Abstract
Background
Somatic mutations in PIK3CA (phosphatidylinositol-4,5-bisphosphonate 3-kinase [PI3K], catalytic subunit alpha gene) activate the PI3K-AKT signaling pathway and contribute to pathogenesis of various malignancies, including colorectal cancer.
Methods
We examined associations of PIK3CA oncogene mutation with relapse, survival, and treatment efficacy in 627 stage III colon carcinoma case subjects within a randomized adjuvant chemotherapy trial (5-fluorouracil and leucovorin [FU/LV] vs irinotecan [CPT11], fluorouracil and leucovorin [IFL]; Cancer and Leukemia Group B 89803 [Alliance]). We detected PIK3CA mutation in exons 9 and 20 by polymerase chain reaction and pyrosequencing. Cox proportional hazards model was used to assess prognostic and predictive role of PIK3CA mutation, adjusting for clinical features and status of routine standard molecular pathology features, including KRAS and BRAF mutations and microsatellite instability (mismatch repair deficiency). All statistical tests were two-sided.
Results
Compared with PIK3CA wild-type cases, overall status of PIK3CA mutation positivity or the presence of PIK3CA mutation in either exon 9 or 20 alone was not statistically significantly associated with recurrence-free, disease-free, or overall survival (log-rank P > .70; P > .40 in multivariable regression models). There was no statistically significant interaction between PIK3CA and KRAS (or BRAF) mutation status in survival analysis (P interaction > .18). PIK3CA mutation status did not appear to predict better or worse response to IFL therapy compared with FU/LV therapy (P interaction > .16).
Conclusions
Overall tumor PIK3CA mutation status is not associated with stage III colon cancer prognosis. PIK3CA mutation does not appear to serve as a predictive tumor molecular biomarker for response to irinotecan-based adjuvant chemotherapy.
Phosphatidylinositol-4,5-bisphosphonate 3-kinase (PI3K) can activate the AKT signaling pathway and facilitate cellular growth, proliferation, and survival (1). Activating mutations in PIK3CA (the phosphatidylinositol-4,5-bisphosphonate 3-kinase, catalytic subunit alpha gene; HGNC ID; HGNC:8975) have been found in various human malignancies, including colon cancers (2). A subset (10%–30%) of colorectal cancers harbor PIK3CA mutations, which have been associated with various clinical and molecular features, including proximal tumor location and KRAS mutation (3–16).
Despite many previous studies (10–24), a prognostic role of overall PIK3CA mutation status in colorectal cancer remains uncertain, although coexistence of PIK3CA mutations in both exons 9 and 20 may be associated with shorter survival (13). Most of these previous studies were underpowered for robust statistical analysis (mostly with total sample size of less than 500 and 10%–20% frequency of PIK3CA mutation). Therefore, additional studies with a large sample size are needed.
A recent study suggests that PIK3CA mutation in colorectal cancer may serve as a molecular biomarker to predict response to aspirin therapy (25). Nonetheless, clinical utility of PIK3CA mutation test on colorectal cancer as a predictive tumor biomarker remains to be fully characterized (22,26–29).
We therefore conducted this study to examine prognostic and predictive roles of PIK3CA mutation in stage III colon cancer patients who enrolled in the National Cancer Institute–sponsored randomized clinical trial comparing postoperative adjuvant 5-fluorouracil (FU)/leucovorin (LV) with irinotecan/FU/LV (IFL) (CALGB 89803 [Alliance]) (30). Because data on postoperative treatment, performance status, and disease stage were carefully recorded in the trial, we could assess prognostic and predictive roles of PIK3CA mutations in colon cancer while controlling for potential confounding by those covariables and key molecular characteristics, including KRAS, BRAF, and microsatellite instability (MSI) status. It is important to consider KRAS, BRAF, and MSI status because these are now routinely assessed in colorectal cancer for patient management.
Methods
Study Population
Patients in this study were participants in the National Cancer Institute–sponsored Cancer and Leukemia Group B (CALGB) phase III adjuvant therapy trial for stage III colon cancer comparing therapy with the weekly Roswell Park regimen of FU/LV with weekly bolus regimen of IFL (CALGB 89803; ClinicalTrials.gov Identifier: NCT00003835) (30). CALGB is now part of the Alliance for Clinical Trials in Oncology. Between April 1999 and May 2001, 1264 patients were enrolled. Patients in the treatment trial (and thus this companion study) were eligible if they underwent a complete surgical resection of the primary tumor within 56 days before study entry and had regional lymph node metastases but no evidence of distant metastases (ie, stage III). Considering the colorectal continuum model, we included cases from cecal cancers to sigmoid cancers (31). Moreover, patients were required to have a baseline Eastern Cooperative Oncology Group performance status of 0 to 2 (ambulatory) and have adequate bone marrow, renal, and hepatic function. Cancer staging was based on Tumor Node Metastatis (TNM) classification (http://www.cancer.org). This analysis represents correlative research based on a subset of patients within the trial and was limited to 627 patients for whom archived formalin-fixed paraffin-embedded tumor tissue and PIK3CA sequencing data were available. All patients signed informed consent, approved by each site’s institutional review board.
We compared baseline characteristics of the patients who were included in this study (with available PIK3CA data: n = 627) with those who were excluded from this study because of unavailability of tissue data (n = 637). We did not detect any statistically significant or substantial difference between these two groups in terms of age, sex, body mass index, family history, tumor location, extent of invasion through bowel wall (pT stage), lymph node involvement (pN stage), performance status, clinical bowel perforation, clinical bowel obstruction, or treatment arm (all P > .05). In addition, recurrence-free (RFS), disease-free (DFS), or overall survival (OS) did not statistically significantly differ in subjects with available PIK3CA data as compared with those without PIK3CA data (multivariable hazard ratio [HR] = 1.05, 95% confidence interval [CI] = 0.87 to 1.27; multivariable HR = 1.10, 95% CI = 0.93 to 1.31; multivariable HR = 1.12, 95% CI = 0.93 to 1.36, respectively).
As part of the quality assurance program of the Alliance, members of the Audit Committee visit all participating institutions at least once every 3 years to review source documents. The auditors verify compliance with federal regulations and protocol requirements, including those pertaining to eligibility, treatment, adverse events, tumor response, and outcome in a sample of protocols at each institution. Such on-site review of medical records was performed for a subgroup of 328 (26%) of the 1264 patients included in the treatment trial. Data quality was also ensured by review of data by the Alliance Statistics and Data Center and by the study chairperson following Alliance policies.
Definitions of Study Endpoints
The study endpoints were 1) RFS, defined as the time from the study enrollment to tumor recurrence or occurrence of a new primary colon cancer; 2) DFS, defined as time from the study enrollment to cancer recurrence, occurrence of a new primary colon cancer, or death from any cause; and 3) OS, defined as the time from the study enrollment to death from any cause. For RFS, patients who died without known cancer recurrence were censored at last documented evaluation by a treating provider.
