Abstract
Background
Long non-coding RNAs (lncRNAs) are prevalently transcribed in the genome yet their potential roles in human cancers are not well understood. The aim of the present study was to determine the lncRNA expression profile in gastric cancer and its potential clinical value.
Methods
The global lncRNA expression profile in gastric cancer was measured by lncRNA microarray. Levels of two representative lncRNAs, H19 and uc001lsz, were confirmed by real-time reverse transcriptase-polymerase chain reaction. The relationship between their levels and clinicopathological factors of patients with gastric cancer was explored. A receiver operating characteristic (ROC) curve was constructed for differentiating gastric cancer from benign gastric diseases.
Results
Total of 135 lncRNAs, which differential expression levels between tumor and non-tumorous tissues were more than twofold, were found (GEO No. GSE47850). The most down-regulated lncRNAs in gastric cancer tissues were FER1L4, uc001lsz, BG491697, AF131784, uc009ycs, BG981369, AF147447, HMlincRNA1600, and AK054588; while the most up-regulated ones were H19, HMlincRNA717, BM709340, BQ213083, AK054978, and DB077273. H19 was found highly expressed in stomach and liver cancer cell lines, while lowly expressed in lung cancer and prostate cancer cell lines. Uc001lsz was lowly expressed in gastric, lung and liver cancer cell lines, while highly expressed in prostate cancer. The areas under ROC curves were up to 0.613, 0.751, and 0.761 for H19, uc001lsz, and the combination, respectively.
Conclusions
The lncRNA expression profile in gastric cancer suggests the potential roles of lncRNAs in gastric cancer occurrence and development. The overexpression of H19 in gastric cancer suggests that H19 may be participated in gastric cancer. The reduced expression of uc001lsz in gastric cancer cell lines and tissues, its associations with TNM stage, and its dysregulation in early cancer and precancerous lesions suggest that uc001lsz may be a potential marker for the diagnosis of early gastric cancer.
Keywords: Gastric cancer, Long non-coding RNA, Expression profile, H19, uc001lsz
Background
The well-studied components in the human genome are those of protein-coding genes. However, the coding exons of these genes account for only 1.5% of the genome [1]. In recent years, it has become increasingly apparent that the non-protein-coding portion of the genome is of crucial functional importance for disease occurrence [2]. The non-coding RNAs (ncRNAs) characterize as three types, long ncRNAs, mid-size ncRNAs and short ncRNAs [1]. Although most studies on ncRNAs are focused on short ncRNAs, such as microRNAs (miRNAs), long non-coding RNAs (lncRNAs) are rapidly gaining prominence recently.
LncRNAs are greater than 200 nucleotides in length [3]. They have emerged recently as major players in governing fundamental biological processes. Aberrant expression of lncRNAs has been associated with cancers [3]. For example, Differential display code 3 (DD3PCA3), a prostate-specific lncRNA, appears to be a marker for early diagnosis of prostate cancer [4]. More important, DD3PCA3 can be detected in urine from patients with prostate cancer [5]. Though Metastasis associated lung adenocarcinoma transcript 1 (MALAT-1) is first found abnormal expressed in metastasizing non-small-cell lung carcinomas [6], it is up-regulated in hepatocarcinoma, breast cancer, pancreatic cancer, colorectal cancer, and prostate cancer [7]. MALAT-1 is not only a potential diagnostic marker, but also a potential prognostic marker [8]. HOX transcript antisense RNA (HOTAIR) is associated with breast cancer and colorectal cancer [9,10]. H19, another famous lncRNA, is frequently involved in pediatric and adult tumors [11].
Gastric cancer is still one of the most frequent causes of mortality in the world [12]. However, traditional strategies based on radical surgery for the treatment of gastric cancer are not yet satisfactory. Therefore, reveal of the mechanisms of occurrence and development of gastric cancer is attracting increased attention in cancer research.
Since the global lncRNA expression profile in gastric cancer is not fully uncovered, in the present study, we explored the lncRNA expression profile in gastric cancer. Then the relationship between the aberrantly expressed-lncRNAs and clinicopathological factors of patients with gastric cancer was explored. Our data provides candidate diagnostic biomarkers of gastric cancer.
Methods
Patients and specimens
Gastric cancer patients’ tissues, including gastric cancer tissues, precancerous lesion and corresponding adjacent non-tumorous tissues were immediately preserved in RNA fixer (Bioteke, Beijing, China) after removal from the body and stored at –80°C until use. Tissue samples were obtained from surgical or biopsy specimens from February 2011 to June 2012 at three cancer centers, Yinzhou People’s Hospital, Ningbo No. 1 Hospital and The Affiliated Hospital of Ningbo University School of Medicine, China. Informed consent was taken from all subjects. The Human Research Ethics Committee of Ningbo University approved all aspects of this study. Tumors were staged according to the tumor-node-metastasis (TNM) staging system of the International Union Against Cancer (5th ed). Histological grade was assessed following the National Comprehensive Cancer Network (NCCN) clinical practice guideline of oncology (V.1.2011). The non-tumorous tissues were 5 cm from the edge of the tumor and there were no obvious tumor cells, as evaluated by a pathologist. There was no radiotherapy, chemotherapy, targeted therapy or Dendritic cell/Cytokine-induced killer (DC/CIK) therapy prior to the upper gastrointestinal endoscopy examination or operation.
