Abstract
Autism is a syndrome characterized by deficits in language and social skills and by repetitive behaviors. We hypothesized that potential quantitative trait loci (QTLs) related to component autism endophenotypes might underlie putative or significant regions of autism linkage. We performed nonparametric multipoint linkage analyses, in 152 families from the Autism Genetic Resource Exchange, focusing on three traits derived from the Autism Diagnostic Interview: “age at first word,” “age at first phrase,” and a composite measure of “repetitive and stereotyped behavior.” Families were genotyped for 335 markers, and multipoint sib pair linkage analyses were conducted. Using nonparametric multipoint linkage analysis, we found the strongest QTL evidence for age at first word on chromosome 7q (nonparametric test statistic [Z] 2.98; P=.001), and subsequent linkage analyses of additional markers and association analyses in the same region supported the initial result (Z=2.85, P=.002; χ2=18.84, df 8, P=.016). Moreover, the peak fine-mapping result for repetitive behavior (Z=2.48; P=.007) localized to a region overlapping this language QTL. The putative autism-susceptibility locus on chromosome 7 may be the result of separate QTLs for the language and repetitive or stereotyped behavior deficits that are associated with the disorder.
Introduction
Autism [MIM accession number 209850] is a neurodevelopmental disorder characterized by language impairments, social and communicative deficits, and repetitive behaviors. The disorder has an onset at <3 years of age and affects 1 per 2,500 individuals (Smalley et al. 1988; Folstein et al. 1998), although this may be an underestimate (Bryson and Smith 1998). The etiology of autism has a genetic component (Bailey et al. 1995) with an unknown mode of inheritance. Twin and family studies suggest that >90% of the risk for the disorder may be due to genes (Bailey et al. 1995), and a sibling of an autistic individual is ∼100 times as likely to have an autism-spectrum disorder compared with an unrelated person (Smalley et al. 1988).
Despite the high heritability of autism, the positional identification of candidate loci has been complicated by several issues, the most important of which is genetic heterogeneity. Formal latent class analysis of family data has led to an estimate of 2–10 interacting genes contributing to autism susceptibility (Pickles et al. 1995). Analysis of a two-stage genome scan in 139 multiplex families showed evidence for many interacting genes (Risch et al. 1999). However, in spite of genetic heterogeneity, several genomewide screens of multiplex families have identified possible susceptibility regions of interest, and several of these have been reported in two or more studies, including loci on chromosomes 1p, 2q, 6q, 7q, 13, 16p, and 19p (International Molecular Genetic Study of Autism Consortium [IMGSAC] 1998, 2001; Barrett et al. 1999; Philippe et al. 1999; Risch et al. 1999; Lamb et al. 2000; Liu et al. 2001). Although X chromosome involvement has been suggested because of the 3:1 to 4:1 ratio of males to females, the X chromosome has been ruled out in several samples (IMGSAC 1998; Risch et al. 1999), whereas in others (Liu et al. 2001) it has been implicated at the level of suggestive linkage. Fine mapping in the original 90 IMGSAC families and in 26 additional families raised the maximum LOD score (MLS) in the 7q32-35 region to 3.6 (Maestrini et al. 2000), very close to genomewide significance, and others have supported the finding in this region in independent samples (Ashley-Koch et al. 1999). Using more-stringent inclusion criteria than those used in the original reports, a recent analysis of the current IMGSAC sample (including data from 89 sib pairs analyzed in previous reports and 69 new sib pairs) identified novel regions of linkage on chromosomes 2q (MLS 3.74) and 17q (MLS 2.34) and confirmed previously reported peaks on chromosomes 7q (MLS 3.20) and 16p (MLS 2.93) (IMGSAC 2001). The 7q chromosomal region (spanning >20 cM) has had at least a nominally significant finding for every scan with a sample size of >50 affected sibling pairs (P<.05, uncorrected for size of the region and number of markers tested; Cook 2001).
Another complicating factor in genetic studies of autism is that the expression of the phenotype is highly variable in affected individuals (Bryson and Smith 1998). A growing body of evidence suggests that component phenotypes, such as language deficits, social or communicative impairments, or rigid behaviors, underlie the disorder; varying degrees of these impairments are observed frequently in first- and second-degree relatives of autistic probands (Landa et al. 1991, 1992; Bolton et al. 1994; Piven et al. 1994; Bailey et al. 1995; Smalley et al. 1995; Folstein et al. 1999). First- and second-degree relatives in multiple-incidence autism families have more language-related problems (i.e., delayed speech onset and reading, spelling, and articulation difficulties) than corresponding relatives from Down syndrome families (Piven et al. 1997). In addition, first-degree relatives with positive histories of language difficulties had lower verbal IQs and reading and spelling scores than relatives of autistic probands without such difficulties (Folstein et al. 1999).
In an attempt to confirm previous linkage findings (Bradford et al. 2001; Buxbaum et al. 2001) and to reduce genetic heterogeneity (Buxbaum et al. 2001), several investigators have stratified affected families according to the proband’s language difficulties, as measured by the age at first phrase item (A13) of the Autism Diagnostic Interview (Lord et al. 1994). Buxbaum et al. (2001) identified a putative autism locus on chromosome 2q in the second stage of a genome scan; most of the evidence for linkage on chromosome 2q was from the families with significant language delay. Bradford et al. (2001) investigated loci on chromosomes 7 and 13 that had previously shown evidence for linkage in their sample; they used language delay as criteria for proband diagnosis and also considered parents as affected if they had a history of language-related difficulties. Again, the language-delayed sample was responsible for most of the evidence for linkage to autism on chromosomes 7 and 13.