DNA Extraction From Tumor, PIK3CA, BRAF, and KRAS sequencing and MSI Analysis
Tumor molecular analyses were performed blinded to patient and outcome data. DNA was extracted from paraffin-embedded colon cancer tissue (32). We marked tumor areas on hematoxylin and eosin–stained slides and dissected tumor tissue by a sterile needle. Polymerase chain reaction (PCR) and pyrosequencing targeted for mutation hotspots in PIK3CA exons 9 and 20 (13), BRAF codon 600 (33), and KRAS codons 12 and 13 were performed, as previously described (32), in the laboratory at the Dana-Farber Cancer Institute. Pyrosequencing assay has been found to be more sensitive than Sanger sequencing and can detect approximately 5% to 10% of mutant alleles among a mixture of mutant and normal alleles (32). MSI was assessed by PCR for 10 microsatellite markers (BAT25, BAT26, D17S250, D5S346, ACTC, D18S55, BAT40, D10S197, BAT34c4, and MycL) (34). Tumors with instability in 50% or more of the loci were classified as MSI-high, and those with instability in 0% to 49% of the loci were classified as microsatellite stable; and the concordance between MSI testing and immunohistochemistry for MLH1 or MSH2 loss was 97% (34). For 28 cases without PCR MSI results, those with loss of MLH1 or MSH2 were classified as MSI-high, and those with intact expression of MLH1 and MSH2 were classified as microsatellite stable.
Statistical Analyses
Detailed statistical methods are described in the Supplementary Methods (available online). All analyses were based on the clinical study database frozen on November 9, 2009. SAS version 9.2 (SAS Institute, Cary, NC) was used for all statistical analyses, and all P values were two-sided. Statistical significance was set at P equal to .05 for main hypothesis testing on PIK3CA status and outcome. For exploratory analyses of interactions (between PIK3CA mutation and each of the variables, including age, sex, etc.) and clinical, pathological, and molecular associations, we adjusted statistical significance level by Bonferroni correction to P equal to .004 (.05 divided by 13) to account for multiple hypothesis testing. Because a vast majority of participants were non-Hispanic whites, we did not perform analyses stratified by ethnic group.
The Kaplan–Meier method was used to estimate the distribution of survival time according to PIK3CA status, and the log-rank test was used to compare survival between subgroups. We used the multivariable Cox proportional hazards model to estimate survival hazard ratio according to tumor PIK3CA status. We conducted power calculations for survival analyses (Supplementary Table 1, available online). The proportionality of hazards assumption was assessed using standard survival plots and by evaluating a time-dependent variable, which was the cross-product of PIK3CA and survival time (P = .46 for RFS; P = .76 for DFS; P = .35 for OS). To assess the potential differential effect of treatment arm according to PIK3CA status, we performed a single multivariable Cox regression analysis, in which we could estimate the effect of treatment arm simultaneously in two strata of PIK3CA status, using a reparameterization of the interaction term(s) (35). Interaction was also assessed by including the cross-product of PIK3CA and another variable of interest (without data-missing cases) in a multivariable model using the Wald test.
Results
PIK3CA Mutation and Patient Survival in Stage III Colon Cancer
Study participants were drawn from a multicenter study of postoperative adjuvant chemotherapy in stage III colon cancer patients who underwent a curative-intent surgical resection (CALGB 89803) (30). We included 627 case subjects in this study based on availability of tumor tissue for sequencing of PIK3CA exons 9 and 20, which detected mutation in 74 (12%) patients. One case subject showed mutations in both exons 9 and 20. Table 1 summarizes baseline characteristics according to PIK3CA mutation status. Overall prevalence of postdiagnosis regular aspirin use (defined as two or more tablets per week) was 7.2% (n = 45 of 627 patients), and there were only four patients who had PIK3CA-mutated tumor and regularly used aspirin after colon cancer diagnosis.
Table 1.
Baseline characteristics according to PIK3CA mutation status in stage III colon cancer*
| Clinical or molecular feature | Total No. | Tumor PIK3CA status | P (wild-type vs exon 9 mutant vs exon 20 mutant) | P (wild-type vs overall mutant) | |||
|---|---|---|---|---|---|---|---|
| Wild-type | Mutation present in only exon 9† | Mutation present in only exon 20† | Overall mutation positive (exon 9 or 20)‡ | ||||
| Total No. | 627 | 553 | 48 | 25 | 74 | ||
| Sex, No. (%) | .001 | .15 | |||||
| Male | 337 (54) | 303 (55) | 15 (31) | 18 (72) | 34 (46) | ||
| Female | 290 (46) | 250 (45) | 33 (69) | 7 (28) | 40 (54) | ||
| Age, y, mean ± SD | 60.2±11.4 | 60.2±11.4 | 59.0±10.7 | 61.9±12.6 | 60.0±11.4 | .59 | .91 |
| Body mass index, kg/m2, No. (%) | .24 | .74 | |||||
| <25 | 162 (30) | 140 (29) | 19 (42) | 3 (15) | 22 (33) | ||
| 25–29 | 183 (34) | 163 (34) | 12 (27) | 8 (40) | 20 (30) | ||
| ≥30 | 197 (36) | 173 (36) | 14 (31) | 9 (45) | 24 (36) | ||
| Family history of colorectal cancer in any first-degree relative, No. (%) | .23 | .97 | |||||
| No | 454 (83) | 399 (83) | 35 (78) | 19 (95) | 55 (83) | ||
| Yes | 92 (17) | 81 (17) | 10 (22) | 1 (5.0) | 11 (17) | ||
| Tumor location, No. (%) | .78 | .90 | |||||
| Proximal, cecum to transverse colon | 359 (58) | 317 (58) | 26 (55) | 16 (64) | 42 (58) | ||
| Distal, splenic flexure to sigmoid | 258 (42) | 227 (42) | 21 (44) | 9 (36) | 31 (42) | ||
| pT stage, No. (%) | .65 | .31 | |||||
| pT1–pT2 | 75 (12) | 66 (12) | 5 (11) | 3 (12) | 9 (13) | ||
| pT3 | 492 (80) | 438 (81) | 35 (76) | 19 (76) | 54 (75) | ||
| pT4 | 49 (8.0) | 40 (7.4) | 6 (13) | 3 (12) | 9 (13) | ||
| pN stage, No. (%) | .94 | .80 | |||||
| pN1 | 381 (62) | 337 (62) | 28 (60) | 15 (60) | 44 (60) | ||
| pN2 | 237 (38) | 208 (38) | 19 (40) | 10 (40) | 29 (40) | ||
| Performance status score | .20 | .07 | |||||
| 0 | 472 (76) | 410 (75) | 40 (85) | 21 (84) | 62 (85) | ||
| 1–2 | 146 (24) | 135 (25) | 7 (15) | 4 (16) | 11 (15) | ||
| Clinical bowel perforation | .54 | .49 | |||||
| No | 578 (95) | 511 (95) | 42 (91) | 24 (96) | 67 (93) | ||
| Yes | 32 (5.3) | 27 (5.0) | 4 (8.7) | 1 (4.0) | 5 (6.9) | ||
| Clinical bowel obstruction | .85 | .63 | |||||
| No | 480 (78) | 425 (78) | 35 (74) | 19 (76) | 55 (75) | ||
| Yes | 139 (22) | 121 (22) | 12 (26) | 6 (24) | 18 (25) | ||
| Microsatellite instability (MSI) status§, No. (%) | .19 | .70 | |||||
| Microsatellite stable | 470 (81) | 420 (81) | 35 (85) | 14 (67) | 50 (79) | ||
| MSI-high | 109 (19) | 96 (19) | 6 (15) | 7 (33) | 13 (21) | ||
| BRAF mutation status, No. (%) | .07 | .048 | |||||
| Wild-type | 486 (84) | 419 (83) | 45 (96) | 21 (84) | 67 (92) | ||
| Mutant (c.1799T>A, p.V600E) | 94 (16) | 88 (17) | 2 (4.3) | 4 (16) | 6 (8.2) | ||
| KRAS mutation status, No. (%) | .07 | .09 | |||||
| Wild-type | 376 (65) | 335 (66) | 24 (50) | 17 (71) | 41 (56) | ||
| Mutant | 203 (35) | 171 (34) | 24 (50) | 7 (29) | 32 (44) | ||
| Treatment arm, No. (%) | .71 | .87 | |||||
| FU/LV | 319 (51) | 282 (51) | 26 (54) | 11 (44) | 37 (50) | ||
| IFL | 308 (49) | 271 (49) | 22 (46) | 14 (56) | 37 (50) | ||
* The percentages indicate the proportion of tumors with a specific clinical or molecular feature among tumors with specific PIK3CA status. There were cases with missing value/status for some of the variables. A χ2 test was used to assess associations, and all P values were two-sided. FU/LV = 5-fluorouracil and leucovorin; IFL = irinotecan, 5-fluorouracil and leucovorin; SD = standard deviation.
