automated correction technique significantly improved segmentation performance for all tissue types except WM (GM: F(2, 20) = 19.19, p = 0.000022, CSF: F(2, 20) = 145.32, p = 1.22 × 10−12, Skull: F(2, 20) = 6.39, p = 0.0072, Scalp: F(2, 20) = 5.68, p = 0.011, Air: F(2, 20) = 54.50, p = 8.02 × 10−9, WH: F(2, 20) = 16.03, p = 0.00007, and WM: F(2, 20) = 0.79, p = 0.47). Indeed, manual correction of WM was performed conservatively (i.e., only changed as a result of changes to other tissues). Figure 8b shows the segmentation deviations computed across the whole head for each subject (H1–H4: healthy subjects, S1–S7: stroke subjects) and the average across all heads (AH). A pairwise t-test shows that the correction procedure improves segmentation performance (C-S vs. NC: t(10) = 6.12, p = 0.00011, and C+S vs. NC: t(10) = 3.71, p = 0.0040), and that smoothing – required only for the FEM generation – does not significantly worsen that result (C−S vs. C+S: t(10) = 0.24, p = 0.81). Also, it is clear from Figure 8b that the segmentation performance on the stroke subjects is degraded (7% for healthy, 18% for stroke heads) as a result of the atypical lesion anatomy. Note that in the stroke cases we have the benefit of using T1 as well as T2 images. Without T2, the performance would have been further degraded as CSF and bone have similar intensities in T1 alone. Figure 8c and 8d show the tissue-specific results separately for the group of healthy and stroke heads.
3.2. Current flow modeling results
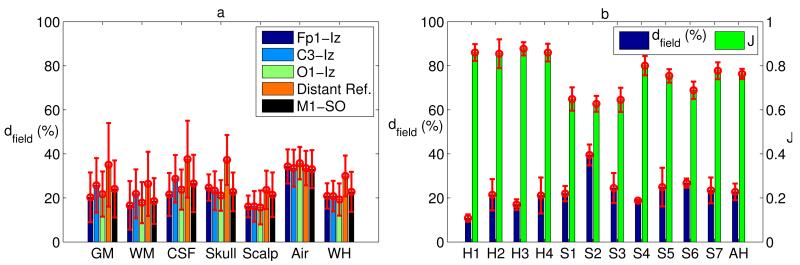
An automated electrode placement program was developed to place all high-definition electrodes on the scalp, as described in Section 2.4. Finite element models were then constructed from both the automatically and manually obtained tissue masks, plus electrodes and gel (Section 2.5). We simulated various electrode configurations with 1 mA current injection (see Section 2.6). As an example, Figure 9 shows the distributions of electric fields from both methods in GM and WM for Head 1 under the M1–SO configuration. Figure 10 shows the deviation of electric field in the automated masks from the results computed with the manually corrected masks (see Eq. 4). Figure 10a is the electric field deviations averaged across all subjects, with the red error bars representing variations among subjects. The average deviation across the entire head for all subjects and configurations is 23% (18% for 4 healthy heads and 26% for 7 stroke heads), and the average deviation in the brain (GM and WM only) is also 23% (12% for healthy and 29% for stroke heads). These values are higher than the overall deviation of segmentation results, indicating that the current flow is sensitive to small structural differences. Figure 10b (blue bars) shows the electric field deviations computed across the whole head for each subject and the average across all heads, with the red error bars indicating variations across different electrode configurations. Note that Head 1 has superior performance due to its high image quality, which will be discussed in detail in Section 3.4. From a clinical point of view, perhaps more important than the overall field intensities are the locations of maximal stimulation. Electrode locations for transcranial stimulation are selected such that “hot-spots” of stimulation coincide with the targeted brain area (Datta et al. 2009, Dmochowski et al. 2011). We determined the voxels in the upper quartile of the induced electric field magnitude as the peak stimulation, and measured how it overlaps with that of manual segmentation by using the Jaccard index (see Eq. 5). Figure 10b (green bars) shows the Jaccard index between the peak patterns in the brain from manual and automated results. An averaged Jaccard value of 0.76 was obtained when averaging over all the heads and all electrode configurations, meaning that 76% of areas in the brain being stimulated remained the same.
Figure 10.