For the present study, rather than stratifying families on the basis of the proband’s language delay and conducting linkage analysis using the diagnosis of autism, we searched for loci that affect potential disease-related quantitative traits or endophenotypes (Stoltenberg and Burmeister 2000). The a priori focus of this investigation was language delay and repetitive behaviors, since linkage (Bradford et al. 2001; Buxbaum et. al. 2001) or family studies in multiplex autism families (e.g., Piven et al. 1994; Folstein et al. 1999) had suggested the relevance of these phenotypes for genetic studies. Prior to the linkage analysis, we estimated measures of sibling resemblance for items from the Autism Diagnostic Interview in data from 152 nuclear families from the Autism Genetic Resource Exchange (AGRE) (Geschwind et al. 2001). The significant sibling correlations that measured age at first word, age at first phrase, and repetitive or stereotyped behaviors were analyzed using nonparametric linkage analysis in sibships (Kruglyak and Lander 1995; Kruglyak et al. 1996) to detect QTLs for these autism-related traits.
Subjects and Methods
Subjects
The sample included 152 nuclear families ascertained for at least two children diagnosed with an autism-spectrum disorder (autism, pervasive developmental disorder [PDD], or Asperger syndrome). Families were recruited through physician referrals and the Cure Autism Now (CAN) foundation, a nonprofit organization established by parents, clinicians, and researchers to fund biomedical research on autism. AGRE has human-subjects approval from the Institutional Review Board of the University of Pennsylvania School of Medicine.
Genotypic and phenotypic family data are publicly available from the AGRE Web site (Geschwind et al. 2001). For the present report, data from a total of 810 individuals (444 children) were available for analysis. Among the 340 affected individuals, the male-female ratio was 3.24 (260 boys and 80 girls), consistent with other reports of an increased prevalence of the disorder in boys (IMGSAC 1998; Barrett et al. 1999; Philippe et al. 1999). The mean age of the children at the time of testing was 7 years. Children with confirmed fragile X syndrome were excluded from the analysis.
Some families from the present report were included in two previous studies of autism. Buxbaum et al. (2001) analyzed data including 82 families from this analysis, and Liu et al. (2001) used the same AGRE sample. However, Liu and colleagues’ qualitative approach required individuals to be diagnosed as affected with autism or PDD for inclusion in their analysis. Thus, for their parametric scan, they excluded the individuals that failed to meet strict autism diagnosis, but who are included in this study. Although largely overlapping samples are used in the current study and in the study by Liu and colleagues, each one used a completely different phenotype and analytic approach.
Measures
The Autism Diagnostic Interview–Revised (ADI-R; Lord et al. 1994) is a semistructured interview for caregivers of individuals that may be affected with autism-spectrum disorders. The ADI-R, based on the ICD-10 and DSM-IV criteria for autism diagnosis, was administered in the family home by a trained tester. As described above, for the present report, we analyzed three items from the ADI-R that measured age at first word (WORD), age at first phrase (PHRASE), and repetitive or stereotyped behavior (RSB): A12, A13, and DD total, respectively. Children from 123 families responded to these three items from the ADI-R. The majority of the families (n=118) had sibships of two, and five families had sibships of three. Data from a total of 251 children were included in the quantitative linkage analysis. From this sample, WORD, PHRASE, and RSB items were available for 236, 192, and 281 individuals, respectively.
Genotyping
The laboratory and genotyping protocols for the AGRE study have been described elsewhere (Liu et al. 2001). In summary, the families, including parents and unaffected siblings, had blood drawn in their homes by a phlebotomist proficient in working with developmentally delayed children. Blood samples of 10–20 μL in volume were collected and shipped to the Rutgers University Cell Repository for transformation (Anderson and Gusella 1984) and storage. DNA was extracted from whole blood or immortalized lymphoblast cell lines by standard proteinase K digestion and salting-out protocols. DNA samples from the parents and offspring were genotyped at Columbia University, using 335 microsatellite markers comprising a modified version of the Weber 8.0 marker set. PCR amplification of microsatellite markers has been described elsewhere (Aita et al. 1999; Liu et al. 2001). The average heterozygosity of markers used in this study was 0.77, and the average density was 10 cM. Information-content plots for all chromosomes are available online, in the electronic version of this article. Information-content plots for all chromosomes are shown in figure 6 (available online only).
Figure 6 (Online Only) .

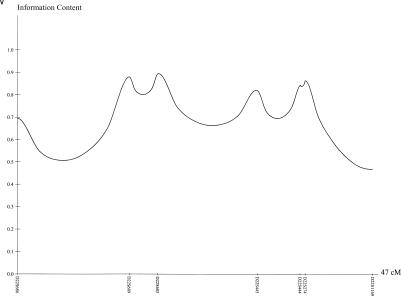
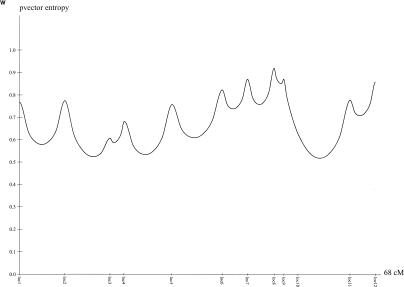
Information-content plots for the genome for 152 nuclear families from AGRE. On average, markers are spaced every 10 cM.