Total RNA preparation
Total RNA was isolated using Trizol reagent (Invitrogen, Karlsruhe, Germany) following the manufacturer’s instructions [13].
LncRNA microarray assay
Three paired biopsy specimens were obtained from patients (55 y and male, 76 y and male, and 88 y and female) with poorly or poorly-and-moderately differentiated gastric cancer. Human lncRNA microarray was manufactured in NimbleGen Hybridization System (Arraystar, Rockville, MD). More than 23 000 lncRNAs were collected from the authoritative data sources including National Center for Biotechnology Information (NCBI) RefSeq, University of California, Santa Cruz (UCSC), lncRNAs from literatures and Ultra Conserved Regions (UCRs) [14]. Data was extracted and normalized using NimbleScan v2.5 software (Roche NimbleGen, Madison, WI). Further Data analysis was performed using Agilent GeneSpring GX 11.5 software (Agilent Technologies, Santa Clara, CA).
Cell culture
Human gastric epithelial cell line (GES-1), gastric cancer cell lines (AGS, MGC-803, and SGC-7901), liver normal cell line (HL-7702), hepatic carcinoma cell lines (HepG2 and SMMC-7721), fetal lung fibroblast cell line (HELF), lung carcinoma cell line (A549), prostate epithelial cell line (RWPE-1), and prostate carcinoma cell lines (Du-145 and PC-3) were obtained from the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). Cells were cultured in culture flasks at 37°C in a humidified atmosphere of 5% CO2[15].
qRT-PCR detection of H19 and uc001lsz
Real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) is the gold standard for data verification. To verify the results of lncRNA microarray, cDNA was generated using the GoScript Reverse Transcription (RT) System (Promega, Madison, WI). Quantitative polymerase chain reaction (qPCR) was achieved by using the GoTaq qPCR Master Mix (Promega, Madison, WI) on an Mx3005P real-time PCR System (Stratagene, La Jolla, CA). The sequences of the PCR primers for H19, uc001lsz, and β-actin were as follows: 5′-ACCAGCCACCACATCATC-3′ (sense) and 5′-TCAGAAACAAAGAGACAGAAGG-3′ (antisense) for H19; 5′-GACGGCACCTACTACACCTT-3′ (sense) and 5′-GCTGACCACCTTGTTGTTGAA-3′ (antisense) for uc001lsz; 5′-AAGCCACCCCACTTCTCTCTAA-3′ (sense) and 5′-AATGCTATCACCTCCCCTGTGT-3′ (antisense) for β-actin. The data were analyzed by the ∆Ct method [16,17]. All results were expressed as the Means ± SD of three independent experiments.
Cloning and sequencing of qRT-PCR products
The qRT-PCR products of lncRNA were first purified using a UNIQ-10 PCR Product Purification Kit and then cloned into the pUCm-T vector (Sangon Biotech, Shanghai, China) following the manufacturer’s instructions. Then, DNA sequencing was performed by Sangon Biotech Co., Ltd.
Serological tumor marker analysis
Serum carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9) were measured using an Elecsys 2010 machine (Roche Diagnostics, Basel, Switzerland). The cutoff values were 5 ng/mL and 35 U/mL for CEA and CA19-9, respectively.
Statistical analysis
All statistical data were analyzed by Statistical Program for Social Sciences (SPSS) 18.0 software (SPSS, Chicago, IL) and GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA). One way analysis of variance test, two-tailed Student’s t-test and rank-sum test were used as appropriate. A receiver operating characteristic (ROC) curve was established to evaluate the diagnostic value. P < 0.05 was considered statistically significant.
Results
LncRNA expression profiles in gastric cancer tissues relative to adjacent non-tumorous tissues
The lncRNA expression patterns between gastric cancer tissues and adjacent non-tumorous tissues were found to be significantly different (Figure 1). Total of 135 lncRNAs, which expression change was more than twofold, were found (GEO accession numbers is 47850; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE47850). Among them, 71 and 64 were up and down expressed in tumor tissues, respectively. The most down-regulated lncRNAs in gastric cancer tissues were FER1L4, uc001lsz, BG491697, AF131784, uc009ycs, BG981369, AF147447, HMlincRNA1600, and AK054588. The most up-regulated lncRNAs in gastric cancer tissues were H19, HMlincRNA717, AI769947, BQ213083, AK054978, and DB077273 (Table 1).
Figure 1.