† One case subject with PIK3CA mutations in both exons 9 and 20 was excluded.
‡ This category includes one case subject with mutations in both exons 9 and 20.
§ For 29 cases without MSI results by polymerase chain reaction, those with loss of MLH1 or MSH2 were classified as MSI-high, and those with intact expression of MLH1 and MSH2 as microsatellite stable because concordance between MSI polymerase chain reaction and immunohistochemistry for MLH1 and MSH2 was very high (97%) among case subjects with both results available (34).
With median follow-up of 7.6 (interquartile range = 7.1–8.1) years among those who were censored for overall survival outcome, there were 225 events for RFS analysis, 258 events for DFS analysis, and 210 events for OS analysis.
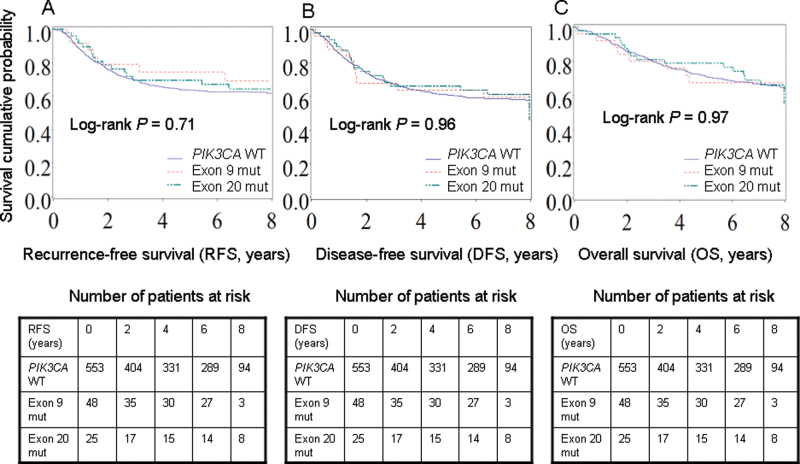
In a Kaplan–Meier analysis (Figure 1), compared with PIK3CA wild-type patients, PIK3CA mutation in either exon 9 or 20 was not statistically significantly associated with RFS, DFS, or OS outcome (log-rank P > .70).
Figure 1.
Kaplan–Meier curves according to PIK3CA mutation in stage III colon cancers for recurrence-free survival (RFS) (A), disease-free survival (DFS) (B), and overall survival (OS) (C). Tables of the numbers of patients at risk are below the graphs.
Exon 9 mut = mutation in only exon 9; Exon 20 mut = mutation in only exon 20; WT = wild-type.
In multivariable Cox regression analysis, we examined the prognostic association of PIK3CA mutation adjusting for other predictors of patient survival (Table 2; Supplementary Table 2, available online, shows data on all variables in the final multivariable models). Compared with PIK3CA wild-type cases, PIK3CA mutation in only exon 9, or only exon 20, or overall PIK3CA mutation status was not statistically significantly associated with RFS, DFS, or OS outcome in univariate or multivariable analysis (P > .40 in multivariable regression models). Multivariable hazard ratios for RFS, DFS, and OS in overall PIK3CA mutated tumors compared with PIK3CA wild-type tumors were 0.84 (95% CI = 0.54 to 1.29), 0.92 (95% CI = 0.62 to 1.35), and 0.95 (95% CI = 0.63 to 1.45), respectively.
Table 2.
PIK3CA mutation status and clinical outcome in stage III colon cancer*
| PIK3CA status | No. | Recurrence-free survival | Disease-free survival | Overall survival | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Five-year survival probability | Univariate HR (95% CI) | Multivariable HR (95% CI) | Five-year survival probability | Univariate HR (95% CI) | Multivariable HR (95% CI) | Five-year survival probability | Univariate HR (95% CI) | Multivariable HR (95% CI) | ||
| Wild-type | 553 | 0.64 | 1 (referent) | 1 (referent) | 0.61 | 1 (referent) | 1 (referent) | 0.71 | 1 (referent) | 1 (referent) |
| Overall mutation positive, exon 9 or 20 † | 74 | 0.71 | 0.84 (0.54 to 1.29) | 0.84 (0.54 to 1.29) | 0.66 | 0.93 (0.63 to 1.37) | 0.92 (0.62 to 1.35) | 0.75 | 0.99 (0.65 to 1.50) | 0.95 (0.63 to 1.45) |
| Mutation in only exon 9‡ | 48 | 0.69 | 0.89 (0.54 to 1.49) | 0.88 (0.53 to 1.46) | 0.66 | 0.94 (0.59 to 1.50) | 0.91 (0.57 to 1.46) | 0.79 | 1.03 (0.63 to 1.70) | 0.98 (0.59 to 1.62) |
| Mutation in only exon 20‡ | 25 | 0.74 | 0.76 (0.36 to 1.62) | 0.76 (0.36 to 1.63) | 0.64 | 0.95 (0.51 to 1.79) | 0.94 (0.50 to 1.78) | 0.67 | 0.93 (0.46 to 1.88) | 0.90 (0.44 to 1.84) |
* The multivariable Cox proportional hazards regression model was used. CI = confidence interval; HR = hazard ratio.
† This category includes one case with mutations in both exons 9 and 20.
‡ One case with PIK3CA mutations in both exons 9 and 20 was excluded.