Statistical and Genetic Analysis
The SAS package (1999) was used to merge data files, calculate descriptive statistics, and prepare the input files for the genetic analyses. The program PedCheck (O'Connell and Weeks 1998) was used to find Mendelian genotype errors. The detectable genotype error rate in this sample was <0.01%. Marker allele frequencies were obtained by counting parental genotypes; however, 93% of these parents were genotyped, and therefore frequencies would not have a major impact on linkage results. Map distances were obtained from the Center for Medical Genetics (Marshfield Medical Research Foundation) and the Genome Database.
Since there is an observed sex difference in the incidence of autism, we examined the descriptive statistics and distributions of each endophenotype separately for boys and girls. We assessed sibling resemblance, using Spearman rank correlations (Conover 1980), among nine ADI-R items that measure two key features of autism: language development and repetitive or stereotyped behaviors (table 1).
Table 1.
Sibling Correlations of Nine Autism Endophenotypes Tested for Familiality
| Endophenotype | ADI-RItem | No. ofSib Pairs | Correlation | Pa |
| Age at first word | A12 | 113 | .32 | .0006 |
| Age at first phrase | A13 | 76 | .26 | .021 |
| Difference between ages at first word and phrase | 74 | .23 | .05 | |
| Language level | A19 | 155 | .03 | NS |
| Repetitive and stereotyped behavior | DD total | 108 | .25 | .01 |
| Circumscribed interests | A70a | 121 | .23 | .001 |
| Unusual preoccupations | A71a | 154 | .14 | NS |
| Repetitive use of objects | A72a | 143 | .02 | NS |
| Difficulties with changes in routines or environment | A73a | 155 | .28 | .0001 |
NS = not significant.
Since the traits were distributed continuously and the model of inheritance for autism was poorly understood, we applied a nonparametric linkage approach to identify QTLs that may contribute to the expression of WORD, PHRASE, and RSB. Linkage analysis was performed using the program Mapmaker/Sibs (Kruglyak and Lander 1995) within the GENEHUNTER 2.1 software package (Kruglyak et al. 1996) that conducts multipoint sib pair linkage analysis of quantitative traits. The “all pairs” option, which includes all sib pairs, was used in the analysis. Nonparametric QTL analyses and Haseman-Elston (HE) regressions were computed for the quantitative sib pair data because multiple linkage analysis methods often do not provide consistent results even when applied to the same data (Alarcón and Cantor 2001). So, in addition to the nonparametric QTL method, we ran a second HE nonparametric test of these data, and we report peaks that were identified in both analyses. Given that the nonparametric QTL method is more robust to violations of the normality assumption than is the HE method, the former linkage method was used in the primary analyses. We reported only peaks with a nonparametric test statistic (Z score) >1.65 and a LOD score >1.0.
The nonparametric QTL statistic is based on the Wilcoxon rank-sum test. Sib pair trait differences are ranked and multiplied by a function of the number of alleles shared IBD; the ratio of the statistic to its standard deviation provides a Z score that follows a standard normal distribution (Conover 1980). A multipoint analysis is implemented in the GENEHUNTER software. The HE method regresses the squared sib pair trait difference on the proportions of alleles shared identical by descent (IBD) at marker loci (Haseman and Elston 1972). Sibling pairs that are IBD at a marker that is close to a locus that influences the trait will have similar phenotypic scores. A negative estimate of the slope of the regression line will indicate linkage between the marker locus and the trait locus. A multipoint extension of the HE method is implemented in Mapmaker/Sibs (Kruglyak and Lander 1995); in addition, the estimation-maximization (EM) algorithm is used when IBD is ambiguous or genotypes are missing. The HE method assumes that the error is normally distributed and uncorrelated with the squared sib pair difference; in contrast, the nonparametric QTL statistic does not have restrictive assumptions about the phenotypic difference distribution (Kruglyak and Lander 1995).
Since GENEHUNTER has not implemented an analysis program to test X linkage on quantitative traits, we used Mapmaker/Sibs on the discrete trait of affection status. We selected those individuals whose trait value exceeded the median for their sex and classified them as “affected.” LOD scores were reported for brother-brother, brother-sister, and sister-sister pairs on a single multipoint plot for each trait.
Follow-up analyses included fine mapping and linkage as well as family-based tests of association in the presence of linkage on chromosome 7. For the association analysis, we used a multiallelic, additive genetic model implemented in the FBAT program (Horvath et al. 2001). This program applies a conditional approach to adjust for the correlation among sibling marker genotypes and does not assume normality of trait distributions (Lake et al. 2000).
Results
Sib Correlation of Language and Repetitive-Behavior Measures
Sibling correlations for nine ADI-R items examined for familiality are presented in table 1. Three of the ADI-R items that assessed language and repetitive behavior had significant sibling correlations: WORD (r=0.32, P<.001), PHRASE (r=0.26, P=.02), and RSB (item DD Total, r=0.25, P=.01), as shown in table 1. The RSB composite included the circumscribed interests item, which itself shows a significant sibling correlation (r=0.23, P=.001). Although the significant sibling correlations suggest the variability in the traits may be genetically influenced, the effects of common sibling environment may also be partially responsible for these results. WORD, PHRASE, and RSB were selected for subsequent linkage analysis because of their significant intersib correlations and because previous studies had also suggested that these traits are transmitted within multiplex autism families (e.g., Le Couteur et al. 1996; Piven et al. 1997).
Language abilities are variable among individuals with autism, and delayed speech onset, although common, is not an absolutely necessary criterion for diagnosis. In this sample, the age at which the probands and their siblings said their first word ranged from 7 to 85 months, and half of the children said their first word at ⩾24 months. In addition, the children in this study said their first phrase between the ages of 9 and 96 months, and half of the sample said their first phrase at >42 months. Children with normal language development, in contrast, typically say their first word at <12 months of age and their first phrase by 24 months of age (Bradford et al. 2001).