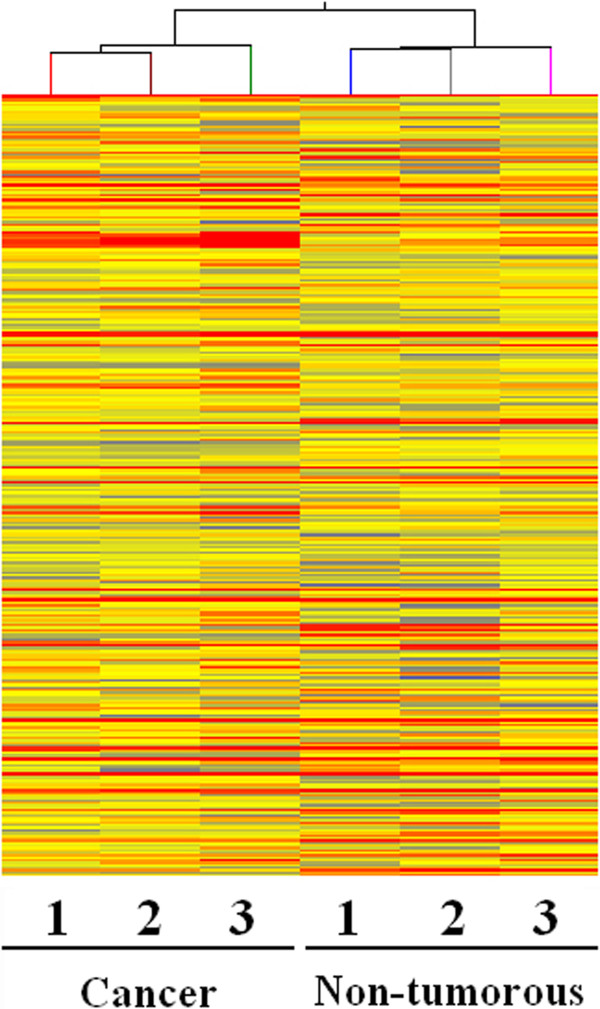
Alterations in lncRNA expression profiles between gastric carcinoma tissues and non-tumorous tissues. The result from Hierarchical Clustering shows distinguishable lncRNA expression profiling among samples. “Red” indicates high relative expression; and “blue” indicates low relative expression.
Table 1.
More than fourfold differentially expressed lncRNAs in gastric cancer tissues comparing with paired non-tumorous tissues
| Name | Chromosome | Regulation | Fold change | Sourcea | P value |
|---|---|---|---|---|---|
| H19 |
11 |
up |
8.91 |
lncRNAdb |
0.020 |
| HMlincRNA717 |
18 |
up |
5.96 |
lincRNA |
0.013 |
|
AI769947 |
7 |
up |
5.51 |
lincRNA |
0.026 |
|
BQ213083 |
2 |
up |
5.45 |
lincRNA |
0.040 |
|
AK054978 |
X |
up |
4.59 |
lincRNA |
0.006 |
|
DB077273 |
2 |
up |
4.11 |
lincRNA |
0.008 |
| FER1L4 |
20 |
down |
9.17 |
refNR |
0.047 |
| uc0 01lsz |
11 |
down |
8.36 |
UCSC_knowngene |
0.020 |
|
BG491697 |
20 |
down |
6.50 |
lincRNA |
0.035 |
|
AF131784 |
18 |
down |
6.25 |
mRNA |
0.019 |
| uc009ycs |
11 |
down |
5.82 |
UCSC_knowngene |
0.009 |
|
BG981369 |
1 |
down |
5.20 |
lincRNA |
0.048 |
|
AF147447 |
16 |
down |
4.76 |
mRNA |
0.038 |
| HMlincRNA1600 |
X |
down |
4.12 |
lincRNA |
0.013 |
| AK054588 | 1 | down | 4.04 | lincRNA | 0.045 |
alncRNAdb: http://lncrnadb.com/Default.aspx; lincRNA: http://genome.ucsc.edu/cgi-bin/hgGateway; refNR: http://genome.ucsc.edu/cgi-bin/hgTables; http://www.ncbi.nlm.nih.gov/RefSeq/; UCSC_knowngene: http://genome.ucsc.edu/cgi-bin/hgTables; mRNA: http://genome.ucsc.edu/cgi-bin/hgTables.
Expression of H19 was up-regulated in gastric carcinoma tissues
Since H19 was found the most up-regulated lncRNA in gastric cancer tissue, up to 8.91-fold change in microarray detection (Table 1), to validate this result, we detected the expression level of H19 in two types of cancer tissues, biopsy tissues and surgical specimens, by qRT-PCR. First, we explored the expression level of H19 in 15 pairs of gastric carcinoma’ biopsy tissues. We found that its level in cancer tissues was significantly higher than that in non-tumorous tissues (Figure 2A). Then we detected H19 level in large number of surgical specimens. As shown in Figure 2B, it was up-regulated in 74.0% (57/77) of gastric cancer tissues (P = 0.029). By sequencing the qRT-PCR product, we found that the sequence of H19 (Figure 2C) was consistent with that from the database (http://www.ncbi.nlm.nih.gov/gene/283120).
Figure 2.

H19 was up-regulated in gastric cancer tissues, gastric cancer cell lines and other common cancer cell lines. Real-time qRT-PCR was used to determine the expression level of H19. The ∆Ct value was determined by subtracting the β-actin Ct value from the target lncRNA Ct value. Smaller ∆Ct value indicates higher expression. Level of H19 in gastric cancer biopsy tissues (n = 15, P = 0.014; A) and gastric cancer tissues (n = 77, P = 0.029; B). Sequencing result of qRT-PCR product of H19 (C). The level of H19 in gastric cancer cell lines (AGS, MGC-803 and SGC-7901) and liver cancer cell lines (SMMC-7721 and HepG2) was higher than that in human gastric epithelial cell line GES-1 and human liver normal cell line HL-7702, respectively; however, its level in lung cancer cell line A549 and prostate cancer cell lines (Du-145 and PC-3) was lower than that in human fetal lung fibroblast cell line HELF cells and human prostate epithelial cell line RWPE-1 (D). All results were expressed as the Means ± SD of three independent experiments. #P < 0.001.