PIK3CA Mutation and Prognosis in Strata of Combined KRAS and BRAF Status
We assessed a prognostic role of PIK3CA mutation in strata of combined KRAS and BRAF status to examine possible effect modification by KRAS or BRAF status on PIK3CA mutation (Table 3). We did not observe statistically significant effect modification by KRAS status. The number (n = 6) of cases with both PIK3CA and BRAF mutations precluded robust assessment of effect modification by BRAF status. There was no statistically significant interaction between PIK3CA and KRAS (or BRAF) status (P interaction > .18). Multivariable hazard ratios for DFS in PIK3CA mutant tumors compared with PIK3CA wild-type tumors were 1.21 (95% CI = 0.71 to 2.06) among the KRAS wild-type BRAF wild-type subtype and 0.76 (95% CI = 0.40 to 1.43) among the KRAS mutant BRAF wild-type subtype. Five-year survival probabilities for DFS among BRAF wild-type tumors were 0.63, 0.62, 0.60, and 0.68 in KRAS wild-type PIK3CA wild-type; KRAS wild-type PIK3CA mutant; KRAS mutant PIK3CA wild-type; and KRAS mutant PIK3CA mutant subtypes, respectively.
Table 3.
PIK3CA mutation status in strata of combined KRAS and BRAF mutation status and clinical outcome in stage III colon cancer*
| Combined KRAS / BRAF status, and PIK3CA status | No. | Recurrence-free survival | Disease-free survival | Overall survival | |||
|---|---|---|---|---|---|---|---|
| Five-year survival probability | Multivariable HR (95% CI) | Five-year survival probability | Multivariable HR (95% CI) | Five-year survival probability | Multivariable HR (95% CI) | ||
| KRAS wild-type BRAF wild-type | |||||||
| PIK3CA wild-type | 245 | 0.65 | 1 (referent) | 0.63 | 1 (referent) | 0.74 | 1 (referent) |
| PIK3CA mutant | 34 | 0.64 | 1.18 (0.65 to 2.09) | 0.62 | 1.21 (0.71 to 2.06) | 0.73 | 1.30 (0.73 to 2.31) |
| KRAS mutant BRAF wild-type | |||||||
| PIK3CA wild-type | 169 | 0.65 | 1 (referent) | 0.60 | 1 (referent) | 0.69 | 1 (referent) |
| PIK3CA mutant | 32 | 0.78 | 0.60 (0.28 to 1.32) | 0.68 | 0.76 (0.40 to 1.43) | 0.77 | 0.76 (0.38 to 1.53) |
| KRAS wild-type BRAF mutant | |||||||
| PIK3CA wild-type | 87 | 0.54 | 1 (referent) | 0.50 | 1 (referent) | 0.62 | 1 (referent) |
| PIK3CA mutant | 6 | 0.83 | 0.27 (0.04 to 2.01) | 0.83 | 0.25 (0.04 to 1.85) | 0.83 | 0.29 (0.04 to 2.12) |
* The multivariable Cox proportional hazards regression model included the same set of covariables that were selected in the final models of the analyses in Table 2, except for stratifying variables KRAS and BRAF mutation status. Cases with mutations in both KRAS and BRAF were excluded. CI = confidence interval; HR = hazard ratio.
Predictive Role of PIK3CA Mutation for IFL-Based Therapy
We assessed the prognostic role of PIK3CA mutation within each treatment arm and the effect of treatment according to PIK3CA status (Table 4). In either treatment arm, PIK3CA mutation was not statistically significantly associated with RFS, DFS, or OS outcome. Multivariable hazard ratios for DFS in PIK3CA mutant tumors compared with PIK3CA wild-type tumors were 0.75 (95% CI = 0.43 to 1.31) among the FU/LV arm and 1.13 (95% CI = 0.65 to 1.96) among the IFL arm.
Table 4.
Stage III colon cancer and clinical outcome according to treatment arm and PIK3CA mutation status*
| Treatment arm and tumor PIK3CA status | No. | Recurrence-free survival | Disease-free survival | Overall survival | |||
|---|---|---|---|---|---|---|---|
| Five-year survival probability | Multivariable HR (95% CI) | Five-year survival probability | Multivariable HR (95% CI) | Five-year survival probability | Multivariable HR (95% CI) | ||
| FU/LV | |||||||
| PIK3CA wild-type | 282 | 0.64 | 1 (referent) | 0.62 | 1 (referent) | 0.72 | 1 (referent) |
| PIK3CA mutant | 37 | 0.71 | 0.70 (0.37 to 1.31) | 0.64 | 0.75 (0.43 to 1.31) | 0.77 | 0.72 (0.39 to 1.32) |
| IFL | |||||||
| PIK3CA wild-type | 271 | 0.63 | 1 (referent) | 0.60 | 1 (referent) | 0.71 | 1 (referent) |
| PIK3CA mutant | 37 | 0.72 | 1.00 (0.55 to 1.84) | 0.67 | 1.13 (0.65 to 1.96) | 0.73 | 1.30 (0.72 to 2.34) |
| P interaction (PIK3CA and treatment arm)† | — | .36 | — | .27 | — | .17 | |
| PIK3CA wild-type | |||||||
| FU/LV | 282 | 0.64 | 1 (referent) | 0.62 | 1 (referent) | 0.72 | 1 (referent) |
| IFL | 271 | 0.63 | 0.90 (0.68 to 1.19) | 0.60 | 0.88 (0.68 to 1.15) | 0.71 | 0.84 (0.62 to 1.12) |
| PIK3CA mutant | |||||||
| FU/LV | 37 | 0.71 | 1 (referent) | 0.64 | 1 (referent) | 0.77 | 1 (referent) |
| IFL | 37 | 0.72 | 1.29 (0.56 to 2.95) | 0.67 | 1.33 (0.64 to 2.78) | 0.73 | 1.51 (0.68 to 3.34) |
* The multivariable Cox proportional hazards regression model included the same set of covariables that were selected in the final models of the analyses in Table 2. CI = confidence interval; FU/LV = 5-fluorouracil and leucovorin; HR = hazard ratio; IFL = irinotecan, 5-fluorouracil and leucovorin.
† Interaction was assessed by including the cross-product of PIK3CA and treatment arm variables in a multivariable model, and the Wald test was used. All P values are two-sided.
In either stratum of patients with PIK3CA mutated or wild-type tumors, IFL treatment was not statistically significantly associated with RFS, DFS, or OS outcome compared with FU/LV treatment (Table 4), and there was no statistically significant interaction between treatment arm and PIK3CA status (P interaction > .16). Multivariable hazard ratios for DFS in the IFL treatment group compared with the FU/LV treatment group were 0.88 (95% CI = 0.68 to 1.15) among the PIK3CA wild-type subtype and 1.33 (95% CI = 0.64 to 2.78) among the PIK3CA mutant subtype.
Interaction Analysis Between PIK3CA and Other Variables
In exploratory analyses, we further examined whether prognostic association of PIK3CA mutation was modified by any other variables, including clinical features and MSI status. We did not observe statistically significant or appreciable effect modification by any of the variables examined for RFS, DFS, or OS outcomes (all P interaction > .04; given multiple hypothesis testing, a P value for statistical significance was adjusted to P = .004).