Trait Distributions
Distributions of WORD, PHRASE, and RSB are reported in figure 1. As shown in table 2, boys have a significantly greater delay in age at first word, which might be expected from the distributions of these traits in the general population. Each of the traits exhibits significant skewing to the right, although they are not kurtotic. This suggests that nonparametric genetic analyses that do not assume distributional normality should be used and that some adjustment for sex differences should be considered. Thus, all genetic analyses had no distributional assumptions, and we ran all analyses using the unadjusted scores. In addition, we reanalyzed the chromosome with the most significant result (chromosome 7) after standardizing the traits within each sex.
Figure 1.
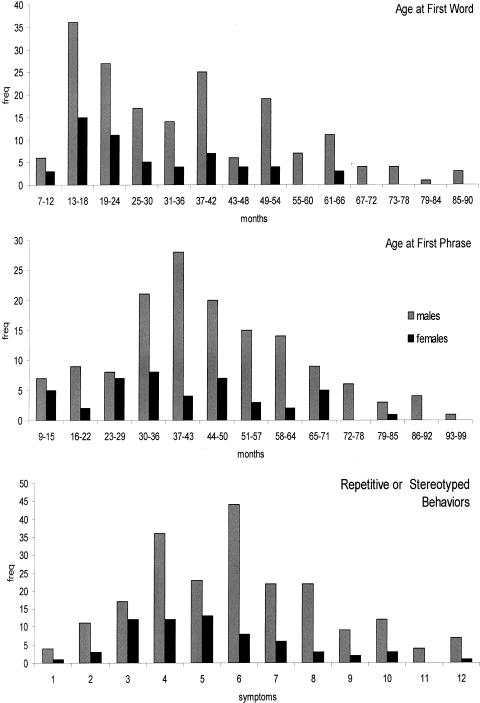
Distributions of autism endophenotypes for each sex
Table 2.
Descriptives for WORD, PHRASE, and RSB as a Function of Sex[Note]
| Measure | n | Mean | SD | Median | Range | Kurtosis | Skewness |
| WORD | |||||||
| Boys | 180 | 30.95 | 18.93 | 27 | 7–85 | −.24 | .77** |
| Girls | 56 | 24.43 | 14.72 | 18 | 8–60 | −.18 | .89** |
| PHRASE | |||||||
| Boys | 145 | 45.92 | 18.67 | 43 | 9–96 | −.04 | .40* |
| Girls | 47 | 41.53 | 18.47 | 39 | 12–84 | −.77 | .26 |
| RSB | |||||||
| Boys | 216 | 5.83 | 2.64 | 6 | 0–12 | −.12 | .24 |
| Girls | 65 | 5.08 | 2.31 | 5 | 0–12 | .62 | .68* |
*P<.05; **P<.01.
Genome Scan of Language and Repetitive Behavior–Related QTL
A complete quantitative multipoint genome scan analysis for autism endophenotypes in nuclear families having at least two children affected with autism-spectrum disorder was performed. Although only phenotypic data from siblings were included in these analyses, the parental genotypes permitted estimation of allele sharing (i.e., IBD). The nonparametric genomewide QTL results for the three quantitative traits are shown in figures 2 and 3. The highest Z score was obtained for WORD, on chromosome 7 between markers D7S1824 and D7S3058: Z=2.98, one-sided P=.001 (fig. 4). The corresponding HE LOD score (derived using the EM algorithm) was 1.14 for WORD. The chromosomal region identified for age at first word in the present study is close to that previously reported as a susceptibility region for autism (IMGSAC 1998; Ashley-Koch et al. 1999; Philippe et al. 1999; Risch et al. 1999) and language-related disorders (Fisher et al. 1998; Lai et al. 2000). As shown in figure 4, the putative QTL for age at first word was <3 cM away from a region linked to autism (IMGSAC 1998) and ∼25 cM away from a locus linked to a severe articulation and language disorder (Fisher et al. 1998; Lai et al. 2000).
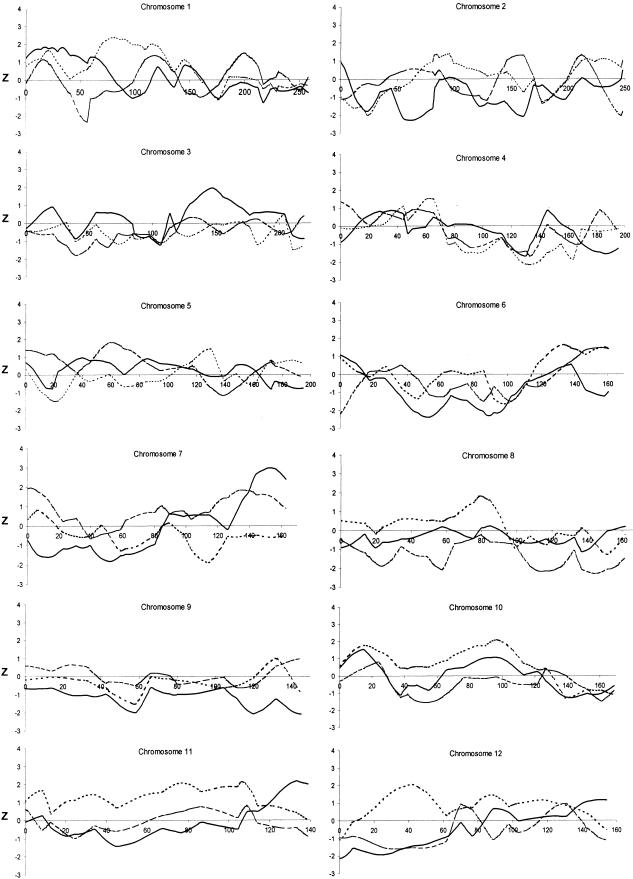
Figure 2.