Expression of H19 in common cancer cell lines
To obtain more information about H19 expression in cancers, we investigated its level in some common cancer cell lines. As shown in Figure 2D, comparing with respective normal cell line, H19 was found highly expressed in stomach cancer cell lines (AGS, MGC-803 and SGC-7901) and hepatocellular carcinoma cell lines (SMMC-7721 and HepG2), while lowly expressed in lung cancer cell line (A549) and prostate cancer cell lines (Du-145 and PC-3).
Expression of uc001lsz was down-regulated in gastric carcinoma tissues
Uc001lsz is a new-found lncRNA, which is transcribed from the forward strand in chromosome 11p15.5. From the microarray results (Table 1), we can see that uc001lsz was the second most down-regulated lncRNA in gastric cancer tissue. Another reason that encouraged us to further study uc001lsz was that it is trans associated with MUC2, which is secreted and forms an insoluble mucous barrier in the gut lumen. To further validate the expression of uc001lsz in gastric cancer, we expanded the sample number. The data indicate that it was significantly down-regulated in 84.4% (65/77) of gastric cancer tissues (Figure 3A, P < 0.001). By sequencing the qRT-PCR product, we found that the sequence of uc001lsz (Figure 3B) was consistent with that from the database (http://genome.ucsc.edu/cgi-bin/hgTables).
Figure 3.
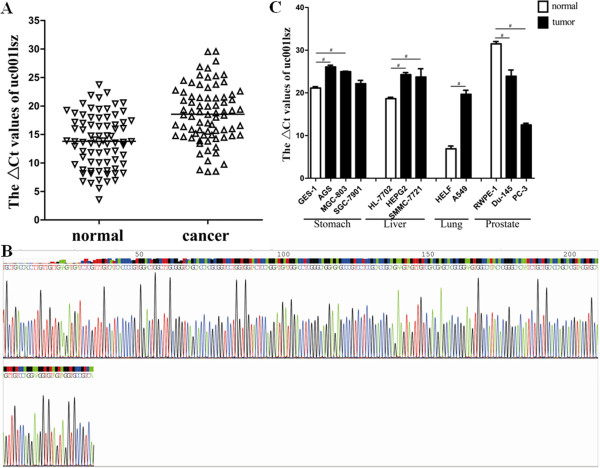
Uc001lsz was down-regulated in gastric cancer tissues, gastric cancer cell lines and other common cancer cell lines. Real-time qRT-PCR was used to determine the expression level of uc001lsz. Three independent experiments were performed. Level of uc001lsz in gastric cancer tissues was significantly lower than that in corresponding non-tumorous tissues (n = 77, P < 0.001; A). Sequencing result of qRT-PCR product of uc001lsz (B). The level of uc001lsz in gastric cancer cell lines (AGS, MGC-803 and SGC-7901), liver cancer cell lines (SMMC-7721 and HepG2) and lung cancer cell line A549 was lower than that in human gastric epithelial cell line GES-1, human liver normal cell line HL-7702 and human fetal lung fibroblast cell line HELF, respectively; however, its level in prostate cancer cell lines (Du-145 and PC-3) was higher than that in human prostate epithelial cell line RWPE-1 (C).
Expression of uc001lsz in common cancer cell lines
To obtain the expression information about uc001lsz in common cancers, we detected its level in several common cancer cell lines. We found that comparing with respective normal cell line, uc001lsz was lowly expressed in gastric cancer (AGS, MGC-803 and SGC-7901), lung cancer (A549) and liver cancer (SMMC-7721 and HepG2) cell lines, while only highly expressed in prostate cancer (Du-145 and PC-3) cell lines (Figure 3C).
Potential diagnostic values of H19 and uc001lsz
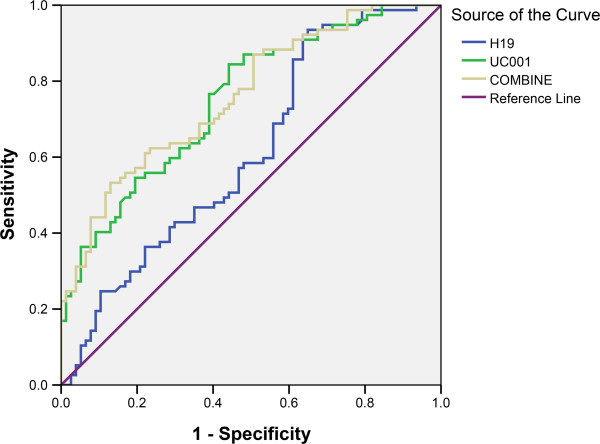
We next performed an analysis to identify whether H19 or uc001lsz expression was associated with the clinicopathological features of gastric cancer. As shown in Table 2, the level of uc001lsz was associated with TNM stages (P = 0.032). The positive detection rates of H19 and uc001lsz are 74.0% and 84.4%, respectively. Both of them are higher than those of common gastric cancer biomarker CEA (64.0%) and CA19-9 (53.3%) (Table 2). The areas under ROC curves were up to 0.613, 0.751, and 0.761 for H19, uc001lsz, and the combination, respectively (Figure 4). The combinative use of H19 and uc001lsz slightly increased the diagnostic value.