(a) Deviations of electric field averaged across all subjects for each tissue type and the whole head (WH) (red error bars indicate standard deviations across subjects). (b) (Blue) Deviations of electric field computed across the whole head for each subject’s head (H1–H4: healthy heads, S1–S7: stroke heads) and the average across all heads (AH). (Green) Change in locations of the peak electric fields in the brain (top 25%) comparing manual and automated results (Jaccard index = 1 means perfect overlap, no change). The red error bars represent standard deviations across different electrode configurations. (Distant Ref.: Cz–Nk1 in H1 and FT9–FT10 in all other heads.)
3.3. Results of optimized targeting
The optimal electrode configuration for each stroke subject is shown in Figure 11. These configurations maximize the electric field intensity at the target (Dmochowski et al. 2013). The target location is indicated as an orange dot in the axial MR image. Electrode montages are depicted schematically (Delorme & Makeig 2004) for the two approaches with anodes and cathodes in red and blue, respectively. An approximate location of the target projected to the scalp is represented by an “X”. For each subject, the results obtained from manual and automated methods are shown in the middle and right plots, respectively.
The optimal configuration is evidently target- and/or subject-dependent due to the individuality in head and lesion anatomy. Given this cross-subject variability, the manually and automatically obtained electrode montages are quite similar for some subjects (the two montages are exactly the same for S1, and same anodes are selected for S2, S4 and S6). The distance between the centroids of anodes in the manual and automated results is 1.3 ± 1.8 cm, and 3.6 ± 3.0 cm for the cathodes. More importantly, when the two montages are applied on the manually obtained head model for each subject, the intensity of the optimized electric field at the target is 0.63 ± 0.16 V/m under the manually obtained montage, and 0.58 ± 0.20 V/m under the automatically obtained montage. A pairwise t-test shows that there is no significant difference between the intensities of electric field at the target achieved from these two techniques (t(6) = 1.64, p = 0.15). Therefore, in terms of electrode montages for optimal current flow targeting, the prescriptions provided by our automated routine can achieve an electric field intensity at the target that is not substantially different from that induced by the montages used in the actual trial (based on careful manual segmentation) (Dmochowski et al. 2013).
3.4. Discussion
Accurate current flow models for HD-tDCS and forward modeling for HD-EEG require detailed anatomical models of the human head as well as placement of a large number of electrodes. Recent modeling work on HD-tDCS (Dmochowski et al. 2012) suggests that individualized modeling can improve stimulation intensity on target by an average of 50% with the same total current delivered. EEG source reconstruction can vary by as much as 66% when a generic standard head is used instead of individualized head models (Vatta et al. 2010). Obtaining models that cover the required tissue types across the entire head (not just brain) relies at present on tedious manual correction of an initial automated segmentation (at least one week of effort to correct an automated result at 1 mm3 resolution). This precludes the widespread adoption of individualized current flow models. In addition, manually placing electrodes and solving the FEM model for a large number of electrode configurations is a time-consuming process.
The tools presented here, which we make freely available, reduce this effort from weeks to hours. The goal of this work is to present these tools and thoroughly evaluate them in terms of segmentation accuracy and the impact this has on current flow models for normal as well as lesioned brain anatomies. The evaluation focused on segmentation performance and current flow results in the context of HD-tDCS. For subjects with normal anatomy, segmentation deviates only by 7%, with that number increasing to 18% for stroke patients with chronic lesions. The predicted electric fields on the brain deviate by 12% and 29% respectively, which is within the spread observed for different modeling choices (see below). Finally, optimized current flow intensities on cortical targets do not differ significantly (see Section 3.3).
There are many open-source packages and commercial software tools for automatic MRI segmentation other than SPM8, e.g., FMRIB Software Library (FSL, (Smith et al. 2004)), FieldTrip (Oostenveld et al. 2011), FreeSurfer (Dale et al. 1999, Fischl et al. 1999), ITK-SNAP (Yushkevich et al. 2006), NFT (Acar & Makeig 2010), ASA (ANT Software BV, Enschede, Netherlands), Curry (Compumedics NeuroScan, Charlotte, NC), BESA (BESA GmbH, Gräfelfing, Germany) and Atropos based on ANTs (Avants et al. 2011). A one-to-one performance comparison with these other segmentation tools was not the purpose of this work. We selected SPM8 primarily for practical considerations: in our hands, none of these other tools were able to automatically segment all the six tissues in the extended FOV as effectively as SPM8 New Segment. FSL Brain Extraction Tool (BET,(Smith 2002)) and FieldTrip can extract the skull and scalp surfaces, but it only operates on the standard FOV (the brain area only). FreeSurfer appears slow in segmentation (22 hours for one head, see (Windhoff et al. 2011)). ITK-SNAP and NFT need user-specified seed point(s) to start segmentation and thus they are not fully automated. ASA and Curry do not provide segmentation for CSF, which is crucial for tDCS and EEG forward modeling. BESA can segment out CSF, but the result is not accurate (with discontinuities). Atropos is a good software for segmentation, but our prior probability image (TPM) was developed under SPM8. Therefore, combined with the improved TPM and the automated correction routine, SPM8 is our preferred choice for automated segmentation.