Nonparametric QTL results of an autosomal genome scan for age at first word (solid line), age at first phrase (dotted line), and the composite measure of repetitive and stereotyped behavior (dashed line). Left, Chromosomes 1–12. Right, Chromosomes 13–22.
Figure 3.

Nonparametric X-linkage results for WORD, PHRASE, and RSB for brother-brother (bold line), brother-sister (solid line), and sister-sister (dotted line) pairs.
Figure 4.
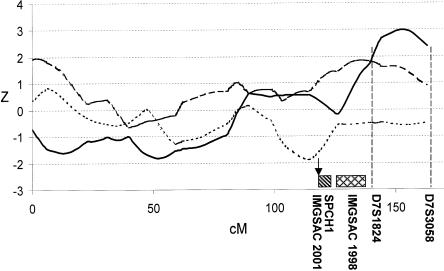
Plots of nonparametric Z scores (calculated at 5-cM intervals) for WORD (solid line), PHRASE (dotted line), and RSB (dashed line) on chromosome 7. Previous loci identified for language deficits or autism are also included in the figure: the rectangle with diagonal stripes represents the region identified in a family with severe speech disorder (Fisher et al. 1998); the rectangle with cross-hatches represents the autism region reported in the initial IMGSAC (1998) study; and the arrow indicates the peak linkage result from an update of the IMGSAC study (IMGSAC 2001).
No evidence for linkage to age at first phrase was found on chromosome 7q; the maximum Z score was 0.84 on the short arm of chromosome 7 at marker D7S3047. The scan did provide evidence for three PHRASE QTL on chromosomes 10 (Z=2.10, P=.018; HE LOD 1.22), 11 (Z=2.19, P=.014; HE LOD 1.35) and 20 (Z=2.29, P=.011; HE LOD 1.21), as shown in figure 2. Moreover, a QTL for WORD may reside 21 cM away from the region linked to PHRASE on chromosome 11 (Z=2.22, P=.013; HE LOD 0.96). Although HE LOD scores had corresponding peaks in the regions identified by the nonparametric analysis, they were all <2.0, roughly equivalent to a P value >.01. Figure 2 shows additional Z score peaks (with nominal P values <.05) that were identified for WORD (on chromosomes 1, 3, 13, 15, 16, and 18) and for PHRASE (on chromosomes 1, 6, 8, 12, and 15). However, none of these peaks were identified in the HE results.
Lastly, results of the initial genome scan do not provide even modest evidence for an RSB QTL. Although there is a nonparametric QTL peak for RSB on chromosome 7q (Z=1.84, P=.033), 15 cM away from the peak for WORD, the corresponding HE LOD score (0.16) does not support the nonparametric evidence for a QTL at this locus.
Fine Mapping of Language-Related QTL on Chromosome 7q
To follow up the initial positive result for WORD on chromosome 7q, an additional 28 microsatellite markers were typed in the 60-cM region between markers D7S1799 and D7S3058. The nonparametric multipoint results were consistent with the initial evidence: the peak nonparametric Z score for WORD (fig. 5) was 2.85 (P=.002), and the corresponding HE LOD score was 0.84. As shown in figure 5, a linkage peak with Z=2.48 (P=.007) was also observed for RSB using the flanking markers; however, the HE LOD score of 0.05 did not support the presence of an RSB peak in the region. Liu et al. (2001) recently analyzed these fine-mapping markers, using the narrow diagnosis of autism, from 160 AGRE families and found a LOD score of 2.13 at marker D7S483, 10 cM from our peak follow-up result for WORD. To fulfill the criteria for a narrow diagnosis of autism, an individual must have language deficits; thus, these results are consistent with the hypothesis of a QTL for language deficits, possibly underlying the autism locus on chromosome 7.
Figure 5.
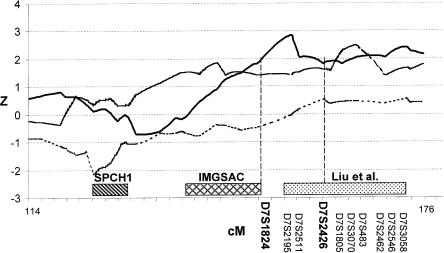
Plots of nonparametric Z scores for linkage analysis of 28 additional markers flanking the chromosome 7q region identified in the quantitative genome scan. Results for WORD (solid line), PHRASE (dotted line), and RSB (dashed line) are shown. The 10 markers included in the association analysis are listed in the figure. The rectangle with diagonal stripes represents the region identified in a family with severe speech disorder (Fisher et al. 1998); the rectangle with cross-hatches represents the autism region reported in the IMGSAC (1998) study; and the rectangle with dots represents the results of a linkage analysis using the narrow diagnosis of autism (Liu et al. 2001). The X-axis tick marks are at 2-cM intervals.