Discussion
In this study, we found that tumor PIK3CA mutation was not statistically significantly associated with recurrence or survival among more than 600 stage III colon cancer patients, who participated in the randomized trial comparing postoperative IFL with FU/LV (CALGB 89803). We found no evidence for a predictive role of PIK3CA mutation status in IFL-based treatment.
It is a challenge to optimize treatment decision-making for patients with colon cancer because of heterogeneity of colon cancer with regard to both biology and clinical response (36). Heterogeneity exists in tumors with different mutations, even in one oncogene such as KRAS (37, 38), and treatment and other host factors can influence (or can be influenced by) tumor characteristics through tumor microenvironment (and vice versa) (39–43). Therefore, studies examining tumor biomarkers and clinical outcome are crucial in colon cancer research (44–49).
Although previous studies (34,50–55) have assessed potential predictive roles of germline genetic or tumor tissue markers for IFL-based chemotherapy, no biomarker has been proven to be useful in predicting response or resistance to IFL-based therapy (54). A previous analysis of patients in the clinical trial studied in this report suggested that MSI-high might predict an improved patient outcome for treatment with IFL relative to FU/LV (34); however, an independent trial of more than 1200 case subjects comparing IFL vs FU/LV failed to prove a predictive value of MSI status (53). Additional studies are necessary to identify and validate predictors for IFL-based chemotherapy against colorectal cancer.
Several other predictive roles for PIK3CA mutation in colorectal cancer have been examined (22,26–28,56). A recent study (25) suggests that PIK3CA mutation in colorectal cancer may predict response to aspirin treatment, and this finding appears to be replicated (56). Challenges include relatively low prevalence (10%–20%) of PIK3CA mutations, possible differential effects of exon 9 and 20 mutations, and potential confounding effect by associated KRAS mutations. A meta-analysis suggested that PIK3CA exon 20 mutation in metastatic colorectal cancer may predict response to anti-EGFR therapy (57). Finally, a recent preclinical study suggested that PIK3CA mutant colorectal cancer may respond to the SRC inhibitor saracatinib (58). Thus, it is possible that PIK3CA mutation status may serve as a predictive biomarker for a number of different therapeutic agents and their combinations, which include aspirin and other drugs. Additional large-scale trials are needed to define predictive roles of PIK3CA mutations in colorectal cancer.
Studies have examined the prognostic significance of PIK3CA mutations in colorectal cancer (10–24). Although two studies linked tumor PIK3CA mutation to poor prognosis (10,17), statistical power in these reports was limited (each total sample size was less than 160). In one study (n = 586) (6), the presence of a mutation in any one of KRAS, BRAF, and PIK3CA oncogenes was associated with inferior prognosis; however, this could be purely because of the effects of BRAF mutations (59–62). Two other studies showed that PIK3CA exon 20 mutations were associated with poor prognosis, whereas PIK3CA exon 9 mutations were not associated with prognosis (20,23). Other reports showed that PIK3CA mutations were associated with neither distant metastasis to liver (63) nor patient survival (11,12,21). The two largest prognostic studies [n = 2091 (14), and n = 1170 (13)] did not support a prognostic role of overall PIK3CA mutation status. In our analysis, neither PIK3CA overall mutation status nor PIK3CA mutation in exon 9 or 20 alone was statistically significantly associated with tumor recurrence or overall survival. Our study further underscores the importance of large, multicenter, collaborative studies; it should be noted that small, underpowered studies with null findings experience higher likelihood of being unwritten and unpublished when compared with small studies with “statistically significant” results. Well-designed, large-scale studies with appropriate statistical power can be published irrespective of positive or null findings, hence being less susceptible to “publication bias.” Thus, more weight should be placed on the data from large-scale studies upon evaluation of published data on the prognostic significance of tumor biomarkers (13).
Interestingly, Liao et al. (13) demonstrated that the coexistence of PIK3CA mutations in both exons 9 and 20 was associated with shorter survival. Experimental evidence suggests that PIK3CA helical and kinase domain mutations differentially activate protein function and that the presence of PIK3CA mutations in both exon 9 and exon 20 results in a synergistic gain of enzymatic function (64). However, in our analysis, there was only one case subject with PIK3CA mutations in both exons 9 and 20, which precluded robust outcome assessment.
A ligand–receptor interaction of EGFR (HGNC ID: HGNC:3236) leads to activation of two main signal transduction pathways, RAS-RAF-MAPK and PI3K-AKT. Activating mutations in KRAS, BRAF, and/or PIK3CA are well-known carcinogenic mechanisms, and there may be interactive effects of KRAS and PIK3CA mutations (65). In our study on colon cancer, we did not observe statistically significant interactive effects of PIK3CA and KRAS (or BRAF) mutations.
This study used the multi-institutional clinical trial of adjuvant chemotherapy and had several strengths. All study subjects were stage III cancer patients, which decreased potential residual confounding by disease stage. Methods of follow-up and treatment were standardized, and the date and nature of recurrence were recorded. Furthermore, integrative database of treatment, behavioral and lifestyle factors, tumor molecular characteristics (including PIK3CA, KRAS, BRAF, and MSI status), and clinical outcomes enable molecular pathological epidemiology research (66,67) and controlling for confounding by lifestyle factors. The paradigm of molecular pathological epidemiology has been widely used (68–76). In colorectal cancer, KRAS, BRAF, and MSI tests are a part of routine clinical practice (77), and PIK3CA test is an emerging clinical test (25,29,56).
We recognize limitations of our study. Patients who enrolled in the clinical trials constituted a selected group of individuals and might differ from the general population. Patients needed to meet enrollment criteria and be motivated to participate and were further selected based on availability of tissue specimens. Nonetheless, demographic, clinical, or prognostic data of the patients selected in this study did not substantially differ from those without available tumor tissue. Because this trial included patients from both academic and community hospitals across the United States and Canada, our results might reflect stage III colon cancers in the general North American population. Finally, because PIK3CA status was not available on all patients, statistical power was attenuated, especially for predictive assessment for response to IFL use.
In conclusion, our current study of stage III colon cancer patients has shown that tumor PIK3CA mutation is not statistically significantly associated with recurrence or survival and that PIK3CA mutation status is not a predictive marker for response to IFL-based chemotherapy. Additional large-scale studies are needed to define predictive roles of PIK3CA mutations in colon cancer.
Funding
The research for CALGB 89803 (Alliance) (ClinicalTrials.gov Identifier: NCT00003835) was supported, in part, by grants from the National Institute of Health (CA031946) to Alliance for Clinical Trials in Oncology (Monica M. Bertagnolli, MD, Chairperson) and to the Alliance Statistics and Data Center (Daniel J. Sargent, PhD, CA33601), as well as support from Pharmacia & Upjohn Company, now Pfizer Oncology. S.O., K.N., D.S., J.A.M. and C.S.F. were supported in part by grants from the National Institute of Health (grant numbers R01 CA151993 [to SO], K07 CA148894 [to KN], R01 CA149222 [to JAM], R01 CA118553 [to CSF], P50 CA127003 [to CSF]). Other authors were supported in part by the following grants: CA33601 (DN); CA77651 (LBS); CO15 (RW); CA32102 (AH); CA46282 (AH); CA17145 (ABB); CA21115 (ABB); CA35415 (RBM); CA077658 (RMG); and CA031946 (MMB).