Automated pipelines exist for tDCS current flow modeling which can be combined with our tools at the front-end of these pipelines (Windhoff et al. 2011, Dannhauer et al. 2012). This may be useful in particular if one wishes to circumvent proprietary softwares (ScanIP and Abaqus) and use free tools such as iso2mesh (Fang & Boas 2009) and GetFEM++ (Renard & Pommier 2010) for meshing and FEM solving. The results provided here are equally applicable to any processing pipeline with our tools at the front end as meshing and FEM solving should give similar, if not identical, solutions. In fact, we have compared our results with iso2mesh and COMSOL (COMSOL Inc., MA) at various mesh resolutions and deviations of results are in the range of 2–4%, which is generally acceptable tolerance for FEM modeling.
We have not numerically evaluated the accuracy of electrode placement in this work. Given the diversity of head sizes and shapes as well as electrode cap sizes, it is a standard practice to adjust the flexible caps to assure a good match of actual positions with the desired locations. Whenever possible one should use commercially available electrode positioning systems to guide this adjustment of the electrode cap (e.g. Polhemus tracker). For this purpose, our software provides the exact 3D coordinates of the electrodes in the original MRI coordinate system. In our experiments, we have provided the 3D rendering (Figure 6a) and electrode locations of the model to the experimenters so as to facilitate proper cap adjustment (Dmochowski et al. 2013).
To the best of our knowledge, there are only a few modeling studies for stroke patients, e.g. (Wagner et al. 2007) and (Datta et al. 2011), and they use conventional manual segmentation. A few automated segmentation algorithms do exist that are specifically intended for lesioned brain anatomies, e.g. (Seghier et al. 2008). We did not test these in detail as we found the results with the proposed method satisfactory.
To evaluate the performance of the automated routine, results from manual method are needed. Since the output from automatic segmentation algorithm includes six tissue types in a large FOV (down to the neck), the manual segmentation results are expected to contain the same number of tissue types. While there are publicly available standard MR image databases (e.g. the Internet Brain Segmentation Repository, IBSR), to the best of our knowledge, none of them provide expert-level manual segmentation for all the six tissue types in the same FOV. Therefore, we did not use any public data to perform the evaluation, but rather relied on our own MRI data. It should be noted that our manual segmentation was done by engineers, not radiologists. So it is by no means a “gold” standard. There is a significant level of ambiguity when segmenting MR images (in particular for voxels on tissue boundaries). Even expert segmenters make different judgments on how to assign ambiguous voxels. In one study (Rajapakse & Kruggel 1998), expert operators segmenting the same brains obtained Jaccard indices of 0.876 and 0.832 for GM and WM, respectively. This means that there is a disagreement of about 15% between two manual segmentations. The disagreement between the automated and manual segmentation obtained here is on average 15%.
It should also be mentioned that the MR image quality and contrast among GM, CSF, skull directly affect the performance of this automated routine. The manual segmentation is more difficult and may be more variable if the image quality is poor, and the automated segmentation algorithm cannot correctly delineate the layers of GM, CSF, and skull around the parietal area if the contrast there is low. Therefore, low image quality and contrast will lead to more differences between manual and automated results in segmentations and electric field distributions. This can be observed from Figure 8 and 10, where it is shown that Head 1 has the best performance, and healthy heads (H1 – H4) generally have a better performance than stroke heads (S1–S7). In fact, Head 1 is the best scan in terms of image quality and contrast. The increased deviation observed for the stroke heads likely results from the bias introduced by the TPM towards normal anatomy, which is beneficial, when normal anatomy is indeed present. Specifically, the present sample represents cases of chronic stroke with large lesions that are filled with CSF in areas predicted by the TPM to contain either GM or WM.