Subsequently, 10 markers within the fine-mapping peak for WORD and the peak linkage region for the narrow diagnosis of autism (between D7S1824 and D7S3058) were tested for association with WORD in the Caucasian families. The markers were, on average, 2.6 cM apart. Two markers showed evidence for association with WORD: D7S1824 (χ2=19.83, P=.031) and D7S2462 (χ2=23.21, P=.003). These results are consistent with a chromosome 7q QTL for language underlying the autism phenotype. Since the two markers were 10.19 cM apart, this suggests that there may be two QTLs for language in the region or that one (or both) of these results is spurious. Interestingly, D7S2462 also showed evidence for association with PHRASE (χ2=18.11, P=.020) and with RSB (χ2=16.49, P=.036).
Since the male and female distributions differ for each of the autism endophenotypes, the analyses were also run on scores standardized within each sex. Although there was a very minor change in the Z score (Z=2.87), the inference that was drawn did not change. Normal scores can also introduce variability into an analysis, so we reported the results derived from the unstandardized data.
Discussion
Nonparametric multipoint linkage analyses of three quantitative autism endophenotypes (age at first word, age at first phrase, and repetitive and stereotyped behaviors) detected four chromosomal regions that may harbor QTLs for these traits. The most significant result was obtained for age at first word, on chromosome 7q. As shown in figure 2, the peak nonparametric Z score for WORD was 2.98 (P=.001), suggesting that a language-related susceptibility locus or QTL may reside on the long arm of chromosome 7, proximal to D7S1824 and D7S3058. Subsequent fine-mapping and association tests in this region support the initial evidence for a QTL for language on 7q35-36. Consistent with this result are reports of a language-specific gene locus in a family with severe speech and language disorder (Fisher et al. 1998; Lai et al. 2000) and in two patients with chromosome rearrangements: the first patient had autism, and the second had specific developmental disorder of speech and language (Warburton et al. 2000). In addition, this region is close to putative autism-susceptibility loci reported in several studies (IMGSAC 1998; Ashley-Koch et al. 1999; Barrett et al. 1999; Philippe et al. 1999; Risch et al. 1999).
Evidence of three possible QTLs for age at first phrase was obtained for chromosomes 10, 11, and 20. The peak Z score for PHRASE, on chromosome 10 (on marker D10S2327), is between two modest peak MLS scores (1.43 for D10S208 and 1.22 for D10S201) from a recent analysis of the IMGSAC sample (2001), suggesting there may be a QTL for autism-related traits in this region. The putative QTL for PHRASE on chromosomes 10 and 20 have not been reported previously as autism susceptibility loci, and they may represent novel QTL for autism endophenotypes or false-positive results.
Interestingly, there was no evidence for linkage of PHRASE to a region overlapping the WORD QTL on chromosome 7q. Although the two traits have a phenotypic correlation of 0.62 (P<.001), their bivariate sibling correlation of 0.26 (P=.011) suggests that their covariation may not be entirely due to genetic effects. Another plausible explanation for the absence of a significant finding of PHRASE on 7q is the possibility that age at first word is recalled with more precision by the caregivers. Although both age at first word and age at first phrase are measures of spoken language onset, the former may be considered a more salient developmental milestone by the parents and, thus, may be recalled more accurately, making that item a more reliable measure than the latter.
Although autism is characterized by deficits in communication, language, and repetitive behaviors, the expression of the disorder is highly variable, and these characteristics may not be present simultaneously. As suggested by Folstein et al. (1999), the individual components of the disorder may be due to distinct and perhaps overlapping genes. In this sample, the chromosome 7q linkage evidence obtained by analyzing the quantitative language item was several-fold stronger than that obtained in the qualitative 10-cM scan based on the narrow or broad autism diagnosis (Liu et al. 2001). It is likely that the present result is due to one or more language loci that underlie or act in conjunction with the weaker autism signal. This result is consistent with the hypothesis that genes residing on chromosome 7q may provide susceptibility to language disorders alone or in combination with the broader autism diagnosis (Folstein and Mankoski 2000). The smaller broad peak observed with RSB on 7q may reflect a distinct locus for the restrictive-repetitive behavior phenotype in this region. That is, separate language-related and RSB loci may exist on chromosome 7q and, in combination, may underlie the autism peak that is consistently observed in this region.
Recently, two groups have successfully used the proband’s language ability, assessed by the ADI-R’s PHRASE, to stratify autistic families prior to linkage analysis (Bradford et al. 2001; Buxbaum et al. 2001), and, in both studies, the evidence for linkage to autism was attributable to the families with language delay. The present analysis differs from these reports by using a QTL approach that investigates the endophenotypes of language and repetitive behaviors in autism families and not the qualitative autism diagnosis. Subsequent extension of this component-phenotype approach to the nonautistic siblings of autistic probands can provide additional analytic power to this study and may provide evidence to confirm the current findings. This is especially important since the items from the ADI-R were not necessarily designed for QTL analysis but for quantitative characterization and diagnosis of autistic individuals. However, it has been suggested recently that data from the ADI-R could be used to quantify features of autism and autism-spectrum disorders for genetic analysis (Lord et al. 2001). More comprehensive study of language performance, such as prospective examination of language and speech development, can be performed in the probands’ nonautistic siblings, and family history information can be obtained from the parents to allow a more-complete analysis of language delay. In this vein, we consider the current results promising but in need of confirmation. Since the etiology of the disorder is so complex, multiple strategies, including stratification and QTL analysis, will be required to help us better understand the mutational basis of autism.
Acknowledgments
We gratefully acknowledge the contributions of the AGRE families who participated in this study and have made the resource possible. Many thanks to Ms. Nancy Hart for her assistance in accessing the AGRE database and to two anonymous reviewers for their useful suggestions on the manuscript. This project was supported by a postdoctoral research award from the M.I.N.D. Institute, University of California, Davis (to M.A.) and by a Young Investigator Award from the Cure Autism Now (CAN) foundation (to J.L.). AGRE was made possible by funds from the CAN foundation. T.C.G. and J.L. also acknowledge support from the generous contribution of Dr. Judith P. Sulzberger.