Supplementary Material
The sponsors did not participate in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institute of Health.
R.Whittom has received honorarium from the speakers bureau of Hoffmann-La Roche and has served as a consultant to Eli-Lilly, Amgen, Novartis, Pfizer, and Boehringer Ingelheim. A. Hantel is a member of Foundation Medicine Advisory Board. A. B. Benson III has received research funding from Pfizer, Imclone, Bristol Myer Squibb, Amgen, and Sanofi Aventis and served as a scientific advisor for Pfizer, Imclone, Bristol Myer Squibb, Amgen, and Sanofi Aventis. R. M. Goldberg has received research funding from Bayer and Sanofi-Aventis and served as a consultant to Bayer, Jennerex, and Sanofi-Aventis. J. A. Meyerhardt has served as a consultant to Bayer. C. S. Fuchs has served as a consultant to Sanofi-Aventis, Pfizer, Genentech, Roche, Bristol Myers Squibb, and Amgen. All other authors declare no conflicts of interest.
We would like to thank the CALGB Pathology Coordinating Office at the Ohio State University for banking and preparing the materials for the study. The following institutions participated in this study: Baptist Cancer Institute CCOP, Memphis, TN (Lee S. Schwartzberg, MD, supported by CA71323); Christiana Care Health Services, Inc. CCOP, Wilmington, DE (Stephen Grubbs, MD, supported by CA45418); Dana-Farber Cancer Institute, Boston, MA (Eric P. Winer, MD, supported by CA32291); Dartmouth Medical School–Norris Cotton Cancer Center, Lebanon, NH (Marc S. Ernstoff, MD, supported by CA04326); Duke University Medical Center, Durham, NC (Jeffrey Crawford, MD, supported by CA47577); Georgetown University Medical Center, Washington, DC (Minetta C. Liu, MD, supported by CA77597); Cancer Centers of the Carolinas, Greenville, SC (Jeffrey K. Giguere, MD, supported by CA29165); Hematology-Oncology Associates of Central New York CCOP, Syracuse, NY (Jeffrey Kirshner, MD, supported by CA45389); Long Island Jewish Medical Center, Lake Success, NY (Kanti R. Rai, MD, supported by CA11028); Massachusetts General Hospital, Boston, MA (Jeffrey W. Clark, MD, supported by CA12449); Memorial Sloan-Kettering Cancer Center, New York, NY (Clifford A. Hudis, MD, supported by CA77651); Missouri Baptist Medical Center, St. Louis, MO (Alan P. Lyss, MD, supported by CA114558-02); Mount Sinai Medical Center, Miami, FL (Rogerio C. Lilenbaum, MD, supported by CA45564); Mount Sinai School of Medicine, New York, NY (Lewis R. Silverman, MD, supported by CA04457); Nevada Cancer Research Foundation CCOP, Las Vegas, NV (John A. Ellerton, MD, supported by CA35421); North Shore-Long Island Jewish Health System, New Hyde Park, NY (Daniel Budman, MD, supported by CA35279); Rhode Island Hospital, Providence, RI (William Sikov, MD, supported by CA08025); Roswell Park Cancer Institute, Buffalo, NY (Ellis Levine, MD, supported by CA02599); Southeast Cancer Control Consortium Inc. CCOP, Goldsboro, NC (James N. Atkins, MD, supported by CA45808); State University of New York Upstate Medical University, Syracuse, NY (Stephen L. Graziano, MD, supported by CA21060); The Ohio State University Medical Center, Columbus, OH (Clara D Bloomfield, MD, supported by CA77658); University of California at San Diego, San Diego, CA (Barbara A. Parker, MD, supported by CA11789); University of California at San Francisco, San Francisco, CA (Alan P. Venook, MD, supported by CA60138); University of Chicago, Chicago, IL (Gini Fleming, MD, supported by CA41287); University of Illinois MBCCOP, Chicago, IL (Lawrence E. Feldman, MD, supported by CA74811); University of Iowa, Iowa City, IA (Daniel A. Vaena, MD, supported by CA47642); University of Maryland Greenebaum Cancer Center, Baltimore, MD (Martin Edelman, MD, supported by CA31983); University of Massachusetts Medical School, Worcester, MA (William V. Walsh, MD, supported by CA37135); University of Minnesota, Minneapolis, MN (Bruce A Peterson, MD, supported by CA16450); University of Missouri/Ellis Fischel Cancer Center, Columbia, MO (Michael C Perry, MD, supported by CA12046); University of Nebraska Medical Center, Omaha, NE (Anne Kessinger, MD, supported by CA77298); University of North Carolina at Chapel Hill, Chapel Hill, NC (Thomas C. Shea, MD, supported by CA47559); University of Tennessee Memphis, Memphis, TN (Harvey B. Niell, MD, supported by CA47555); University of Vermont, Burlington, VT (Hyman B. Muss, MD, supported by CA77406); Wake Forest University School of Medicine, Winston-Salem, NC (David D Hurd, MD, supported by CA03927); Walter Reed Army Medical Center, Washington, DC (Thomas Reid, MD, supported by CA26806); Washington University School of Medicine, St. Louis, MO (Nancy Bartlett, MD, supported by CA77440); Weill Medical College of Cornell University, New York, NY (John Leonard, MD, supported by CA07968).