The proposed automated correction technique contains a few free parameters which can affect its performance (see Section 2.3): for instance, the size of the structural element used in the process of filling-in CSF discontinuities. We assume that the brain is entirely engulfed in CSF, but imaging noises combined with the limited resolution lead to discontinuities (CSF thickness in some locations is close to the sampling resolution of 1 mm3). To be conservative in preserving the original anatomy, the size of the structure element should be small. However, if it is too small, then remaining discontinuities will lead to significant differences in the electric field predictions. This is the origin of the >50 % deviations in CSF (Figure 10a). Since we do not have a large population of MRI data to perform systematic parameter selection, we determined all parameters by hand. Future work may attempt to automatically determine free parameters in the automated correction technique based on image quality.
It should be noted that to avoid convergence problems when generating the FEM in ScanIP, all of the heads in this study were smoothed to some extent. Noisy MR images typically required more smoothing to even out rough tissue surfaces resulting from image noise. At the same time, smoothing disrupts some details of the segmentation, and as such was used conservatively. Indeed, the change in segmentation performance as a result of smoothing was not statistically significant in this sample. Future work will aim to integrate smoothing and segmentation into a single step by merging Markov Random Field models (Roche et al. 2011) with the New Segment formalism (Ashburner & Friston 2005).
Skull bone is an inhomogeneous three-layered structure. The outer layers consist of compact cortical bone while the middle layer is spongy bone containing fatty marrow. Current segmentation algorithms do not make this distinction, primarily because no prior probability map is yet available at this level of detail. This differentiation is important because these two types of bones have different electrical conductivities (Dannhauer et al. 2011). Fortunately, there is evidence that a more detailed three-layer bone model does not significantly change electric field estimates at the level of the brain as compared to a single-layer model (Rampersad et al. 2012). However, there is nothing that prevents the present approach to incorporate this level of details once a corresponding TPM becomes available.
There are a number of approaches to convert diffusion tensor imaging (DTI) data to conductivity tensor data thereby incorporating anisotropy in the modeling for tDCS (Windhoff et al. 2011, Neuling et al. 2012, Suh et al. 2012, Shahid et al. 2013). However, in clinical practice DTI may be less viable due to the longer acquisition time and the cumbersome software tools for combining DTI with FEM modeling. More importantly, the exact relationship between electric and DTI anisotropy has not yet been established experimentally, and thus there is a certain level of arbitrariness to the different choices made by these techniques (from 3% to 58% difference in current flow distribution compared to isotropic modeling (Suh et al. 2012, Shahid et al. 2013)). In this context, the average 23% disagreement in our work between automated and manual methods seems to be well within variability resulting from specific choices that the modelers have to make.
The most severe limitation, not only to this study but to all tDCS modeling studies, is that to date there is no direct experimental validation of any of the estimated field intensities inside the head. Initial efforts to validate the models have been limited to voltage recordings on the surface of the scalp (Datta et al. 2013) and indirect measurements using functional MRI (Antal et al. 2012). It would seem that the level of precision in modeling has advanced to a point where further meaningful improvement is not possible without empirical confirmation.
Acknowledgments
The authors would like to thank Marom Bikson and René Kempe of the City College of New York for valuable discussions. We also thank Jessica Richardson and Julius Fridriksson of University of South Carolina for providing the MRI data and target locations for the stroke subjects. A number of students from Department of Biomedical Engineering at CCNY contributed significant time and effort to manually improve the segmentation of these MRI scans. This work was partially supported by NIH-NINDS grant R41 NS076123 “Targeted transcranial electrotherapy system to accelerate stroke recovery” and a CUNY-CAT matching grant supported by NYSTAR.
Footnotes
SPM8 also implements an extended version of Unified Segmentation that can use information from multiple co-registered volumes, e.g. T1, T2, PD, CT, etc., and incorporates skull and scalp. In the SPM8 Manual this routine is called “New Segment”, and is employed in this work.
Strictly speaking, air is not a tissue. However, the air inside the cavities has dramatically different conductivity from other tissues (see Section 2.5), and thus it is crucial to model the air explicitly to obtain realistic current flow distributions.
All durations in this work were measured on a PC with Intel® Quad Core Xeon® W5580 CPU at 3.2 GHz.
Strictly speaking, this type of error is in fact a bug from New Segment, since its output is a probability distribution for each voxel that should be summed to 1.