Table 2.
The relationship of H19 and uc001lsz expression levels (ΔCt) in cancer tissues with clinicopathological factors of patients with gastric cancera
| Characteristics |
No. of patients (%) |
H19 |
uc001lsz |
||
|---|---|---|---|---|---|
| Mean ± SD | P value | Mean ± SD | P value | ||
| Age (y) |
|
|
|
|
|
| ≥ 60 |
59 (76.6) |
8.74 ± 4.67 |
0.588 |
18.85 ± 4.94 |
0.240 |
| < 60 |
18 (23.4) |
8.08 ± 3.99 |
|
17.28 ± 4.85 |
|
| Gender |
|
|
|
|
|
| Male |
57 (74.0) |
8.66 ± 4.81 |
0.823 |
18.19 ± 5.01 |
0.355 |
| Female |
20 (26.0) |
8.40 ± 3.44 |
|
19.37 ± 4.58 |
|
| Diameter (cm)b |
|
|
|
|
|
| ≥ 5 |
39 (52.0) |
7.98 ± 4.44 |
0.282 |
18.49 ± 5.03 |
0.682 |
| < 5 |
36 (48.0) |
9.10 ± 4.53 |
|
18.01 ± 5.11 |
|
| CEAb |
|
|
|
|
|
| Positive |
48 (64.0) |
8.45 ± 4.21 |
0.882 |
18.28 ± 4.54 |
0.593 |
| Negative |
27 (36.0) |
8.61 ± 5.01 |
|
18.92 ± 5.71 |
|
| CA19-9b |
|
|
|
|
|
| Positive |
40 (53.3) |
8.68 ± 3.87 |
0.720 |
18.65 ± 4.64 |
0.792 |
| Negative |
35 (46.7) |
8.31 ± 5.14 |
|
18.35 ± 5.37 |
|
| Differentiationc |
|
|
|
|
|
| Well |
3 (4.9) |
8.28 ± 1.92 |
0.635 |
15.34 ± 5.40 |
0.371 |
| Moderate |
33 (54.1) |
9.05 ± 4.40 |
|
19.37 ± 4.85 |
|
| Poor |
25 (41.0) |
7.84 ± 5.47 |
|
19.54 ± 4.91 |
|
| Lymphatic metastasisc |
|
|
|
|
|
| N0 |
20 (32.8) |
8.95 ± 3.17 |
0.926 |
18.55 ± 6.01 |
0.472 |
| N1&N2&N3 |
41 (67.2) |
9.07 ± 5.25 |
|
19.53 ± 4.39 |
|
| Distal metastasisc |
|
|
|
|
|
| M0 |
58 (95.1) |
8.87 ± 4.70 |
0.258 |
19.13 ± 5.02 |
0.617 |
| M1 |
3 (4.9) |
12.00 ± 1.30 |
|
20.62 ± 3.74 |
|
| Invasionc |
|
|
|
|
|
| Tis&T1-T3 |
20 (32.8) |
10.04 ± 3.86 |
0.237 |
17.73 ± 4.72 |
0.105 |
| T4 |
41 (67.2) |
8.53 ± 4.95 |
|
19.93 ± 4.95 |
|
| TNM stagec |
|
|
|
|
|
| 0&I&II |
24 (39.3) |
9.40 ± 3.86 |
0.620 |
17.81 ± 5.52 |
0.032 |
| III&IV | 37 (60.7) | 8.79 ± 5.14 | 20.49 ± 3.98 | ||
aAll samples are consist of 16 endoscopic biopsy samples and 61 surgery samples from patients with gastric cancer.
bNot detected for 2 patients who were performed endoscopy examination.
cOnly includes 61 surgery patients.
Figure 4.
The ROC curve.
Expression of uc001lsz was aberrant in early cancer and precancerous lesions
At last, to observe the possible early diagnostic values of lncRNA, we measured the level of uc001lsz in early cancer and precancerous lesions. We found that its level was remarkable lower in these lesions compared with those of corresponding adjacent non-tumorous tissues (Figure 5A). The level of uc001lsz in normal tissues was significantly higher than that in precancerous lesions and early cancer tissues. Besides, its level in precancerous lesions was conspicuous higher than that in early cancer tissues (Figure 5B).
Figure 5.

Expression of uc001lsz in early cancer and precancerous lesions. The level of uc001lsz in early pathological change tissue is lower than that in paired normal tissue (n = 14, P = 0.0016; A). The level of uc001lsz in normal tissue is significantly higher than that in precancerous lesions and early cancers (B). #P < 0.001; *P < 0.05.
Discussion
Studies have shown that ~18% of the protein coding genes that produce lncRNAs are associated with cancer, whereas only 9% of all human protein coding genes are associated with cancer [18]. The relationship between lncRNAs and tumors has currently become one of the focuses of cancer studies.