References
- 1. Samuels Y, Diaz LA, Jr, Schmidt-Kittler O, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7(6):561–573. [DOI] [PubMed] [Google Scholar]
- 2. Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. [DOI] [PubMed] [Google Scholar]
- 3. Velho S, Oliveira C, Ferreira A, et al. The prevalence of PIK3CA mutations in gastric and colon cancer. Eur J Cancer. 2005;41(11):1649–1654. [DOI] [PubMed] [Google Scholar]
- 4. Baldus SE, Schaefer KL, Engers R, Hartleb D, Stoecklein NH, Gabbert HE. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res. 2010;16(3):790–799. [DOI] [PubMed] [Google Scholar]
- 5. Benvenuti S, Frattini M, Arena S, et al. PIK3CA cancer mutations display gender and tissue specificity patterns. Hum Mutat. 2008;29(2):284–288. [DOI] [PubMed] [Google Scholar]
- 6. Barault L, Veyries N, Jooste V, et al. Mutations in the RAS-MAPK, PI(3)K (phosphatidylinositol-3-OH kinase) signaling network correlate with poor survival in a population-based series of colon cancers. Int J Cancer. 2008;122(10):2255–2259. [DOI] [PubMed] [Google Scholar]
- 7. Yamauchi M, Morikawa T, Kuchiba A, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61(6):847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Whitehall VL, Rickman C, Bond CE, et al. Oncogenic PIK3CA mutations in colorectal cancers and polyps. Int J Cancer. 2012;131(4):813–820. [DOI] [PubMed] [Google Scholar]
- 9. Voutsina A, Tzardi M, Kalikaki A, et al. Combined analysis of KRAS and PIK3CA mutations, MET and PTEN expression in primary tumors and corresponding metastases in colorectal cancer. Mod Pathol. 2013;26(2):302–313. [DOI] [PubMed] [Google Scholar]
- 10. Kato S, Iida S, Higuchi T, et al. PIK3CA mutation is predictive of poor survival in patients with colorectal cancer. Int J Cancer. 2007;121(8):1771–1778. [DOI] [PubMed] [Google Scholar]
- 11. Abubaker J, Bavi P, Al-Harbi S, et al. Clinicopathological analysis of colorectal cancers with PIK3CA mutations in Middle Eastern population. Oncogene. 2008;27(25):3539–3545. [DOI] [PubMed] [Google Scholar]
- 12. Souglakos J, Philips J, Wang R, et al. Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. Br J Cancer. 2009;101(3):465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liao X, Morikawa T, Lochhead P, et al. Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res. 2012;18(8):2257–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gavin P, Colangelo LH, Fumagalli D, et al. Mutation profiling and microsatellite instability in stage II and III colon cancer: an assessment of their prognostic and oxaliplatin predictive value. Clin Cancer Res. 2012;18(23):6531–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rosty C, Young JP, Walsh MD, et al. PIK3CA activating mutation in colorectal carcinoma: associations with molecular features and survival. PLoS One. 2013;8(6):e65479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Day FL, Jorissen RN, Lipton L, et al. PIK3CA and PTEN gene and exon mutation-specific clinicopathological and molecular associations in colorectal cancer. Clin Cancer Res. 2013;19(12):3285–3296. [DOI] [PubMed] [Google Scholar]
- 17. Sartore-Bianchi A, Martini M, Molinari F, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69(5):1851–1857. [DOI] [PubMed] [Google Scholar]
- 18. Ogino S, Nosho K, Kirkner GJ, et al. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol. 2009;27(9):1477–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. He Y, Van’t Veer LJ, Mikolajewska-Hanclich I, et al. PIK3CA mutations predict local recurrences in rectal cancer patients. Clin Cancer Res. 2009;15(22):6956–6962. [DOI] [PubMed] [Google Scholar]
- 20. De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11(8):753–762. [DOI] [PubMed] [Google Scholar]
- 21. Tol J, Dijkstra JR, Klomp M, et al. Markers for EGFR pathway activation as predictor of outcome in metastatic colorectal cancer patients treated with or without cetuximab. Eur J Cancer. 2010;46(11):1997–2009. [DOI] [PubMed] [Google Scholar]
- 22. Prenen H, De Schutter J, Jacobs B, et al. PIK3CA mutations are not a major determinant of resistance to the epidermal growth factor receptor inhibitor cetuximab in metastatic colorectal cancer. Clin Cancer Res. 2009;15(9):3184–3188. [DOI] [PubMed] [Google Scholar]
- 23. Farina Sarasqueta A, Zeestraten EC, van Wezel T, et al. PIK3CA kinase domain mutation identifies a subgroup of stage III colon cancer patients with poor prognosis. Cell Oncol (Dordr). 2011;34(6):523–531. [DOI] [PubMed] [Google Scholar]
- 24. Phipps AI, Makar KW, Newcomb PA. Descriptive profile of PIK3CA-mutated colorectal cancer in postmenopausal women [published online ahead of print June 1, 2013]. Int J Colorectal Dis. 2013. 10.1007/s00384-00013-01715-00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liao X, Lochhead P, Nishihara R, et al. Aspirin use, tumor PIK3CA mutation status, and colorectal cancer survival. N Engl J Med. 2012;367(17):1596–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. De Roock W, Vriendt VD, Normanno N, Ciardiello F, Tejpar S. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol. 2011;12(6):594–603. [DOI] [PubMed] [Google Scholar]
- 27. Lievre A, Blons H, Laurent-Puig P. Oncogenic mutations as predictive factors in colorectal cancer. Oncogene. 2010;29(21):3033–3043. [DOI] [PubMed] [Google Scholar]
- 28. Febbo PG, Ladanyi M, Aldape KD, et al. NCCN Task Force report: evaluating the clinical utility of tumor markers in oncology. J Natl Compr Canc Netw. 2011;9(Suppl 5):S1–32; quiz S33. [DOI] [PubMed] [Google Scholar]
- 29. Ogino S, Lochhead P, Giovannucci E, Meyerhardt JA, Fuchs CS, Chan AT. Discovery of colorectal cancer PIK3CA mutation as potential predictive biomarker: power and promise of molecular pathological epidemiology [published online ahead of print June 24, 2013]. Oncogene. 2013; 10.1038/onc.2013.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saltz LB, Niedzwiecki D, Hollis D, et al. Irinotecan fluorouracil plus leucovorin is not superior to fluorouracil plus leucovorin alone as adjuvant treatment for stage III colon cancer: results of CALGB 89803. J Clin Oncol. 2007;25(23):3456–3461. [DOI] [PubMed] [Google Scholar]
- 31. Yamauchi M, Lochhead P, Morikawa T, et al. Colorectal cancer: a tale of two sides or a continuum? Gut. 2012;61(6):794–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ogino S, Kawasaki T, Brahmandam M, et al. Sensitive sequencing method for KRAS mutation detection by pyrosequencing. J Mol Diagn. 2005;7(3):413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8(5):582–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bertagnolli MM, Niedzwiecki D, Compton CC, et al. Microsatellite instability predicts improves response to adjuvant therapy with irinotecan, fluorouracil, and leucovorin in stage III colon cancer: Cancer and Leukemia Group B protocol 89803. J Clin Oncol. 2009;27 ( 11):1814–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nosho K, Irahara N, Shima K, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS ONE. 2008;3(11):e3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ogino S, Fuchs CS, Giovannucci E. How many molecular subtypes? Implications of the unique tumor principle in personalized medicine. Expert Rev Mol Diagn. 2012;12(6):621–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Imamura Y, Morikawa T, Liao X, et al. Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF-wild-type colorectal cancers. Clin Cancer Res. 2012;18(17):4753–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen CC, Er TK, Liu YY, et al. Computational analysis of KRAS mutations: implications for different effects on the KRAS p.G12D and p.G13D mutations. PLoS One. 2013;8(2):e55793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Straussman R, Morikawa T, Shee K, et al. Tumor microenvironment contributes to innate RAF-inhibitor resistance through HGF secretion. Nature. 2012;487(7408):500–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ogino S, Galon J, Fuchs CS, Dranoff G. Cancer immunology—analysis of host and tumor factors for personalized medicine. Nat Rev Clin Oncol. 2011;8(12):711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dahlin AM, Henriksson ML, Van Guelpen B, et al. Colorectal cancer prognosis depends on T-cell infiltration and molecular characteristics of the tumor. Mod Pathol. 2011;24(5):671–682. [DOI] [PubMed] [Google Scholar]