Code available at http://bme.ccny.cuny.edu/faculty/lparra/autosegment/
A cluster of voxels that are connected together by 26-connectivity (Rosenfeld & Kak 1982).
Available from http://www.easycap.de/easycap/
Code available at http://bme.ccny.cuny.edu/faculty/lparra/autosegment/
For the MRI volume which is not in RAS orientation, the electrode placement program can automatically detect its orientation and convert it into RAS.
Available at http://bme.ccny.cuny.edu/faculty/lparra/autosegment/
This is equivalent to comparing current flow distributions, since the current flow is the electric field multiplied by the conductivity.
The air mask was not included in the “whole head” computation under NC condition, because the air outside of the head was removed after manual and automated correction, but it is included in the initial automated results leading to artificially large differences.
Available from http://www.cma.mgh.harvard.edu/ibsr/data.html
References
- Acar ZA, Makeig S. Journal of Neuroscience Methods. 2010;190(2):258–270. doi: 10.1016/j.jneumeth.2010.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akalin Acar Z, Makeig S. Brain topography. 2013;26(3):378–396. doi: 10.1007/s10548-012-0274-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antal A, Bikson M, Datta A, Lafon B, Dechent P, Parra LC, Paulus W. NeuroImage. 2012 doi: 10.1016/j.neuroimage.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. NeuroImage. 2005;26(3):839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Wu J, Cook PA, Gee JC. Neuroinformatics. 2011;9(4):381–400. doi: 10.1007/s12021-011-9109-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JM, Rorden C, Fridriksson J. Stroke. 2010;41(6):1229–1236. doi: 10.1161/STROKEAHA.109.576785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caparelli-Daquer EM, Zimmermann TJ, Mooshagian E, Parra LC, Rice JK, Datta A, Bikson M, Wassermann EM. 34th Annual International Conference of the IEEE Engineering in Medicine and Biology Society; San Diego, CA. 2012; pp. 735–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. NeuroImage. 1999;9(2):179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Dannhauer M, Brooks D, Tucker D, MacLeod R. 34th Annual International Conference of the IEEE Engineering in Medicine and Biology Society; San Diego, CA. 2012; pp. 5486–5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannhauer M, Lanfer B, Wolters CH, Knsche TR. Human Brain Mapping. 2011;32(9):1383–1399. doi: 10.1002/hbm.21114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Baker JM, Bikson M, Fridriksson J. Brain Stimulation. 2011;4(3):169–174. doi: 10.1016/j.brs.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Bansal V, Diaz J, Patel J, Reato D, Bikson M. Brain Stimulation. 2009;2(4):201–207. doi: 10.1016/j.brs.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Bikson M, Fregni F. NeuroImage. 2010;52(4):1268–1278. doi: 10.1016/j.neuroimage.2010.04.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Truong D, Minhas P, Parra LC, Bikson M. Frontiers in Psychiatry. 2012;3:91. doi: 10.3389/fpsyt.2012.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Zhou X, Su Y, Parra LC, Bikson M. Journal of Neural Engineering. 2013;10(3):036018. doi: 10.1088/1741-2560/10/3/036018. [DOI] [PubMed] [Google Scholar]
- Delorme A, Makeig S. Journal of Neuroscience Methods. 2004;134(1):9–21. doi: 10.1016/j.jneumeth.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Dmochowski JP, Bikson M, Parra LC. Physics in Medicine and Biology. 2012;57(20):6459–6477. doi: 10.1088/0031-9155/57/20/6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmochowski JP, Datta A, Bikson M, Su Y, Parra LC. Journal of Neural Engineering. 2011;8(4):046011. doi: 10.1088/1741-2560/8/4/046011. [DOI] [PubMed] [Google Scholar]
- Dmochowski JP, Datta A, Huang Y, Richardson JD, Bikson M, Fridriksson J, Parra LC. NeuroImage. 2013;75:12–19. doi: 10.1016/j.neuroimage.2013.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Q, Boas D. IEEE International Symposium on Biomedical Imaging: From Nano to Macro, 2009. ISBI ’09.2009. pp. 1142–1145. [Google Scholar]