There is mounting evidence that lncRNAs are relation to digestive system tumors [19]. Three lncRNAs, H19, HOTAIR and lncRNA highly upregulated in liver cancer (HULC), were found overexpression in human hepatocellular carcinomas (HCC) [20-22]. In addition, HULC expression is not only confined to HCC, but is also expressed in colorectal carcinomas that metastasize to the liver [23].
Researches about lncRNAs related to stomach are limited. Sun et al. found that the expression level of gastric cancer-associated transcript 1 (GACAT1), or AC096655.1-002, was significantly correlated with lymph node metastasis, distant metastasis, TNM stages, and differentiation [17]. Mei et al. reported that ubiquitin-like modifier (SUMO) 1 pseudogene 3, SUMO1P3, might be a potential biomarker in the diagnosis of gastric cancer [16]. Niinuma et al. found that overexpression of HOTAIR was markedly associated with high-risk gastrointestinal stromal tumors [24]. RNA, 7SK small nuclear (RN7SK) can indirectly regulate gastric tumorigenesis via positive transcription elongation factor-b (p-TEFb) [25]. H19 may play an important role in gastric cancer by loss of imprinting and other mechanisms [26,27]. Recent, Cao et al. used bioinformatics methods to screen lncRNA expression profiles associated with gastric cancer [28]. First, two publicly available human exon arrays for gastric cancer and data for the corresponding normal tissue were downloaded from the GEO. Then, the probes of the human exon arrays were re-annotated. Finally, the probes uniquely mapping to lncRNAs at the gene level were retained. Total of 88 lncRNAs that were differentially expressed in gastric cancer were identified [28].
Here, the approaches for the screening of gastric cancer–associated lncRNAs were different from those used by Cao et al. [28]. To identify the remarkably down-regulated or up-regulated lncRNAs in gastric cancer, we first collected gastric cancer and adjacent non-tumorous tissue samples from upper gastrointestinal endoscopy examination. From the lncRNA expression profiles obtained from lncRNA microarray analysis (Figure 1), we found that among the significantly different expressed lncRNAs (Table 1), only H19 has been found in other cancers [11,20,26,27]. As a result, it is crucial to further clarify the clinical signatures of lncRNAs in the diagnosis and treatment of gastric cancer.
The growing studies of functionally characterized lncRNAs reveals that these transcripts are important in different physiological processes, including embryonic stem cell differentiation [29], T-cell differentiation [30], keratinocyte differentiation [31], especially, the altered expression of lncRNAs could result in cancer [32].
In the present study, we focused on two lncRNAs, H19 and uc001lsz. The expression level of H19 in gastric cancer tissues was found to be evidently higher than that in non-tumor tissues (Figure 2A and B). Together with the increased expression of H19 in gastric cancer cell lines (Figure 2D), we suggest that H19 may play an important role in gastric cancer pathogenesis. Interesting, H19 expression is not increased in every types of tumor. As showed in Figure 2D, H19 expression is decreased in hepatocarcinoma and prostate cancer. These results further certify that H19 acts not only as an oncogene but also as a tumor suppressor [20,27,33]. Matouk et al. found that the knocking-down of H19 RNA resulted in nearly complete attenuation of p57kip2 induction in response to hypoxic stress [20]. They further found that H19 was associated with angiopoietin (ANG) and fibroblast growth factor-18 (FGF-18), whose functions are involved in tumor growth and proliferation [20]. To understand the molecular mechanism by which H19 increases gastric cancer cell growth, Yang et al. examined whether H19 affects the function of the tumor suppressor p53 [27]. They found that H19 was associated with p53, and that this association resulted in partial p53 inactivation [27].
Contrary to H19, uc001lsz expression level in gastric cancer tissues was found to be markedly lower (Figure 3A). As showed in Figure 3C, the expression of uc001lsz in gastric cancer cell lines (AGS and MGC-803) is lower than that in gastric epithelial cell (GES-1), but there was no significant different between SGC-7901 and GES-1. Maybe the low grade malignancy of SGC-7901 leads to this result. More importantly, a greater association between uc001lsz expression and TNM stage was found (Table 2). These results confirmed uc001lsz as an important player in inhibiting the development of gastric cancer. As we known, many lncRNAs are transcribed close to or within protein-coding loci, which has strengthened the hypothesis that lncRNAs may have cis-acting effects within these loci. But uc001lsz may have trans-acting effect within its adjacent protein-coding loci. MUC2, a member of the mucin protein family of genes, is located next to UC001LSZ. The MUC2 is secreted onto mucosal surfaces, where it is secreted from goblet cells in the epithelial lining into the lumen of the stomach [34]. As reported, MUC2 was high expression in gastric cancer [35]. Although uc001lsz seems playing as tumor suppressor gene in gastric cancer and many other types of tumors, it may play different role in prostate cancer where uc001lsz was highly expressed (Figure 3C).
Molecular tumor biomarkers are vital diagnostic and prognostic tools. Our data show that the expression of uc001lsz was aberrant in early gastric cancer and gastric precancerous lesions (Figure 5A). And the extraordinary changes maybe appear in the precancerous lesions (Figure 5B). This investigation indicates that uc001lsz may be a candidate biomarker of gastric cancer.