- 42. Nosho K, Baba Y, Tanaka N, et al. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer and prognosis: cohort study and literature review. J Pathol. 2010;222(4):350–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Galon J, Franck P, Marincola FM, et al. Cancer classification using the Immunoscore: a worldwide task force. J Transl Med. 2012;10(1):205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8(12):686–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Goel A, Boland CR. Epigenetics of colorectal cancer. Gastroenterology. 2012;143(6):1442–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Colussi D, Brandi G, Bazzoli F, Ricciardiello L. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. Int J Mol Sci. 2013;14:16365–16385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bardhan K, Liu K. Epigenetics and colorectal cancer pathogenesis. Cancers. 2013;5:676–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kunzmann AT, Murray LL, Cardwell CR, McShane CM, McMenamin UC, Cantwell MM. PTGS2 (Cyclooxygenase-2) expression and survival amongst colorectal cancer patients: a systematic review. Cancer Epidemiol Biomarkers Prev. 2013;22(9):1490–1497. [DOI] [PubMed] [Google Scholar]
- 49. Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58(1):90–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dopeso H, Mateo-Lozano S, Elez E, et al. Aprataxin tumor levels predict response of colorectal cancer patients to irinotecan-based treatment. Clin Cancer Res. 2010;16(8):2375–2382. [DOI] [PubMed] [Google Scholar]
- 51. Glimelius B, Garmo H, Berglund A, et al. Prediction of irinotecan and 5-fluorouracil toxicity and response in patients with advanced colorectal cancer. Pharmacogenomics J. 2011;11(1):61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vallbohmer D, Iqbal S, Yang DY, et al. Molecular determinants of irinotecan efficacy. Int J Cancer. 2006;119(10):2435–2442. [DOI] [PubMed] [Google Scholar]
- 53. Tejpar S, Bosman F, Delorenzi M, et al. Microsatellite instability (MSI) in stage II and III colon cancer treated with 5FU-LV or 5FU-LV and irinotecan (PETACC 3-EORTC 40993-SAKK 60/00 trial). J Clin Oncol. 2009;27(15s):abstract 4001. [Google Scholar]
- 54. Ogino S, Shima K, Meyerhardt J, et al. Predictive and prognostic roles of BRAF mutation in stage III colon cancer: results from intergroup trial CALGB 89803. Clin Cancer Res. 2012;18(3):890–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ogino S, Meyerhardt JA, Irahara N, et al. KRAS mutation in stage III colon cancer and clinical outcome following intergroup trial CALGB 89803. Clin Cancer Res. 2009;15(23):7322–7329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Domingo E, Church DN, Sieber O, et al. Evaluation of PIK3CA mutation as a predictor of benefit from NSAID therapy in colorectal cancer. J Clin Oncol. 2013. [DOI] [PubMed] [Google Scholar]
- 57. Mao C, Yang ZY, Hu XF, Chen Q, Tang JL. PIK3CA exon 20 mutations as a potential biomarker for resistance to anti-EGFR monoclonal antibodies in KRAS wild-type metastatic colorectal cancer: a systematic review and meta-analysis. Ann Oncol. 2012;23(6):1518–1525. [DOI] [PubMed] [Google Scholar]
- 58. Arcaroli JJ, Quackenbush KS, Powell RW, et al. Common PIK3CA mutants and a novel 3’ UTR mutation are associated with increased sensitivity to saracatinib. Clin Cancer Res. 2012;18(9):2704–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Phipps AI, Buchanan DD, Makar KW, et al. BRAF mutation status and survival after colorectal cancer diagnosis according to patient and tumor characteristics. Cancer Epidemiol Biomarkers Prev. 2012;21(10):1792–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zlobec I, Bihl MP, Schwarb H, Terracciano L, Lugli A. Clinicopathological and protein characterization of BRAF- and K-RAS-mutated colorectal cancer and implications for prognosis. Int J Cancer. 2010;127(2):367–380. [DOI] [PubMed] [Google Scholar]
- 61. Popovici V, Budinska E, Tejpar S, et al. Identification of a poor-prognosis BRAF-mutant-like population of patients with colon cancer. J Clin Oncol. 2012;30(12):1288–1295. [DOI] [PubMed] [Google Scholar]
- 62. Lochhead P, Kuchiba A, Imamura Y, et al. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J Natl Cancer Inst. 2013;105(15):1151–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bruin SC, He Y, Mikolajewska-Hanclich I, et al. Molecular alterations associated with liver metastases development in colorectal cancer patients. Br J Cancer. 2011;105(2):281–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A. 2008;105(7):2652–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kim A, Lee JE, Lee SS, et al. Coexistent mutations of KRAS and PIK3CA affect the efficacy of NVP-BEZ235, a dual PI3K/MTOR inhibitor, in regulating the PI3K/MTOR pathway in colorectal cancer. Int J Cancer. 2013;133(4):984–996. [DOI] [PubMed] [Google Scholar]
- 66. Ogino S, Stampfer M. Lifestyle factors and microsatellite instability in colorectal cancer: the evolving field of molecular pathological epidemiology. J Natl Cancer Inst. 2010;102(6):365–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60(3):397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Curtin K, Slattery ML, Samowitz WS. CpG island methylation in colorectal cancer: past, present and future. Pathol Res Int. 2011;2011:902674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hughes LA, Khalid-de Bakker CA, Smits KM, et al. The CpG island methylator phenotype in colorectal cancer: progress and problems. Biochim Biophys Acta. 2012;1825(1):77–85. [DOI] [PubMed] [Google Scholar]
- 70. Ku CS, Cooper DN, Wu M, et al. Gene discovery in familial cancer syndromes by exome sequencing: prospects for the elucidation of familial colorectal cancer type X. Mod Pathol. 2012;25(8):1055–1068. [DOI] [PubMed] [Google Scholar]
- 71. Gay LJ, Mitrou PN, Keen J, et al. Dietary, lifestyle and clinico-pathological factors associated with APC mutations and promoter methylation in colorectal cancers from the EPIC-Norfolk Study. J Pathol. 2012;228(3):405–415. [DOI] [PubMed] [Google Scholar]
- 72. Chia WK, Ali R, Toh HC. Aspirin as adjuvant therapy for colorectal cancer-reinterpreting paradigms. Nat Rev Clin Oncol. 2012;9(10):561–570. [DOI] [PubMed] [Google Scholar]
- 73. Spitz MR, Caporaso NE, Sellers TA. Integrative cancer epidemiology—the next generation. Cancer Disc. 2012;2(12):1087–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rosty C, Young JP, Walsh MD, et al. Colorectal carcinomas with KRAS mutation are associated with distinctive morphological and molecular features. Mod Pathol. 2013;26(6):825–834. [DOI] [PubMed] [Google Scholar]
- 75. Buchanan DD, Win AK, Walsh MD, et al. Family history of colorectal cancer in BRAF p.V600E mutated colorectal cancer cases. Cancer Epidemiol Biomarkers Prev. 2013;22(5):917–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Burnett-Hartman AN, Newcomb PA, Potter JD, et al. Genomic aberrations occuring in subsets of serrated colorectal lesions but not conventional adenomas. Cancer Res. 2013;73(9):2863–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Funkhouser WK, Lubin IM, Monzon FA, et al. Relevance, pathogenesis, and testing algorithm for mismatch repair-defective colorectal carcinomas: a report of the Association for Molecular Pathology. J Mol Diagn. 2012;14(2):91–103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.