AGRE Consortium: Daniel H. Geschwind, University of California at Los Angeles, Los Angeles; Maja Bucan, University of Pennsylvania, Philadelphia; W. Ted Brown, New York State Institute for Basic Research in Developmental Disabilities, Staten Island; Joseph D. Buxbaum, Mt. Sinai School of Medicine, New York; T. Conrad Gilliam, Columbia Genome Center, New York; David A. Greenberg, Mt. Sinai Medical Center, New York; David H. Ledbetter, University of Chicago, Chicago; Bruce L. Miller, University of California at San Francisco, San Francisco; Stanley F. Nelson, University of California at Los Angeles School of Medicine, Los Angeles; Jonathan Pevsner, Kennedy Krieger Institute, Baltimore; Jerome I. Rotter, Cedars-Sinai Medical Center, Los Angeles; Gerard D. Schellenberg, University of Washington and Veterans Affairs Medical Center, Seattle; Carole Samango-Sprouse, Children’s National Medical Center, Baltimore; Rudolph E. Tanzi, Massachusetts General Hospital, Boston; and Kirk C. Wilhelmsen, University of California at San Francisco, San Francisco.
Electronic-Database Information
- AGRE, http://agre.org/
- CAN, http://canfoundation.org/
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/
- Genome Database, The, http://gdbwww.gdb.org/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for autism disorder [MIM accession number 209850])
References
- Aita VM, Christiano AM, Gilliam TC (1999) Mapping complex traits in diseases of the hair and skin. Exp Dermatol 8:439–452 [DOI] [PubMed] [Google Scholar]
- Alarcón M, Cantor RM (2001) QTL mapping of serum IgE in an isolated Hutterite population. Genet Epidemiol 21:S224–S229 [DOI] [PubMed] [Google Scholar]
- Anderson MA, Gusella JF (1984) Use of cyclosporin A in establishing Epstein-Barr virus–transformed human lymphoblastoid cell lines. In Vitro 20:856–858 [DOI] [PubMed] [Google Scholar]
- Ashley-Koch A, Wolpert CM, Menold MM, Zaeem L, Basu S, Donnelly SL, Ravan SA, Powell CM, Qumsiyeh MB, Aylsworth AS, Vance JM, Gilbert JR, Wright HH, Abramson RK, DeLong GR, Cuccaro ML, Pericak-Vance MA (1999) Genetic studies of autistic disorder and chromosome 7. Genomics 61:227–236 [DOI] [PubMed] [Google Scholar]
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M (1995) Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med 25:63–77 [DOI] [PubMed] [Google Scholar]
- Barrett S, Beck JC, Bernier R, Bisson E, Braun TA, Casavant TL, Childress D, et al (1999) An autosomal genomic screen for autism: collaborative linkage study of autism. Am J Med Genet 88:609–615 [DOI] [PubMed] [Google Scholar]
- Bolton P, Macdonald H, Pickles A, Rios P, Goode S, Crowson M, Bailey A, Rutter M (1994) A case-control family history study of autism. J Child Psychol Psychiatry 35:877–900 [DOI] [PubMed] [Google Scholar]
- Bradford Y, Haines J, Hutcheson H, Gardiner M, Braun T, Sheffield V, Cassavant T, Huang W, Wang K, Vieland V, Folstein S, Santangelo S, Piven J (2001) Incorporating language phenotypes strengthens evidence of linkage to autism. Am J Med Genet 105:539–547 [PubMed] [Google Scholar]
- Bryson SE, Smith IM (1998) Epidemiology of autism: prevalence, associated characteristics, and implications for research and service delivery. Ment Retard 4:97–103 [Google Scholar]
- Buxbaum JD, Silverman JM, Smith CJ, Kilifarski M, Reichert J, Hollander E, Lawlor BA, Fitzgerald M, Greenberg DA, Davis KL (2001) Evidence for a susceptibility gene for autism on chromosome 2 and for genetic heterogeneity. Am J Hum Genet 68:1514–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover WJ (1980) Practical nonparametric statistics. John Wiley and Sons, New York [Google Scholar]
- Cook EH Jr (2001) Genetics of autism. Child Adolesc Psychiatr Clin N Am 10:333–350 [PubMed] [Google Scholar]
- Fisher SE, Vargha-Khadem F, Watkins KE, Monaco AP, Pembrey ME (1998) Localisation of a gene implicated in a severe speech and language disorder. Nat Genet 18:168–170 [erratum {1998}, Nat Genet 18:298] [DOI] [PubMed] [Google Scholar]
- Folstein SE, Bisson E, Santangelo SL, Piven J (1998) Finding specific genes that cause autism: a combination of approaches will be needed to maximize power. J Autism Dev Disord 28:439–445 [DOI] [PubMed] [Google Scholar]
- Folstein SE, Mankoski RE (2000) Chromosome 7q: where autism meets language disorder? Am J Hum Genet 67:278–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folstein SE, Santangelo SL, Gilman SE, Piven J, Landa R, Lainhart J, Hein J, Wzorek M (1999) Predictors of cognitive test patterns in autism families. J Child Psychol Psychiatry 40:1117–1128 [PubMed] [Google Scholar]
- Geschwind DH, Sowinski J, Lord C, Iversen P, Shestack J, Jones P, Ducat L, Spence SJ, AGRE Steering Committee (2001) The autism genetic resource exchange: a resource for the study of autism and related neuropsychiatric conditions. Am J Hum Genet 69:463–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseman JK, Elston RC (1972) The investigation of linkage between a quantitative trait and a marker locus. Behav Genet 2:3–19 [DOI] [PubMed] [Google Scholar]