- Fischl B, Sereno MI, Dale AM. NeuroImage. 1999;9(2):195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Griffiths DJ. Introduction to electrodynamics. 3rd ed. edn Prentice Hall Upper Saddle River; NJ: 1999. [Google Scholar]
- Gllmar D, Haueisen J, Reichenbach JR. NeuroImage. 2010;51(1):145–163. doi: 10.1016/j.neuroimage.2010.02.014. [DOI] [PubMed] [Google Scholar]
- Jaccard P. Bulletin de la Socit Vaudoise des Sciences Naturelles. 1901;37:241–272. [Google Scholar]
- Klem GH, Lders HO, Jasper HH, Elger C. Electroencephalography and clinical neurophysiology. 1999;52(Supplement):3–6. [PubMed] [Google Scholar]
- Koessler L, Benhadid A, Maillard L, Vignal JP, Felblinger J, Vespignani H, Braun M. NeuroImage. 2008;41(3):914–923. doi: 10.1016/j.neuroimage.2008.02.039. [DOI] [PubMed] [Google Scholar]
- Lanfer B, Scherg M, Dannhauer M, Knsche T, Burger M, Wolters C. NeuroImage. 2012;62(1):418–431. doi: 10.1016/j.neuroimage.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Logan DL. A First Course in the Finite Element Method. 4th ed. edn Nelson Toronto; 2007. [Google Scholar]
- Mendonca ME, Santana MB, Baptista AF, Datta A, Bikson M, Fregni F, Araujo CP. The Journal of Pain: Official Journal of the American Pain Society. 2011;12(5):610–617. doi: 10.1016/j.jpain.2010.12.015. [DOI] [PubMed] [Google Scholar]
- Minhas P, Bikson M, Woods AJ, Rosen AR, Kessler SK. 34th Annual International Conference of the IEEE Engineering in Medicine and Biology Society; San Diego, CA. 2012; pp. 859–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuling T, Wagner S, Wolters CH, Zaehle T, Herrmann CS. Frontiers in Psychiatry. 2012;3:83. doi: 10.3389/fpsyt.2012.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostenveld R, Fries P, Maris E, Schoffelen JM. Computational Intelligence and Neuroscience. 2011:1–9. doi: 10.1155/2011/156869. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapakse JC, Kruggel F. Image and Vision Computing. 1998;16(3):165–180. [Google Scholar]
- Rampersad S, Stegeman D, Oostendorp T. IEEE Transactions on Neural Systems and Rehabilitation Engineering. 2012;PP(99):1. doi: 10.1109/TNSRE.2012.2206829. [DOI] [PubMed] [Google Scholar]
- Renard Y, Pommier J. 2010 http://download.gna.org/getfem/html/homepage/index.html.
- Rice JK, Rorden C, Little JS, Parra LC. NeuroImage. 2013;64:476–484. doi: 10.1016/j.neuroimage.2012.09.041. [DOI] [PubMed] [Google Scholar]
- Roche A, Ribes D, Bach-Cuadra M, Krger G. Medical Image Analysis. 2011;15(6):830–839. doi: 10.1016/j.media.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Rosenfeld A, Kak AC. Digital Picture Processing. 2nd ed. edn Academic Press, Inc; Orlando, FL: 1982. [Google Scholar]
- Seghier ML, Ramlackhansingh A, Crinion J, Leff AP, Price CJ. NeuroImage. 2008;41(4-3):1253–1266. doi: 10.1016/j.neuroimage.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid S, Wen P, Ahfock T. Computer Methods and Programs in Biomedicine. 2013;109(1):48–64. doi: 10.1016/j.cmpb.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Smith SM. Human Brain Mapping. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM. NeuroImage. 2004;23(Suppl 1):S208–219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Suh HS, Lee WH, Kim TS. Physics in Medicine and Biology. 2012;57(21):6961–6980. doi: 10.1088/0031-9155/57/21/6961. [DOI] [PubMed] [Google Scholar]
- Vatta F, Meneghini F, Esposito F, Mininel S, Salle FD. Computational Intelligence and Neuroscience. 2010:1–11. doi: 10.1155/2010/972060. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T, Fregni F, Fecteau S, Grodzinsky A, Zahn M, Pascual-Leone A. NeuroImage. 2007;35(3):1113–1124. doi: 10.1016/j.neuroimage.2007.01.027. [DOI] [PubMed] [Google Scholar]
- Windhoff M, Opitz A, Thielscher A. Human Brain Mapping. 2011;34(4):923–935. doi: 10.1002/hbm.21479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. NeuroImage. 2006;31(3):1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]