Conclusions
In summary, we depict an lncRNA expression profile that associated with gastric cancer. The overexpression of H19 in gastric cancer cell lines and tissues suggests that H19 may be participated in gastric cancer. The reduced expression of uc001lsz in gastric cancer cell lines and tissues, its associations with TNM stage, and its dysregulation in early cancer and precancerous lesions suggest that uc001lsz may have an important role in gastric cancer occurrence and be a potential biomarker for the diagnosis of early gastric cancer.
Abbreviations
ANG: Angiopoietin; CA19-9: Carbohydrate antigen 19-9; CEA: Carcinoembryonic antigen; Ct: Threshold cycle; DC/CIK: Dendritic cell/Cytokine-induced killer; FGF-18: Fibroblast growth factor-18; GACAT1: Gastric cancer-associated transcript 1; H19: The reciprocally imprinted partner of Igf2; HCC: Hepatocellular carcinomas; HOTAIR: HOX transcript antisense RNA; HOX: Homeobox; HULC: Highly upregulated in liver cancer; IGF2: Insulin-like growth factor 2; IGF2-AS: IGF-IR and IGF-IIR antisense; IGF-IR: Insulin-like growth factor type I receptor; IGF-IIR: Insulin-like growth factor type II receptor; lncRNA: Long non-coding RNA; MALAT-1: Metastasis associated lung adenocarcinoma transcript 1; miRNA: microRNA; MUC2: Mucin 2; NCCN: National comprehensive cancer network; ncRNA: Non-coding RNA; PCR: Polymerase chain reaction; p-TEFb: Positive transcription elongation factor-b; qPCR: Quantitative polymerase chain reaction; RN7SK: RNA, 7SK small nuclear; RT: Reverse transcription; SPSS: Statistical program for social sciences; SUMO1P3: Ubiquitin-like modifier (SUMO) 1 pseudogene 3; TNM: Tumor-node-metastasis.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
HS conceived of and carried out experiments, analysed and interpreted data and drafted the manuscript; BX and JG conceived of experiments, analysed and interpreted data and wrote the manuscript; WS, GY, XD, ZL, SZ, TX, YX analysed and interpreted data. All authors read and approval the final manuscript.
Contributor Information
Haojun Song, Email: 061050225@163.com.
Weiliang Sun, Email: msfsyk@163.com.
Guoliang Ye, Email: ndfyygl@163.com.
Xiaoyun Ding, Email: dyyyding@126.com.
Zhong Liu, Email: liuzhong1954@163.com.
Sijie Zhang, Email: zhangsijie0500@sina.com.
Tian Xia, Email: summer_wo@wo.com.cn.
Bingxiu Xiao, Email: xiaobingxiu@nbu.edu.cn.
Yang Xi, Email: xiyang@nbu.edu.cn.
Junming Guo, Email: guojunming@nbu.edu.cn.
Acknowledgements
This work was supported by National Natural Science Foundation of China (No. 81171660), Natural Science Foundation of Ningbo (No. 2012A610207), the Scientific Innovation Team Project of Ningbo (No. 2011B82014), the Project of Key Disciplines in Ningbo (No. XKL11D2127 and No. XKL11D2128), the Outstanding (Postgraduate) Dissertation Growth Foundation of Ningbo University (No. PY2012004), The Postgraduate Innovation Projects of Zhejiang Province (No. YK2011050), and The K. C. Wong Magna Fund in Ningbo University.
References
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Klecka J, Holubec L, Pesta M, Topolcan O, Hora M, Eret V, Finek J, Chottova-Dvorakova M, Babjuk M, Novak K, Stolz J. Differential display code 3 (DD3/PCA3) in prostate cancer diagnosis. Anticancer Res. 2010;30:665–670. [PubMed] [Google Scholar]
- Shen M, Chen W, Yu K, Chen Z, Zhou W, Lin X, Weng Z, Li C, Wu X, Tao Z. The diagnostic value of PCA3 gene-based analysis of urine sediments after digital rectal examination for prostate cancer in a Chinese population. Exp Mol Pathol. 2011;90:97–100. doi: 10.1016/j.yexmp.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Müller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, WU LM, Chen LM, Zheng SS. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- DeBaun MR, Niemitz EL, McNeil DE, Brandenburg SA, Lee MP, Feinberg AP. Epigenetic alterations of H19 and LIT1 distinguish patients with Beckwith-Wiedemann syndrome with cancer and birth defects. Am J Hum Genet. 2002;70:604–611. doi: 10.1086/338934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, Wang Y. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24:652–657. doi: 10.1111/j.1440-1746.2008.05666.x. [DOI] [PubMed] [Google Scholar]
- Zhou H, Guo JM, Lou YR, Zhang XJ, Zhong FD, Jiang Z, Cheng J, Xiao BX. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using microRNA as a marker. J Mol Med (Berl) 2010;88:709–717. doi: 10.1007/s00109-010-0617-2. [DOI] [PubMed] [Google Scholar]