- Horvath S, Xu X, Laird N (2001) The family based association test method: strategies for studying general genotype-phenotype associations. Eur J Hum Genet 9:301–306 [DOI] [PubMed] [Google Scholar]
- IMGSAC (1998) A full genome screen for autism with evidence for linkage to a region on chromosome 7q. International Molecular Genetic Study of Autism Consortium. Hum Mol Genet 7:571–578 [DOI] [PubMed] [Google Scholar]
- ——— (2001) A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am J Hum Genet 69:570–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Lander ES (1995) Complete multipoint sib-pair analysis of qualitative and quantitative traits. Am J Hum Genet 57:439–454 [PMC free article] [PubMed] [Google Scholar]
- Lai CS, Fisher SE, Hurst JA, Levy ER, Hodgson S, Fox M, Jeremiah S, Povey S, Jamison DC, Green ED, Vargha-Khadem F, Monaco AP (2000) The SPCH1 region on human 7q31: genomic characterization of the critical interval and localization of translocations associated with speech and language disorder [see comments]. Am J Hum Genet 67:357–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake SL, Blacker D, Laird NM (2000) Family-based tests of association in the presence of linkage. Am J Hum Genet 67:1515–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JA, Moore J, Bailey A, Monaco AP (2000) Autism: recent molecular genetic advances. Hum Mol Genet 9:861–868 [DOI] [PubMed] [Google Scholar]
- Landa R, Folstein SE, Isaacs C (1991) Spontaneous narrative-discourse performance of parents of autistic individuals. J Speech Hear Res 34:1339–1345 [DOI] [PubMed] [Google Scholar]
- Landa R, Piven J, Wzorek MM, Gayle JO, Chase GA, Folstein SE (1992) Social language use in parents of autistic individuals. Psychol Med 22:245–254 [DOI] [PubMed] [Google Scholar]
- Le Couteur A, Bailey A, Goode S, Pickles A, Robertson S, Gottesman I, Rutter M (1996) A broader phenotype of autism: the clinical spectrum in twins. J Child Psychol Psychiatry 37:785–801 [DOI] [PubMed] [Google Scholar]
- Liu J, Nyholt DR, Magnussen P, Parano E, Pavone P, Geschwind DH, Lord C, Iversen P, Hoh J, Autism Resource Exchange Consortium, Ott J, Gilliam TC (2001) A genomewide screen for autism susceptibility loci. Am J Hum Genet 69:327–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Leventhal BL, Cook EH (2001) Quantifying the phenotype in autism spectrum disorders. Am J Med Genet 105:36–38 [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A (1994) Autism Diagnostic Interview–Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord 24:659–685 [DOI] [PubMed] [Google Scholar]
- Maestrini E, Paul A, Monaco AP, Bailey A (2000) Identifying autism susceptibility genes. Neuron 28:19–24 [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe A, Martinez M, Guilloud-Bataille M, Gillberg C, Råstam M, Sponheim E, Coleman M, Zappella M, Aschauer H, van Malldergerme L, Penet C, Feingold J, Brice A, Leboyer M (1999) Genome-wide scan for autism susceptibility genes: Paris Autism Research International Sibpair Study. Hum Mol Genet 8:805–812 [DOI] [PubMed] [Google Scholar]
- Pickles A, Bolton P, Macdonald H, Bailey A, Le Couteur A, Sim CH, Rutter M (1995) Latent-class analysis of recurrence risks for complex phenotypes with selection and measurement error: a twin and family history study of autism. Am J Hum Genet 57:717–726 [PMC free article] [PubMed] [Google Scholar]
- Piven J, Palmer P, Jacobi D, Childress D, Arndt S (1997) Broader autism phenotype: evidence from a family history study of multiple-incidence autism families. Am J Psychiatry 154:185–190 [DOI] [PubMed] [Google Scholar]
- Piven J, Wzorek M, Landa R, Lainhart J, Bolton P, Chase GA, Folstein S (1994) Personality characteristics of the parents of autistic individuals. Psychol Med 24:783–95 [DOI] [PubMed] [Google Scholar]
- Risch N, Spiker D, Lotspeich L, Nouri N, Hinds D, Hallmayer J, Kalaydjieva L, et al. (1999) A genomic screen of autism: evidence for a multilocus etiology. Am J Hum Genet 65:493–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley SL, Asarnow RF, Spence A (1988) Autism and genetics. Arch Gen Psychiatr 45:953–961 [DOI] [PubMed] [Google Scholar]
- Smalley SL, McCracken J, Tanguay P (1995) Autism, affective disorders, and social phobia. Am J Med Genet 60:19–26 [DOI] [PubMed] [Google Scholar]
- Stoltenberg SF, Burmeister M (2000) Recent progress in psychiatric genetics-some hope but no hype. Hum Mol Genet 9:927–935 [DOI] [PubMed] [Google Scholar]
- Warburton P, Baird G, Chen W, Morris K, Jacobs BW, Hodgson S, Docherty Z (2000) Support for linkage of autism and specific language impairment to 7q3 from two chromosome rearrangements involving band 7q31. Am J Med Genet 96:228–234 [DOI] [PubMed] [Google Scholar]