- Han L, Zhang K, Shi Z, Zhang J, Zhu J, Zhu S, Zhang A, Jia Z, Wang G, Yu S, Pu P, Dong L, Kang C. LncRNA profile of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. Int J Oncol. 2012;40:2004–2012. doi: 10.3892/ijo.2012.1413. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Guo J, Xiao B, Miao Y, Huang R, Li D, Zhang Y. Increased expression of miR-421 in human gastric carcinoma and its clinical association. J Gastroenterol. 2010;45:17–23. doi: 10.1007/s00535-009-0135-6. [DOI] [PubMed] [Google Scholar]
- Mei D, Song H, Wang K, Lou Y, Sun W, Liu Z, Ding X, Guo J. Up-regulation of SUMO1 pseudogene 3 (SUMO1P3) in gastric cancer and its clinical association. Med Oncol. 2013;30:709. doi: 10.1007/s12032-013-0709-2. [DOI] [PubMed] [Google Scholar]
- Sun W, Wu Y, Yu X, Liu Y, Song H, Xia T, Xiao B, Guo J. Decreased expression of long noncoding RNA AC096655.1-002 in gastric cancer and its clinical significance. Tumour Biol. 2013. DOI 10.1007/s13277-013-0821-0. [DOI] [PubMed]
- Khachane AN, Harrison PM. Mining mammalian transcript data for functional long non-coding RNAs. PLoS ONE. 2010;5:e10316. doi: 10.1371/journal.pone.0010316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva J, Bardag-Gorce F, French BA, Li J, French SW. The regulation of non-coding RNA expression in the liver of mice fed DDC. Exp Mol Pathol. 2009;87:12–9. doi: 10.1016/j.yexmp.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, Hochberg A, Galun E. The H19 non-coding RNA is essential for human tumor growth. PLoS ONE. 2007;2:e84. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011;39:2119–2128. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder R, Trauner M, Zatloukal K. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- Matouk IJ, Abbasi I, Hochberg A, Galun E, Dweik H, Akkawi M. Highly upregulated in liver cancer noncoding RNA is overexpressed in hepatic colorectal metastasis. Eur J Gastroenterol Hepatol. 2009;21:688–692. doi: 10.1097/MEG.0b013e328306a3a2. [DOI] [PubMed] [Google Scholar]
- Niinuma T, Suzuki H, Nojima M, Nosho K, Yamamoto H, Takamaru H, Yamamoto E, Maruyama R, Nobuoka T, Miyazaki Y, Nishida T, Bamba T, Kanda T, Ajioka Y, Taguchi T, Okahara S, Takahashi H, Nishida Y, Hosokawa M, Hasegawa T, Tokino T, Hirata K, Imai K, Toyota M, Shinomura Y. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 2012;72:1126–1136. doi: 10.1158/0008-5472.CAN-11-1803. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Jin Z, Agarwal R, Ma K, Yang J, Ibrahim S, Olaru AV, David S, Ashktorab H, Smoot DT, Duncan MD, Hutcheon DF, Abraham JM, Meltzer SJ, Mori Y. LARP7 is a potential tumor suppressor gene in gastric cancer. Lab Inves. 2012;92:1013–1019. doi: 10.1038/labinvest.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Lu P, Zhu Z, Xu H, Zhu X. Loss of imprinting of insulin-like growth factor 2 is associated with increased risk of lymph node metastasis and gastric corpus cancer. J Exp Clin Cancer Res. 2009;28:125. doi: 10.1186/1756-9966-28-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]
- Cao WJ, Wu HL, He BS, Zhang YS, Zhang ZY. Analysis of long non-coding RNA expression profiles in gastric cancer. World J Gastroenterol. 2013;19:3658–3664. doi: 10.3748/wjg.v19.i23.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, Sunkin SM, Crowe ML, Grimmond SM, Perkins AC, Mattick JS. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang KC, Dinger ME, Mercer TR, Malquori L, Grimmond SM, Chen W, Mattick JS. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J Immunol. 2009;182:7738–7748. doi: 10.4049/jimmunol.0900603. [DOI] [PubMed] [Google Scholar]
- Mazar J, Sinha S, Dinger ME, Mattick JS, Perera RJ. Protein-coding and non-coding gene expression analysis in differentiating human keratinocytes using a three-dimensional epidermal equivalent. Mol Genet Genomics. 2010;284:1–9. doi: 10.1007/s00438-010-0543-6. [DOI] [PubMed] [Google Scholar]
- Hall PA, Russell SH. New perspectives on neoplasia and the RNA world. Hematol Oncol. 2005;23:49–53. doi: 10.1002/hon.748. [DOI] [PubMed] [Google Scholar]
- Yoshimizu T, Miroglio A, Ripoche MA, Gabory A, Vernucci M, Riccio A, Colnot S, Godard C, Terris B, Jammes H, Dandolo L. The H19 locus acts in vivo as a tumor suppressor. Proc Natl Acad Sci USA. 2008;105:12417–12422. doi: 10.1073/pnas.0801540105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen A, Hutton DA, Pearson JP. The MUC2 gene product: a human intestinal mucin. Int J Biochem Cell Biol. 1998;30:797–801. doi: 10.1016/S1357-2725(98)00028-4. [DOI] [PubMed] [Google Scholar]
- Khattab AZ, Nasif WA, Lotfy M. MUC2 and MUC6 apomucins expression in human gastric neoplasm: an immunohistochemical analysis. Med Oncol. 2011;28:S207–S213. doi: 10.1007/s12032-010-9699-5. [DOI] [PubMed] [Google Scholar]